Abstract
SHP2 is a tyrosine phosphatase involved in the activation of the Ras/ERK signaling pathway downstream of a number of receptor tyrosine kinases. One of the proposed mechanisms involving SHP2 in this context is to dephosphorylate and inactivate inhibitors of the Ras/ERK pathway. Two protein families bearing a unique, common domain, Sprouty and SPRED proteins, are possible candidates because they have been reported to inhibit the Ras/ERK pathway upon FGF activation. We tested whether any of these proteins are likely substrates of SHP2. Our findings indicate that Sprouty2 binds to the C-terminal tail of SHP2, which is an unlikely substrate binding site, whereas SPRED proteins bind to the tyrosine phosphatase domain that is known to be the binding site for its substrates. Overexpressed SHP2 was able to dephosphorylate SPREDs but not Sprouty2. Finally, we found two tyrosine residues on SPRED1 that are required, when phosphorylated, to inhibit Ras/ERK activation and identified Tyr-420 as a specific dephosphorylation target of SHP2. The evidence obtained indicates that SPRED1 is a likely substrate of SHP2, whose tyrosine dephosphorylation is required to attenuate the inhibitory action of SPRED1 in the Ras/ERK pathway.
Keywords: ERK, Growth Factors, MAP Kinases (MAPKs), Protein Phosphorylation, Tyrosine Protein Phosphatase (Tyrosine Phosphatase), FGFR Pathway, SHP2, SPRED, Sprouty
Introduction
The Ras/MAPK pathway was the first to be delineated to a degree of completeness and despite the discovery of other pathways maintains a central role in cell signaling. Historically, research was focused on the EGF pathway as a prototypical example of the pathway downstream of a receptor tyrosine kinase; upon cross-linking of EGF receptors by the EGF ligand, the cytoplasmic tails of each receptor pair become tyrosine phosphorylated on key residues and create binding sites for proteins bearing phosphotyrosine-binding domains. Grb2 is an adaptor protein with a Src homology 2 (SH2)2 and two SH3 domains that constitutively binds to Son of Sevenless, a Ras-Guanine nucleotide exchange factor protein that activates the small membrane-located GTPase Ras. Ras activates Raf, a kinase that activates a downstream kinase MEK that in turn activates ERK. ERK phosphorylates substrates in the cytosol, including additional kinases, as well as traverses the nuclear membrane to activate transcription factors such as Elk (1). An indication of the physiological importance of this central pathway is the accumulated data that show a number of the core component proteins in the Ras/MAPK pathway to be overexpressed or mutated in a large number and diverse array of cancers (2).
After the discovery of the core components of the pathway, a number of proteins have been characterized to position, modify, up-regulate, or down-regulate the pathway at various levels. Our laboratory has been involved in characterizing the Ras/ERK pathway downstream of FGF receptor activation. The FGF pathway differs slightly from the EGF pathway in that a docker protein FRS2 acts as a surrogate receptor tail and associates with FGFR. A number of tyrosine residues on FRS2 become phosphorylated and attract either Grb2 or the protein tyrosine phosphatase non-receptor type 11 (PTPN11 or SHP2) via their respective SH2 domains (3). The majority of the signaling to ERK goes through the SHP2 connection, even though SHP2 can also bind to Grb2/Son of Sevenless. In FGFR signaling, SHP2 acts as an adaptor as well as a tyrosine phosphatase, and its phosphatase activity is necessary for a functioning Ras/ERK pathway (4–6).
There has been considerable interest in identifying the signaling substrate protein of SHP2 in the context of a fully active Ras/ERK pathway. A recent review has comprehensively summarized the theories and suspect proteins that may be the sought-after SHP2 substrates (7). Such potential substrates include Src family kinases and their regulators, signal regulatory protein α (SIRPα), major vault protein, Csk (via Cbp/PAG), p120-RasGAP, the inhibitor proteins Sprouty (Spry) and Sprouty-related Ena/vasodilator-stimulated phosphoprotein homology 1-domain-containing protein (SPRED).
As a corollary of studying the FGFR/Ras/ERK pathway, we have also been studying the mechanism of action of both Spry and SPRED proteins, which share a common cysteine-rich domain (8). Spry was first identified in Drosophila as one of the gene products that regulates the ramifying tubular network of the tracheal system (9). Four mammalian isoforms have been described since, and Sprouty2 (Spry2) was deemed to be the most similar to the larger Drosophila protein in that it plays a similar role in forming the branching of alveolae (10, 11). Several groups reported that Spry2 may be a SHP2 substrate (12, 13), but such data are not universally accepted. The most likely SHP2 substrate should ideally be an inhibitor of the Ras/ERK pathway when tyrosine-phosphorylated but should not inhibit when active SHP2 dephosphorylates specific tyrosine residue(s). Tyrosine 55 on human Spry2 needs to be phosphorylated so that Spry2 can behave as a Ras/ERK pathway inhibitor (14). The exact mechanism has not yet been fully elucidated and the only conclusive data show that phosphorylated Tyr-55 is contained within a canonical binding site for the phosphotyrosine binding domain of c-Cbl, a scaffold and ubiquitin E3 ligase protein (15). Although Spry2 would be a likely candidate as the sought-after SHP2 substrate if the tyrosine phosphorylated Y55 were a target for active SHP2, there is no compelling evidence for this, currently.
SPRED proteins have been also characterized as Ras/ERK pathway inhibitors. The location of this inhibition has been better characterized for SPRED1, through binding to Raf and disruption of the Ras/Raf interaction (16). Although SPRED proteins have a number of tyrosine residues, it is currently not established whether the phosphorylation of any of these residues has an impact on the inhibitory action of the protein in the Ras/ERK pathway.
SHP2 consists of a pair of N-terminal SH2 domains, a catalytic phosphatase domain, and a pair of C-terminal tyrosine residues that are important for its function. SHP2 has a low basal activity due to the interaction between the N-SH2 and the phosphatase domain that keeps SHP2 in a “closed” conformation. The active form of the enzyme requires an open conformation that it is acquired when the SH2 domains bind to phosphotyrosines on target or interacting proteins (6, 7). Such validated partners include the insulin receptor, the scaffold protein IRS-1, and the large adapter protein Gab-1. Evidence indicates that SHP2 is a somewhat novel tyrosine phosphatase as it promotes activation as well as down-regulation of certain pathways. There has been some controversy over the role of the two C-terminal tyrosines Tyr-542 and Tyr-580, which are contained in a canonical Grb2 SH2 binding sequence. Previous studies suggest that the Tyr-542 site is the dominant Grb2 binding residue (17, 18).
In various organisms the inactivation of the PTPN11 gene results in major developmental defects, often similar to receptor tyrosine kinase loss-of-function mutations. Mutations of PTPN11, the gene encoding SHP2 in mammals, have been found to be responsible for cancers, most often leukemias (19–22), as well as developmental diseases, such as Noonan and LEOPARD syndromes (23–25). Noonan syndrome is caused by stimulatory mutations of PTPN11 or other key elements of the Ras/ERK pathway, including Son of Sevenless, K-Ras, or Raf1 (26–29). It is interesting to note that loss-of-function mutations in SPRED proteins also occur in patients with a variant neurofibromatosis/Noonan-like syndrome (30, 31).
Our area of interest encompasses the Ras/ERK pathway downstream of FGFR, with a current emphasis on the function of Spry and SPRED proteins and the mechanism involving the positive function of SHP2 in the Ras/ERK pathway. In this study, we aimed to examine whether Spry or SPRED were likely substrates of SHP2 and its critical role in the activation of this central signaling pathway.
EXPERIMENTAL PROCEDURES
Plasmid and Expression Vectors
Full-length human SHP2 (NM_002834) obtained from Addgene plasmid 8381 (Cambridge, MA) kindly provided by Dr. Ben Neel (Ontario Cancer Institute), was subcloned into pXJ40-HA vector for mammalian expression and pGEX-4T-1 vector for bacterial expression. SHP2 deletion mutations were PCR-amplified with primers containing BamHI (5′ primer) and XhoI (3′ primer) restriction sites, and the amplified products were cloned into pXJ40-HA and pGEX-4T1 vectors. FGFR1 (NM_023110), FLAG-tagged human Spry1 (NM_005841), Spry2 (NM_005842), Spry4 (NM_030964), mouse SPRED1 (NM_033524) and SPRED2 (NM_033523) full-length constructs have been described previously (32). myc-Raf1 was a gift from Dr. R. Jackson and Dr. Li Dan (Institute of Molecular and Cell Biology). H-RasV12 was kindly provided by Dr. E. Manser. Mutations were introduced by PCR-based site-directed mutagenesis using the proofreading Pfu DNA polymerase (Promega, Madison, WI). SIRPα (ΝΜ_001040022) was purchased from Thermo Fisher Scientific (Waltham, MA).
Antibodies and Reagents
Mouse and rabbit anti-FLAG (F3164, F7425), agarose-conjugated anti-FLAG M2 beads (F2426), rabbit anti-HA (H6908), mouse anti-β-tubulin-Cy3 (C4585), and FITC-conjugated goat anti-rabbit IgG (F0382) were purchased from Sigma. Mouse and rabbit anti-SHP2 (sc-7384, sc-280), rabbit anti-FGFR1 (sc-121), rabbit anti-c-Myc (sc-789), rabbit anti-H-Ras (sc-68742), mouse and rabbit anti-SIRPα (sc-17803, sc-11374), and mouse anti-GST (sc-138) were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-pan ERK (610124) and mouse HRP anti-PY20 (610012) were from BD Transduction Laboratories. Mouse anti-phospho ERK1/2 (9106) was purchased from Cell Signaling Technology (Beverly, MA). Glutathione-Sepharose 4B was from GE Healthcare, and the SHP2 inhibitor NSC-87877 was from TOCRIS Bioscience (Bristol, UK).
Cell Lines and Transfection
HEK293 and PC12 cell lines were purchased from the American Type Culture Collection (Manassas, VA). HEK293 cells were maintained in RPMI 1640 medium, supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 2 mm l-glutamine. PC12 cells were maintained in DMEM (4.5 g of glucose/ml) supplemented with 10% fetal bovine serum, 5% horse serum, 2 mm l-glutamine, 10 mm HEPES (pH 7.4), and 1% penicillin/streptomycin. For experiments, cells were seeded in 10-cm dishes, and all transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Immunoprecipitation and Immunoblotting
Immunoprecipitation and immunoblotting were carried out essentially as described previously (33). Cells were harvested 24 h post-transfection in HEPES lysis buffer (20 mm HEPES (pH 7.4), 137 mm NaCl, 1 mm EGTA, 1.5 mm MgCl2, 10% (v/v) glycerol, 1% Triton X-100, a protease inhibitors mixture (Complete protease inhibitor, Roche Applied Science), and 1 mm Na3VO4). Cell lysates were then used for immunoprecipitation, subsequent SDS-PAGE, and immunoblotting. Blots were scanned and quantified using NIH ImageJ software (http://rsbweb.nih.gov/ij/). All Western blot data shown are representatives of at least three separate individual experiments, unless otherwise stated.
GST Pulldown
Escherichia coli BL21-Gold cells bearing GST-SHP2 plasmids were induced overnight with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 25 °C. Cells were centrifuged, resuspended in PBS (0.1% Triton X-100, 20 mm DTT, and protease inhibitors mixture), and subjected to sonication. The lysates were centrifuged, and the supernatants were incubated with glutathione-Sepharose 4B beads for 4 h at 4 °C. The beads were washed with PBS (0.1% Triton X-100 and protease inhibitors mixture) and 20 mm Tris-HCl (pH 8, 0.1% Triton X-100, and protease inhibitors mixture). HEK293 cells transfected with FLAG-Spry and SPRED proteins were lysed and incubated with GST-SHP2 beads overnight at 4 °C. The beads were then washed with HEPES lysis buffer, and bound proteins were separated in SDS-PAGE and analyzed by immunoblotting.
Immunofluorescence Microscopy
For the neurite outgrowth assay, PC12 cells were transfected with the various SPRED constructs. 48 h post-transfection, the cells were stimulated with basic fibroblast growth factor (50 ng/ml), every second day for 5 days. PC12 cells were fixed, stained, and visualized essentially as described (33) with the following modifications. Permeabilized cells were blocked 1 h with phosphate-buffered saline supplemented with 1 mm each of CaCl2 and MgCl2, 2% bovine serum albumin, and 7% fetal bovine serum. Primary and secondary antibodies were diluted in blocking buffer and incubated at room temperature for 1 h each. The images were captured using a confocal microscope Zeiss LSM510 META (Carl Zeiss Microimaging).
RESULTS
Spry2 Binds to C-terminal Tail of SHP2
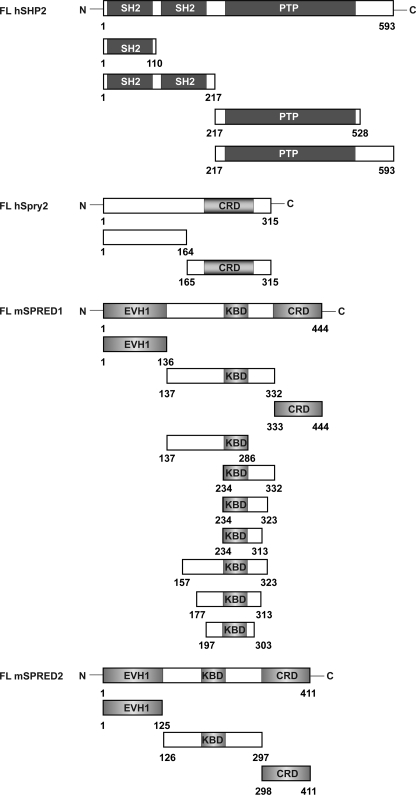
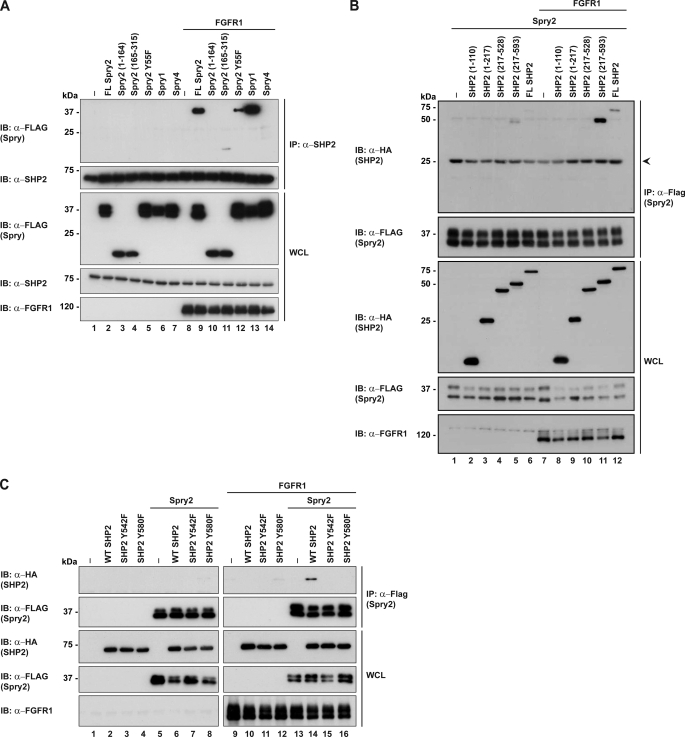
A schematic diagram of the domains and full-length proteins used in the subsequent experiments is shown in Fig. 1. To first ascertain whether and how the Spry proteins bind to SHP2, FLAG-tagged Spry1, Spry2 or Spry4 were transfected into HEK293 cells and activated by overexpressing FGFR1. Immunoprecipitations and Western blotting were carried out as shown in Fig. 2A. It is apparent that Spry1 and Spry2 bound to SHP2 upon FGFR1 activation, but Spry4 did not. At this point, we did not pursue the binding of Spry1 to SHP2 because we have previously shown that Spry1 does not inhibit the Ras/ERK pathway downstream of FGFR1 activation (32). Next, we performed a similar experiment that included the N- and C-terminal halves of Spry2 to determine where SHP2 bound (Fig. 2A). From the results, we concluded that SHP2 bound to the C-terminal half of Spry2 containing the cysteine-rich domain (CRD residues 165–315), albeit to a lower extent when compared with the full-length protein. It is also noteworthy that Spry2Y55F showed a decreased binding. In the reverse binding experiment, to map the SHP2 binding site, we observed that full-length Spry2 bound to the C-terminal fragment of SHP2 comprising residues 217–593 (Fig. 2B). To further define the site of binding of Spry2, two point mutations of SHP2 were included and tyrosines Tyr-542 and Tyr-580 were individually mutated to phenylalanine. The rationale for this is that, Spry2 upon activation binds the N-SH3 domain of Grb2 and the SH2 domain of Grb2 binds either of the two tyrosines on SHP2, both of which contain the Grb2 binding signature (18, 32). Binding of Spry2 was abolished when either tyrosine was mutated to phenylalanine, as shown in Fig. 2C. Previous evidence detailed that both tyrosines bind the SH2 domain of Grb2, therefore suggesting that Spry2 binds to SHP2 via Grb2. As mentioned above, this region is an unlikely binding site for a potential substrate and hence negates Spry2 as a SHP2 substrate.
FIGURE 1.
Schematic representation of the domain structures of SHP2, Spry2, SPRED1, and SPRED2. FL hSHP2, full-length human SHP2, SH2, and PTP. FL hSpry2, full-length human Spry2, CRD. FL mSPRED1, full-length mouse SPRED1 and SPRED2, Ena-vasodilator-stimulated phosphoprotein homology-1 domain (EVH1), KBD, and CRD. These constructs were used in the subsequent figures.
FIGURE 2.
Spry2 binds to the C-terminal tail of SHP2. A, HEK293 cells were transfected with the indicated plasmids (FLAG-Sprys, FGFR1) or the pXJ40 vector control. 24 h post-transfection, cell lysates were subjected to immunoprecipitation (IP) with anti-SHP2. Immunoprecipitates were separated in SDS-PAGE and immunoblotted (IB) with the antibodies indicated on the left. Whole cell lysates (WCL) were immunoblotted to verify equal protein expression levels. B, deletion mutants of SHP2 were co-expressed with Spry2 and FGFR1 on lanes 7–12. Lysates were immunoprecipitated with anti-FLAG, and blots were tested with the antibodies indicated on the left. C, FLAG-Spry2 and HA-SHP2 mutants were co-expressed in HEK293 cells. Lysates were immunoprecipitated with anti-FLAG and immunoblotted with anti-HA to probe for interactions with SHP2. Arrow indicates the immunoglobulin light chain.
SPRED1 and SPRED2 Interact with SHP2 through a Sequence between KBD and Ena-VASP Homology-1 Domain
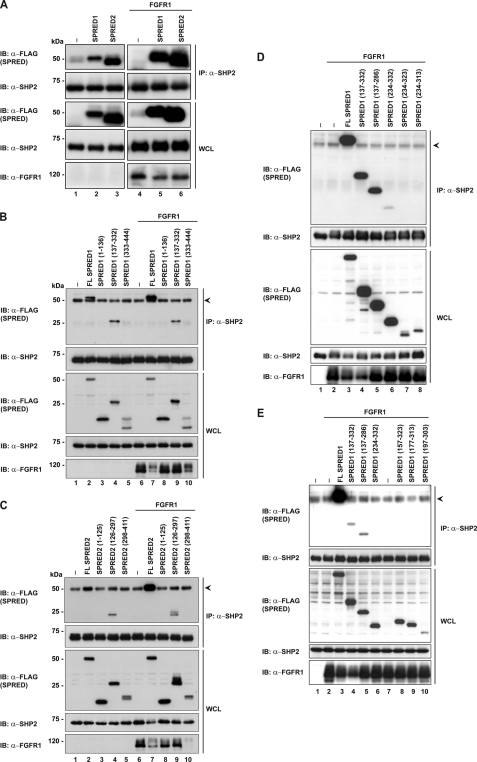
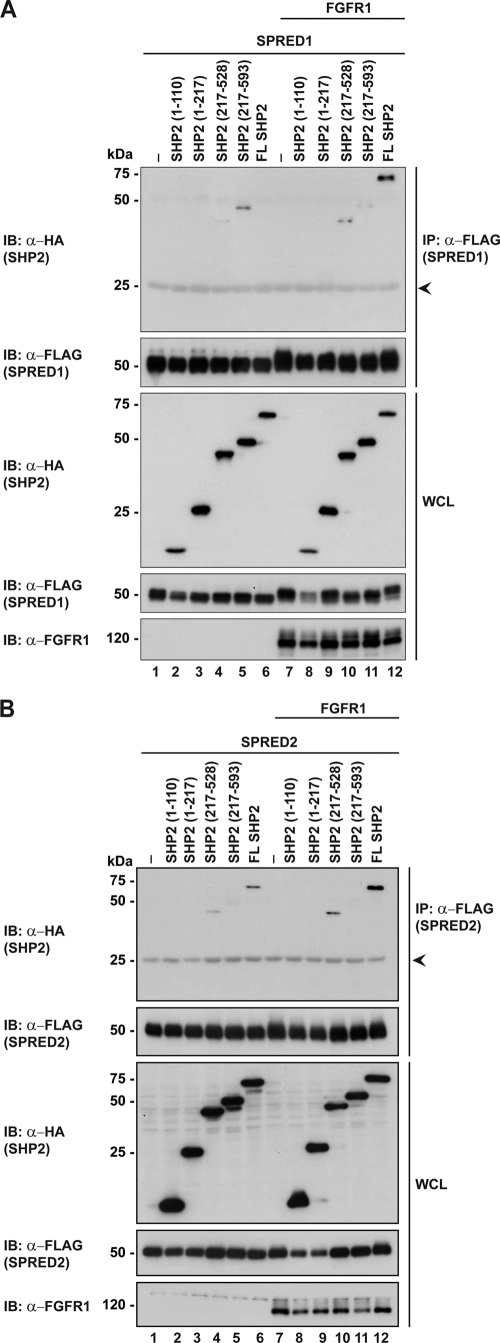
Because the binding of SHP2 to Spry2 is through the CRD which is conserved in SPRED proteins, we designed the following experiments to investigate the binding of SPRED1 and SPRED2 to SHP2. Experiments were performed in a similar manner to those described in Fig. 2. SPRED1 or SPRED2 were transfected into HEK293 cells with or without FGFR1 activation. The endogenous SHP2 was immunoprecipitated, and the results in Fig. 3A demonstrated that both SPRED1 and SPRED2 interact with SHP2 in unstimulated cells, and to a greater extent in activated cells. In vitro pulldown assays with GST-SHP2 also confirmed its binding to SPRED1 and SPRED2 (supplemental Fig. 1). To map the region of SPREDs that interact with SHP2, a series of FLAG-tagged truncation constructs of SPRED1 and SPRED2 were prepared (Fig. 1) and used in co-immunoprecipitation assays with SHP2. The data indicates that both SPRED1 and SPRED2 bound to SHP2 via the extended c-Kit binding domains (KBD) (Fig. 3, B and C). Interestingly the binding did not happen through the CRD domain, as with Spry2 (Fig. 2A). The binding region of SHP2 on the KBD domain was further narrowed down using various truncated mutants of these 200 residues as shown in Fig. 1. The binding of SHP2 requires a sequence between the KBD and Ena-VASP homology-1 domains from amino acids 137 to 157 (Fig. 3, D and E). Experiments to determine the domain of SHP2 where the SPRED proteins bound are shown in Fig. 4, A (SPRED1) and B (SPRED2). Both SPRED1 and SPRED2 bound to the protein tyrosine phosphatase (PTP) domain of SHP2. Although it is too early to draw any conclusions, it would be expected from precedent literature that any substrate of SHP2 would bind to either the SH2 or the PTP domains (7).
FIGURE 3.
SPRED1 and SPRED2 interact with SHP2 through via amino acids 135–157. A, endogenous SHP2 was immunoprecipitated in cells overexpressing FLAG-tagged SPRED1 and SPRED2 with or without FGFR1 overexpressed. Lysates were processed as described in Fig. 2A. Immunoblots were incubated with anti-FLAG to prove for interactions with SHP2. B and C, FLAG-SPRED1 and FLAG-SPRED2, respectively, full-length or deletion mutants were transfected in HEK293 cells, and endogenous SHP2 was immunoprecipitated with anti-SHP2. Immunoprecipitates were separated in SDS-PAGE. D and E, HEK293 lysates expressing SPRED1 mutants as indicated on the panels were immunoprecipitated with anti-SHP2 to pull down endogenous SHP2. Immunoprecipitates were separated in SDS-PAGE and probed with anti-FLAG to detect binding. Arrowheads indicate the immunoglobulin heavy chain. IP, immunoprecipitation; IB, immunoblot; WCL, whole cell lysate.
FIGURE 4.
SPRED1 and SPRED2 bind to the PTP domain of SHP2. A, FLAG-SPRED1 was co-expressed with HA-SHP2 full-length or deletion mutants. Lysates were immunoprecipitated with anti-FLAG and evaluated by Western blotting. B, FLAG-SPRED2 was co-expressed together with HA-SHP2 full-length or deletion mutants, and samples were immunoprecipitated with anti-FLAG and analyzed by Western blotting with the indicated antibodies on the left. Arrowheads indicate the immunoglobulin light chain. IP, immunoprecipitation; IB, immunoblot; WCL, whole cell lysate.
SHP2 Dephosphorylates SPRED1 and SPRED2 but Not Spry2
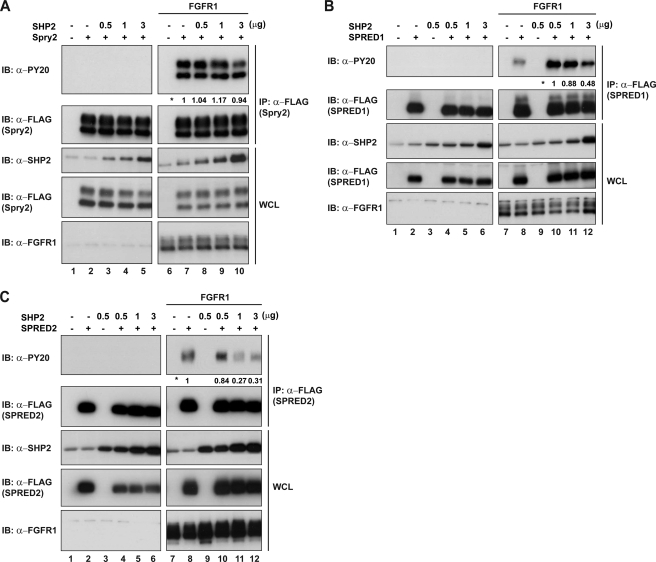
Historically, it has proven difficult to demonstrate whether proteins are substrates of tyrosine phosphatases (34). In vitro experiments often show false positives, and such a system overrides the often intricate sequence of phosphatase activation. Initially, we evaluated the ability of SHP2 to dephosphorylate Spry and SPRED proteins. We observed the tyrosine phosphorylation status of the prospective substrates while we progressively increased the expression of SHP2. Such experiments were carried out with Spry2 (Fig. 5A), SPRED1 (Fig. 5B), and SPRED2 (Fig. 5C). Spry2 only showed a slight decrease on tyrosine phosphorylation at the higher dose of SHP2, SPRED1 showed a modest but steady decrease in tyrosine phosphorylation with increased SHP2 (∼50% decrease when comparing lanes 10 and 12 Fig. 5B), whereas the decrease with SPRED2 was profound (∼75% decrease when comparing lanes 8 and 11 Fig. 5C). These results suggested that the SPREDs may be SHP2 substrates but that Spry2 was unlikely to be. Previous reports showed that Spry2 was a likely substrate of SHP2, by using the above approach although often a single “dose” of SHP2 or a constitutively active SHP2 was employed. The above experiment does not demonstrate categorically that SPRED proteins are direct substrates of SHP2. It is also possible that SHP2 may indirectly affect another phosphatase that dephosphorylates SPREDs. In addition, the mechanism of SHP2 is complex because it acts upstream as a scaffold protein, what may have an opposite effect compared with the effect that the phosphatase activity may have on a particular substrate.
FIGURE 5.
SHP2 dephosphorylates SPRED1 and SPRED2 but not Spry2. A, HEK293 cells were transfected with FLAG-Spry2 and increasing concentrations of SHP2 with or without FGFR1 activation. Lysates were immunoprecipitated with anti-FLAG, and immunoblots were probed with anti-PY20 to detect tyrosine phosphorylation levels. B, cell lysates co-transfected with FLAG-SPRED1 and SHP2 were treated as mentioned in A. C, cell lysates co-transfected with FLAG-SPRED2 and SHP2 were treated as mentioned in A. Numbers next to the asterisk indicate quantification results of tyrosine phosphorylation levels on the PY20 blot. SHP2 band on the anti-SHP2 blots (A–C) on lanes 1 and 2 represent endogenous SHP2 detected by the antibody. IP, immunoprecipitation; IB, immunoblot; WCL, whole cell lysate.
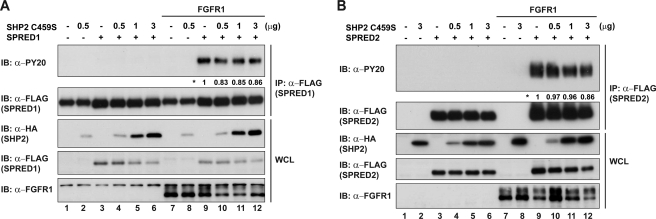
In an endeavor to rule out the non-phosphatase effects of SHP2 and to provide additional evidence for SPREDs as likely SHP2 substrates, we repeated the above experiments overexpressing SPRED1 or SPRED2 together with the dominant negative mutant SHP2C459S instead of WT SHP2. Increasing concentrations of SHP2C459S caused no reduction in the phosphorylation status of SPRED1 or SPRED2 (Fig. 6, A and B). Moreover, we confirmed that the absence of dephosphorylation on SPREDs was due to the catalytically inactive SHP2 and not to the lack of binding because we observed that SHP2C459S still maintained the binding to SPREDs (supplemental Fig. 2). Because SHP2C459S was capable of acting as a scaffold protein with its own SH2 domain, and its tyrosine phosphorylation sites were fully functional, it appears that the dephosphorylation by SHP2 is the most likely reason for the reduced tyrosine signal on SPRED proteins (Fig. 5, B and C).
FIGURE 6.
SHP2C459S does not dephosphorylate SPRED. A, lysates co-expressing FLAG-SPRED1 and increasing concentrations of HA-SHP2C459S were immunoprecipitated with anti-FLAG. Immunoblots were probed with anti-PY20 to detect tyrosine phosphorylation levels. B, cells co-expressing FLAG-SPRED2 and HA-SHP2C459S were treated as mentioned in A. Numbers next to the asterisk indicate quantification of the tyrosine phosphorylation levels on the PY20 blot. IP, immunoprecipitation; IB, immunoblot; WCL, whole cell lysate.
Another experiment commonly used to identify PTP substrates is to knock down the phosphatase and examine the effect on the tyrosine phosphorylation levels of the candidate substrate. However when we did a knockdown of SHP2 to ∼85%, the expression of SPREDs was also reduced to ∼85%, and thus, we were unable to evaluate the effect on the tyrosine phosphorylation levels of SPREDs. A possible explanation may be that SHP2 knockdown eliminated not only the PTP activity but also the scaffold function of SHP2, which might affect the entire function of the Ras/ERK pathway and any downstream gene expression. In a similar manner, it has been shown that the expression of Spry2 in mammalian cells is dependent on the activation level of the Ras/ERK pathway (14).
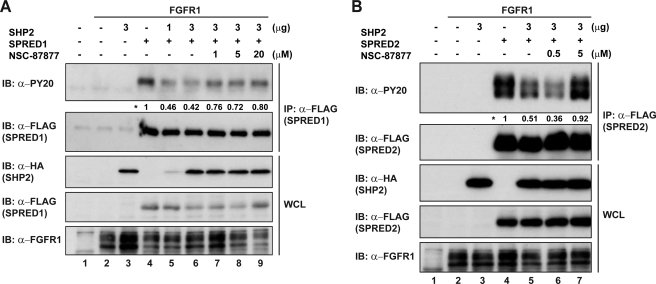
To obtain additional evidence of the effect of SHP2 on the phosphorylation status of SPRED1 and SPRED2, we studied the effect of adding increasing doses of the SHP1/2 phosphatase inhibitor NSC-87877, under conditions similar to those showed in Fig. 5. Molecular modeling and site-directed mutagenesis studies suggest that NSC-87877 can bind to the catalytic cleft of SHP2 PTP domain and cross-inhibit SHP1 in vitro, but it is selective for SHP2 over other PTPs (PTP1B, HePTP, DEP1, CD45, and Leukocyte antigen-related phosphatase) (35). As shown in Fig. 7, A and B, the addition of NSC-87877 reversed the SHP2-induced tyrosine dephosphorylation on SPRED1 and more profoundly on SPRED2.
FIGURE 7.
SHP2 phosphatase inhibition rescues tyrosine phosphorylation of SPRED. A, HEK293 cells were transfected with FLAG-SPRED1 and HA-SHP2. 24 h post-transfection, cells were treated with SHP2 inhibitor NSC-87877 for 2 h (lanes 7–9). Lysates were immunoprecipitated with anti-FLAG, subjected to SDS-PAGE, and immunoblotted as indicated. B, FLAG-SPRED2 and HA-SHP2 overexpressed in HEK293 cells were treated with NSC-87877 (lanes 5–7) for 2 h before harvesting. Lysates were treated as mentioned in A. Numbers next to the asterisk indicate quantification of tyrosine phosphorylation levels on the PY20 blot. IP, immunoprecipitation; IB, immunoblot; WCL, whole cell lysate.
SHP2 Dephosphorylates Tyr-420 on SPRED1
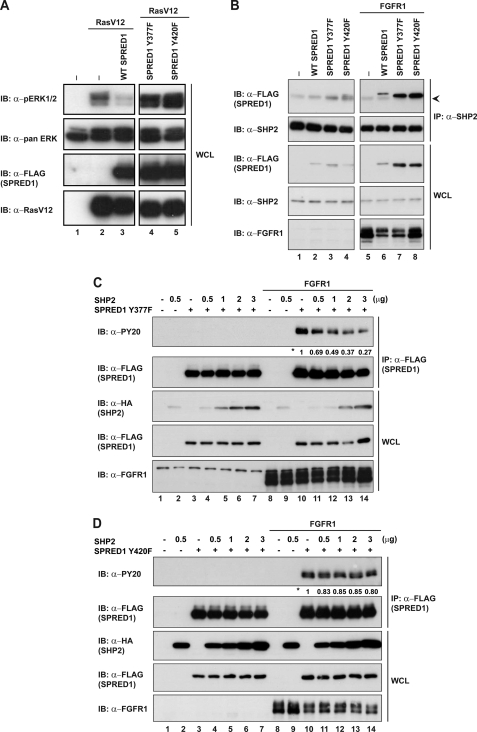
Although there is a general consensus that Tyr-55 of Spry2 needs to be phosphorylated to inhibit the Ras/ERK pathway, little is known about the tyrosine phosphorylation status of SPRED proteins in relation to their ability to inhibit the Ras/ERK pathway. It has been shown that SPRED1 is a potent inhibitor of the Ras/ERK pathway downstream of various activators, whereas SPRED2 is only a weak inhibitor (supplemental Fig. 3). After mutating all of the tyrosine residues of SPRED1 to phenylalanine, we found that Y377F and Y420F abrogated the Ras/ERK inhibitory action of SPRED1 downstream of the RasV12 activating mutant (Fig. 8A). It has been described that SPRED1 binds to the Ras-Raf complex to inhibit the Ras/ERK pathway (16). We tested whether the abrogation of Ras/ERK inhibition by SPRED1Y377F and Y420F was due to the inability of SPRED1 mutants to bind to the Ras-Raf complex. HEK293 lysates overexpressing FLAG-tagged WT SPRED1, SPRED1Y377F, and Y420F were immunoprecipitated with anti-FLAG and probed for Ras and Raf1 binding (supplemental Fig. 4). We detected SPRED1 point mutants binding to Ras and Raf1 to the same degree as WT SPRED1, which indicates their inhibitory effect is not via a binding disruption to the SPRED-Ras-Raf1 complex. This implies that the ability of SPRED1 to inhibit the Ras/ERK pathway depends directly on the phosphorylation status of Tyr-377 and Tyr-420.
FIGURE 8.
SHP2 dephosphorylates Tyr-420 on SPRED1. A, HEK293 cells were transfected with the indicated plasmids (FLAG-SPRED and RasV12) or the pXJ40 vector control. Whole cell lysates (WCL) were analyzed by Western blotting with the indicated antibodies on the left. B, endogenous SHP2 was immunoprecipitated, with anti-SHP2, in cells overexpressing FLAG-SPRED1 WT or respective mutants. Immunoprecipitates were resolved by SDS-PAGE. C, FLAG-SPRED1Y377F was co-transfected with increasing concentrations of HA-SHP2 (0.5–3 μg), and lysates were immunoprecipitated with anti-FLAG. Immunoblots were probed with the antibodies indicated on the left. D, cells overexpressing FLAG-SPRED1Y420F and HA-SHP2 were treated as mentioned in A. Numbers next to the asterisk indicate quantification of the tyrosine phosphorylation levels on the PY20 blot. The arrowhead indicates the light band at 50 kDa corresponding to the immunoglobulin heavy chain.
Furthermore, we tested whether any of these tyrosines could be the dephosphorylation site for SHP2. First, we evaluated the interaction between SPRED1 mutants and SHP2. Both SPRED1Y337F and Y420F mutants maintained the binding to SHP2 (Fig. 8B). Next, we analyzed whether SHP2 could decrease the tyrosine phosphorylation in any of these mutants. Interestingly, SPRED1Y377F showed a progressive dephosphorylation with increasing amounts of transfected SHP2, similar to SPRED1 WT (Fig. 8C). However, SPRED1Y420F did not show any dephosphorylation, suggesting that Tyr-420 is a possible dephosphorylation site (Fig. 8D).
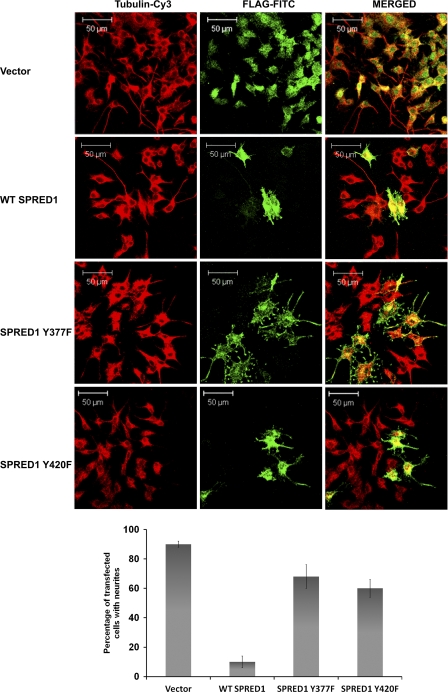
Tyrosines 377 and 420 of SPRED1 Are Required to Inhibit PC12 Differentiation
To translate the binding studies into a more physiological context, we evaluated the effect of the SPRED1Y377F and Y420F point mutants on their ability to inhibit the ERK-driven neurite outgrowth in PC12 cells. Previously, Wakioka et al. (16) described that SPRED1 inhibited NGF-induced differentiation of PC12 cells. As shown in Fig. 9, WT SPRED1 inhibits PC12 differentiation following FGF stimulation with respect to the strong retardation of neurite outgrowth. SPRED1Y377F and Y420F mutants while causing some decrease in neurite number and length essentially have a greatly reduced inhibition when compared with WT SPRED1. This is further evidence that the phosphorylation status of Tyr-377 and Tyr-420 determines the degree that SPRED1 can inhibit the Ras/ERK pathway. Taken together, these results suggest that SPRED1 may be a substrate of SHP2 and, specifically, the phosphorylation status of Tyr-420 might control the activation status of the Ras/ERK pathway in the context of FGFR1 signaling.
FIGURE 9.
Tyrosines 377 and 420 of SPRED1 are required for inhibition of basic fibroblast growth factor-induced neurite outgrowth in PC12 cells. PC12 cells grown on poly-l-lysine-coated coverslips were transfected with FLAG-SPRED1, FLAG-SPRED1Y377F, or FLAG-SPRED1Y420F, after transfection cells were stimulated with basic fibroblast growth factor as described in “Experimental Procedures.” The SPRED1 protein was stained with FLAG-FITC, and the cells were counterstained with Cy3-conjugated anti-tubulin. Scale bar, 50 μm. The average percentage of transfected cells bearing neurites from a minimum of three independent experiments is graphically shown in the bottom panels.
DISCUSSION
In our search to characterize the function of Spry and SPRED proteins, we evaluated whether they were possible substrates of SHP2. We observed that Spry2 bound to the C-terminal tail of SHP2, which is an unlikely binding site for a substrate. Because this region contains a binding site for Grb2 (non-substrate of SHP2) we hypothesized that Spry2 might bind to SHP2 through Grb2. On the other hand, the SPREDs did bind to the PTP domain of SHP2, where the dephosphorylation event occurs. The ability of SHP2 to dephosphorylate SPREDs but not Spry2 led us to further study the SPREDs as possible substrates. Subsequent experiments proved that SHP2 dominant negative mutant did not affect tyrosine phosphorylation on SPREDs. Similarly, the chemical inhibition of SHP2 phosphatase activity rescued SPRED tyrosine phosphorylation from previous dephosphorylation by SHP2. Finally, we identified two tyrosines on SPRED1 that are necessary for Ras/ERK inhibition and for the inhibitory effect of SPRED1 in PC12 neurite outgrowth assay. Further investigation revealed that Tyr-420 but not Tyr-377 is a likely SHP2 substrate site.
Although this investigation may not be an exhaustive study of the sought-after SHP2 substrate that has been the target of a number of studies and a comprehensive review (7), we have addressed whether Spry2 or the SPRED proteins are the more likely Ras/ERK controlling SHP2 substrate. Within the recognized restrictions of characterizing tyrosine phosphatase substrates, we have provided evidence that Spry2 is an unlikely substrate, and it is more likely that SPRED1 is a candidate for the elusive SHP2 substrate.
As well as providing evidence as to the most likely SHP2 substrate in the context of the Ras/ERK signaling pathway, we have provided a plausible mechanism of action for SPRED1 via the tyrosine phosphorylation of Tyr-420 and Tyr-377. It is possible another tyrosine phosphatase is specific for the dephosphorylation of Tyr-377. Evidence suggests that the mechanism of action of SPRED2 has additional factors to consider, and our laboratory is currently addressing these factors.
Supplementary Material
Acknowledgments
We thank Chye Yun Yu and Nur Farehan Mohamed Asgar for technical assistance, Dr. Chow Soah Yee for critical review of the manuscript, and members of the Graeme R. Guy laboratory for helpful discussions.
This work was supported by the Agency of Science, Technology, and Research (A-STAR), Singapore.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- SH2
- Src homology 2
- SHP2
- protein tyrosine phosphatase non-receptor type 11
- FGFR
- FGF receptor
- CRD
- cysteine-rich domain
- KBD
- c-Kit binding domain
- PTP
- protein tyrosine phosphatase.
REFERENCES
- 1. Schlessinger J. (2002) Cell 110, 669–672 [DOI] [PubMed] [Google Scholar]
- 2. Malumbres M., Barbacid M. (2003) Nat. Rev. Cancer 3, 459–465 [DOI] [PubMed] [Google Scholar]
- 3. Eswarakumar V. P., Lax I., Schlessinger J. (2005) Cytokine Growth Factor Rev. 16, 139–149 [DOI] [PubMed] [Google Scholar]
- 4. Hadari Y. R., Kouhara H., Lax I., Schlessinger J. (1998) Mol. Cell. Biol. 18, 3966–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Araki T., Nawa H., Neel B. G. (2003) J. Biol. Chem. 278, 41677–41684 [DOI] [PubMed] [Google Scholar]
- 6. Neel B. G., Gu H., Pao L. (2003) Trends Biochem. Sci. 28, 284–293 [DOI] [PubMed] [Google Scholar]
- 7. Dance M., Montagner A., Salles J. P., Yart A., Raynal P. (2008) Cell Signal 20, 453–459 [DOI] [PubMed] [Google Scholar]
- 8. Guy G. R., Jackson R. A., Yusoff P., Chow S. Y. (2009) J. Endocrinol. 203, 191–202 [DOI] [PubMed] [Google Scholar]
- 9. Hacohen N., Kramer S., Sutherland D., Hiromi Y., Krasnow M. A. (1998) Cell 92, 253–263 [DOI] [PubMed] [Google Scholar]
- 10. Tefft J. D., Lee M., Smith S., Leinwand M., Zhao J., Bringas P., Jr., Crowe D. L., Warburton D. (1999) Curr. Biol. 9, 219–222 [DOI] [PubMed] [Google Scholar]
- 11. de Maximy A. A., Nakatake Y., Moncada S., Itoh N., Thiery J. P., Bellusci S. (1999) Mech. Dev. 81, 213–216 [DOI] [PubMed] [Google Scholar]
- 12. Hanafusa H., Torii S., Yasunaga T., Matsumoto K., Nishida E. (2004) J. Biol. Chem. 279, 22992–22995 [DOI] [PubMed] [Google Scholar]
- 13. Jarvis L. A., Toering S. J., Simon M. A., Krasnow M. A., Smith-Bolton R. K. (2006) Development 133, 1133–1142 [DOI] [PubMed] [Google Scholar]
- 14. Sasaki A., Taketomi T., Wakioka T., Kato R., Yoshimura A. (2001) J. Biol. Chem. 276, 36804–36808 [DOI] [PubMed] [Google Scholar]
- 15. Wong E. S., Fong C. W., Lim J., Yusoff P., Low B. C., Langdon W. Y., Guy G. R. (2002) EMBO J. 21, 4796–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wakioka T., Sasaki A., Kato R., Shouda T., Matsumoto A., Miyoshi K., Tsuneoka M., Komiya S., Baron R., Yoshimura A. (2001) Nature 412, 647–651 [DOI] [PubMed] [Google Scholar]
- 17. Bennett A. M., Tang T. L., Sugimoto S., Walsh C. T., Neel B. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7335–7339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li W., Nishimura R., Kashishian A., Batzer A. G., Kim W. J., Cooper J. A., Schlessinger J. (1994) Mol. Cell. Biol. 14, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tartaglia M., Niemeyer C. M., Fragale A., Song X., Buechner J., Jung A., Hählen K., Hasle H., Licht J. D., Gelb B. D. (2003) Nat. Genet. 34, 148–150 [DOI] [PubMed] [Google Scholar]
- 20. Tartaglia M., Martinelli S., Iavarone I., Cazzaniga G., Spinelli M., Giarin E., Petrangeli V., Carta C., Masetti R., Aricò M., Locatelli F., Basso G., Sorcini M., Pession A., Biondi A. (2005) Br. J. Haematol. 129, 333–339 [DOI] [PubMed] [Google Scholar]
- 21. Bentires-Alj M., Paez J. G., David F. S., Keilhack H., Halmos B., Naoki K., Maris J. M., Richardson A., Bardelli A., Sugarbaker D. J., Richards W. G., Du J., Girard L., Minna J. D., Loh M. L., Fisher D. E., Velculescu V. E., Vogelstein B., Meyerson M., Sellers W. R., Neel B. G. (2004) Cancer Res. 64, 8816–8820 [DOI] [PubMed] [Google Scholar]
- 22. Loh M. L., Reynolds M. G., Vattikuti S., Gerbing R. B., Alonzo T. A., Carlson E., Cheng J. W., Lee C. M., Lange B. J., Meshinchi S. (2004) Leukemia 18, 1831–1834 [DOI] [PubMed] [Google Scholar]
- 23. Tartaglia M., Mehler E. L., Goldberg R., Zampino G., Brunner H. G., Kremer H., van der Burgt I., Crosby A. H., Ion A., Jeffery S., Kalidas K., Patton M. A., Kucherlapati R. S., Gelb B. D. (2001) Nat. Genet. 29, 465–468 [DOI] [PubMed] [Google Scholar]
- 24. Tartaglia M., Martinelli S., Stella L., Bocchinfuso G., Flex E., Cordeddu V., Zampino G., Burgt I., Palleschi A., Petrucci T. C., Sorcini M., Schoch C., Foa R., Emanuel P. D., Gelb B. D. (2006) Am. J. Hum. Genet. 78, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kontaridis M. I., Swanson K. D., David F. S., Barford D., Neel B. G. (2006) J. Biol. Chem. 281, 6785–6792 [DOI] [PubMed] [Google Scholar]
- 26. Razzaque M. A., Nishizawa T., Komoike Y., Yagi H., Furutani M., Amo R., Kamisago M., Momma K., Katayama H., Nakagawa M., Fujiwara Y., Matsushima M., Mizuno K., Tokuyama M., Hirota H., Muneuchi J., Higashinakagawa T., Matsuoka R. (2007) Nat. Genet. 39, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 27. Gelb B. D., Tartaglia M. (2006) Hum. Mol. Genet. 15, R220–226 [DOI] [PubMed] [Google Scholar]
- 28. Tartaglia M., Pennacchio L. A., Zhao C., Yadav K. K., Fodale V., Sarkozy A., Pandit B., Oishi K., Martinelli S., Schackwitz W., Ustaszewska A., Martin J., Bristow J., Carta C., Lepri F., Neri C., Vasta I., Gibson K., Curry C. J., Siguero J. P., Digilio M. C., Zampino G., Dallapiccola B., Bar-Sagi D., Gelb B. D. (2007) Nat. Genet. 39, 75–79 [DOI] [PubMed] [Google Scholar]
- 29. Pandit B., Sarkozy A., Pennacchio L. A., Carta C., Oishi K., Martinelli S., Pogna E. A., Schackwitz W., Ustaszewska A., Landstrom A., Bos J. M., Ommen S. R., Esposito G., Lepri F., Faul C., Mundel P., López Siguero J. P., Tenconi R., Selicorni A., Rossi C., Mazzanti L., Torrente I., Marino B., Digilio M. C., Zampino G., Ackerman M. J., Dallapiccola B., Tartaglia M., Gelb B. D. (2007) Nat. Genet. 39, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 30. Brems H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., Kato R., Somers R., Messiaen L., De Schepper S., Fryns J. P., Cools J., Marynen P., Thomas G., Yoshimura A., Legius E. (2007) Nat. Genet. 39, 1120–1126 [DOI] [PubMed] [Google Scholar]
- 31. Spurlock G., Bennett E., Chuzhanova N., Thomas N., Jim H. P., Side L., Davies S., Haan E., Kerr B., Huson S. M., Upadhyaya M. (2009) J. Med. Genet. 46, 431–437 [DOI] [PubMed] [Google Scholar]
- 32. Lao D. H., Chandramouli S., Yusoff P., Fong C. W., Saw T. Y., Tai L. P., Yu C. Y., Leong H. F., Guy G. R. (2006) J. Biol. Chem. 281, 29993–30000 [DOI] [PubMed] [Google Scholar]
- 33. Lao D. H., Yusoff P., Chandramouli S., Philp R. J., Fong C. W., Jackson R. A., Saw T. Y., Yu C. Y., Guy G. R. (2007) J. Biol. Chem. 282, 9117–9126 [DOI] [PubMed] [Google Scholar]
- 34. Tiganis T., Bennett A. M. (2007) Biochem. J. 402, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen L., Sung S. S., Yip M. L., Lawrence H. R., Ren Y., Guida W. C., Sebti S. M., Lawrence N. J., Wu J. (2006) Mol. Pharmacol. 70, 562–570 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.