Abstract
The peptide transporter (PTR) family represents a group of proton-coupled secondary transporters responsible for bulk uptake of amino acids in the form of di- and tripeptides, an essential process employed across species ranging from bacteria to humans. To identify amino acids critical for peptide transport in a prokaryotic PTR member, we have screened a library of mutants of the Escherichia coli peptide transporter YdgR using a high-throughput substrate uptake assay. We have identified 35 single point mutations that result in a full or partial loss of transport activity. Additional analysis, including homology modeling based on the crystal structure of the Shewanella oneidensis peptide transporter PepTso, identifies Glu56 and Arg305 as potential periplasmic gating residues. In addition to providing new insights into transport by members of the PTR family, these mutants provide valuable tools for further study of the mechanism of peptide transport.
Keywords: Amino Acid Transport, Membrane Proteins, Protein Structure, Proton Transport, Site-directed Mutagenesis, Peptide Transport
Introduction
Amino acid uptake provides a critical source of nitrogen, sulfur, and metabolic intermediates for many organisms. Amino acids are often transported into the cell in the form of short peptides to efficiently mediate their bulk uptake. Although several different systems mediating peptide transport exist in bacteria and fungi, only the peptide transporters (PTR),6 also called proton oligopeptide transporters, are conserved in higher organisms, including humans (1, 2). Like many proton-coupled transporters, the PTR family is a subdivision of the major facilitator superfamily (MFS) (3).
Sequence alignment among members of the PTR family across species reveals three conserved motifs in the protein sequence (4) (supplemental Fig. S1). The first such motif is found in the first predicted transmembrane helix, whereas the second is found in loop 2-3 and corresponds to the motif GXXX(D/E)(R/K)[X]G[X](R/K)(R/K), where [X] is either one or two amino acids, which is also found in many MFS transporters (5, 6). The third motif, the PTR signature motif FXXFYXXINXGS, is located in the fifth predicted transmembrane helix of PTR members, and mutation of these residues leads to reduced transport activity or changes in substrate selectivity (7, 8). In addition, chimeric studies on human PTR members PEPT1 and PEPT2 have revealed that helices 2–5 as well as 7 contain residues that mediate substrate recognition (9). This is supported by cysteine scanning mutagenesis of residues in helices 5 and 7 of PEPT1, which suggests that residues in these helices line the substrate translocation pathway (8, 10).
PTR family members possess a large number of possible substrates, including essentially all 400 possible di- or 8000 tripeptides. For human PEPT1 and PEPT2, the most well characterized PTRs, transport is stereoselective for the l-enantiomers of amino acids, although d-amino acids are accepted in the N-terminal position. Substrate affinity varies depending on the sequence of the particular di- or tripeptide (11). Furthermore, these transporters are able to take up peptidomimetics such as β-lactam antibiotics, some angiotensin-converting enzyme inhibitors, and some antiviral pro-drugs, such as valaciclovir (12). Due to the presence of PEPT1 in the brush-border membrane of the small intestine, and PEPT2 in the kidney and blood-brain barrier, these proteins significantly influence the systemic availability of these and other pharmacological compounds (12).
The Escherichia coli PTR protein YdgR, also called DtpA, is one of the few prokaryotic PTR members to have been characterized and shows substrate selectivity similar to that of PEPT1 (13, 14). Recently, the crystal structure of another prokaryotic PTR family member from Shewanella oneidensis, referred to as PepTso, was determined and revealed a fold very similar to that of other MFS transporters, including LacY, GlpT, and EmrD (15–18). The substrate binding cavity is lined with conserved residues, many of which were previously identified as critical for function (17). However, several functional residues, including His61 and Glu316, which are suggested to be essential for protonation and/or periplasmic gating, are not conserved between PepTso and other bacterial PTRs, including YdgR.
To identify additional residues critical for the function of YdgR and its homologues, we have performed random mutagenesis on this transporter and utilized a high-throughput substrate uptake assay to identify mutants exhibiting loss-of-function phenotypes. The results of this assay combined with additional analysis allowed us to identify 15 mutations leading to a complete loss-of-function phenotype, and 12 further mutations showing partial loss-of-function. In addition, 6 mutations were identified that exhibit this phenotype only in one of the uptake assays. Mapping of these residues onto a homology model of YdgR based on the PepTso structure and additional comparison to the known structure of the lactose permease LacY (15) allows for prediction of the potential functions of several of these residues, including Glu56, Phe289, Met295, and Arg305. Further analysis of Glu56 and Arg305 reveals that these residues may function as periplasmic gating residues in YdgR and other close bacterial homologues.
EXPERIMENTAL PROCEDURES
Molecular Cloning
The full-length ydgR gene was amplified from E. coli genomic DNA and cloned into the pCS19 vector (kind gift from Michael Ehrmann (19)) using the NcoI and BglII restriction sites. Site-directed mutagenesis was performed as described in Ref. 20.
Random Mutagenesis
Random mutations were introduced into regions of the ydgR gene encompassing helices 2–4 (H2–4; base pairs 130–444) and helices 6–8 (H6–8; base pairs 535–1000) using the GeneMorph II Random Mutagenesis Kit (Stratagene). To determine the mutation frequency in each library, the resulting PCR products were inserted into the pJET cloning vector using the GeneJET Cloning Kit (Fermentas). At least 10 different clones for each library were sequenced to estimate a mutation frequency of 0.35% for library H2–4 and 0.65% for library H6–8. The PCR products were then used as megaprimers with pCS19-YdgR as a template, to create two libraries of mutants in the pCS19 background.
Alafosfalin Growth Assay
The mutant libraries were transformed into E. coli BL21(DE3) and individual colonies were grown in 96-well plates under two conditions, one containing 200 μl of Luria-Bertani (LB) medium plus the antibiotic ampicillin, and one additionally containing 200 μg/ml of the antibiotic alafosfalin (Sigma). A600 was monitored 1 and 5 h after induction of ydgR expression. Clones exhibiting a growth advantage when compared with native YdgR, as determined by visual inspection of the growth curves, were sequenced within the entire open reading frame to identify the mutation(s). Several clones of each mutant identified in this manner were then re-tested in the growth assay to eliminate false positives.
Isolation of E. coli Membrane Fractions
Cell pellets from 50-ml cultures of C43(DE3) cells transformed with mutant or control constructs were harvested and membranes were isolated as described in Ref. 21. Equal concentrations of membranes from each construct were analyzed by SDS-PAGE and Western blot using an α-His-HRP-conjugated antibody (Qiagen). The intensity of each band was quantified using the ImageJ software (22).
Uptake of β-Ala-Lys(AMCA)
Uptake of β-Ala-Lys-AMCA (Biotrend) was measured essentially as described in Ref. 14, however, a 2.5 mm β-Ala-Lys-AMCA stock solution was used. Uptake was measured in 25 mm Tris buffer at pH 7.5 containing 140 mm NaCl, 5.4 mm KCl, 1.8 mm CaCl2, 0.8 mm MgSO4, and 5 mm glucose. Fluorescence measurements were additionally normalized to A600. Competition assays were carried out in the presence of 0 to 75 mm alafosfalin or alanyl-alanine (Bachem AG). IC50 values were determined by nonlinear regression analysis using the software program GraphPad Prism 5.0.
Homology Modeling
The HHPred-generated alignment of YdgR with PepTso (PDB code 2XUT) was used as the basis for the alignment input into the program Modeller (9 version 8) (23, 31). Evaluation of this model with PROCHECK resulted in 93.8% of the residues in the core region, 5.2% in the allowed region, 0.5% in the generously allowed region, and 0.5% disallowed (24). Figs. 5 and 6 were generated using the software package MOE (Molecular Operating Environment, version 2009.10, Chemical Computing Group, Montreal, Canada). For Fig. 6, the LacY (PDB code 2CFQ) and PepTso structures and the YdgR homology model were aligned using the Superposition tool in MOE.
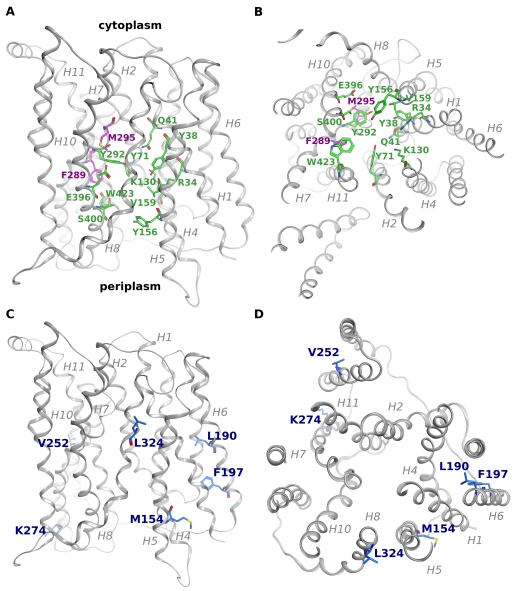
FIGURE 5.
Homology modeling reveals putative functional residues. A homology model of YdgR was generated based on the structure of the PepTso transporter (PDB code 2XUT). The YdgR backbone is colored gray and shown in ribbon representation with amino acids shown in stick representation. The transmembrane helices are labeled in gray. A, view perpendicular to the membrane. Conserved amino acids lining the substrate binding cavity are colored green, CPK. Phe289 and Met295 are shown in magenta, CPK. B, view parallel to the membrane from the cytoplasmic side. Colored as in A. C, view perpendicular to the membrane. Residues resulting in selectivity against alafosfalin are colored blue, CPK. D, view parallel to the membrane from the periplasmic side, colored as in C.
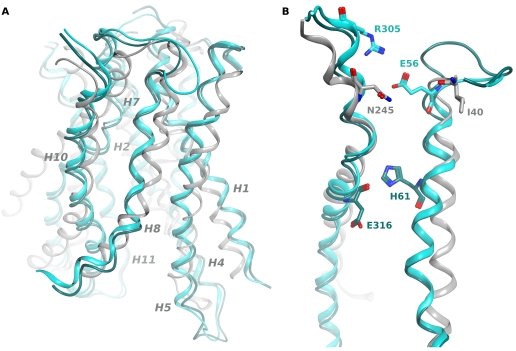
FIGURE 6.
Alignment of the YdgR model with LacY and PepTso identifies potential periplasmic gating residues. The YdgR homology model was aligned with the crystal structures of LacY (PDB code 2CFQ) and PepTso (PDB code 2XUT) to compare the positions of the periplasmic gating residues from LacY (Ile40, Asn245) and PepTso (His61, Glu316) with Glu56 and Arg305. A, view perpendicular to the membrane. The backbone of the YdgR homology model (cyan) is aligned with that of the LacY structure (light gray) and that of PepTso (dark cyan). For clarity only helices lining the substrate cavity are shown, and are labeled in gray. B, the isolated helices 2 and 7 of the aligned structures, colored as in A. Gating residues from each protein are shown in stick conformation.
RESULTS
Isolation of YdgR Loss-of-function Mutants
To identify residues critical for peptide transport in the E. coli YdgR protein, we used PCR mutagenesis to generate mutant libraries in two regions of the ydgR gene. The first library, encompassing helices 2–4 (H2–4), spans amino acids 44–148 of the ydgR gene (base pairs 130–444), and the second library, encompassing helices 6–8 (H6–8), spans amino acids 179–334 (base pairs 535–1000) (supplemental Fig. S1).
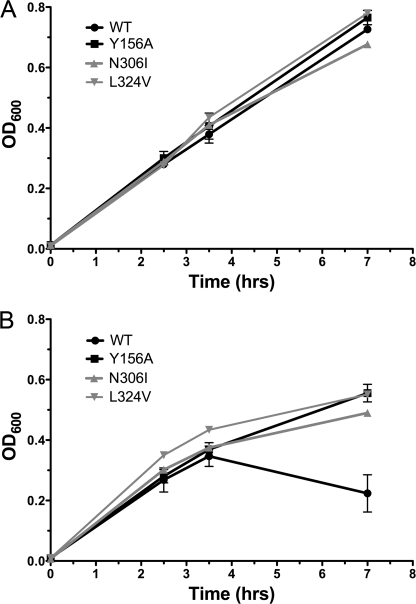
These two libraries were then tested for functionality based on the ability to transport the phosphonopeptide antibiotic alafosfalin, which is a specific substrate of prokaryotic PTRs, transport of which leads to inhibition of cell growth (25). Growth in the absence and presence of alafosfalin was monitored by measuring A600 at several time points after induction of ydgR expression (Fig. 1). Cells expressing transporters with mutations leading to a loss-of-function (LOF) phenotype are expected to grow normally in the presence of alafosfalin, whereas cells expressing the native transporter or transporters with mutations that do not affect transport ability will fail to survive, due to the toxic effect of the antibiotic. The Y156A mutant, which represents the homologous mutation of Tyr167 from PepT1 and results in a non-functional transporter (26), was used as a positive control. Representative data for 2 LOF mutants isolated in this assay, the native transporter and the Y156A mutant indicates that the mutants show a clear growth advantage as compared with the native transporter in the presence of alafosfalin (Fig. 1).
FIGURE 1.
Selection of mutants based on the ability to survive in the presence of alafosfalin. Cells expressing individual clones from each mutant library were grown in the absence (A) and presence (B) of alafosfalin and protein expression was induced at 2.5 h. A600 was measured at 1 h and 4.5 h after induction. Representative primary data are shown for the native transporter (WT), the Y156A mutant (Y156A), and the clones later identified as mutants N306I and L324V. Reported A600 values are from measurements in 96-well plates, and are ∼2.5 times lower than A600 determined using a 1-cm path length. The means of triplicate measurements of A600 for the native transporter and the Y156A loss-of-function mutant in the presence of alafosfalin from three individual experiments were compared using a paired t test within the Graphpad Prism 5.0 software, and were found to have a p value <0.0024, indicating that the difference is statistically significant.
Approximately 1600 mutants were screened in this assay, 600 from library H2–4 and 1000 from H6–8. From library H2–4, 101 LOF mutants were isolated. After sequencing the YdgR open reading frame, these consisted of 21 single amino acid mutations and 12 with double amino acid mutations. The remaining mutants contained single base pair deletions or insertions resulting in a frameshift, silent mutations, stop codons, or point mutations resulting in 3 or more amino acid changes. The mutant M154K lies outside of the mutagenized region, and therefore must have arisen independently during library generation. From library H6–8, 170 LOF mutants were identified. Initial sequencing revealed the frequent occurrence of mutants containing a long insertion rather than single point mutations, most likely occurring during the megaprimer amplification step. Therefore clones were pre-screened by colony PCR to eliminate clones containing the insertion. The remaining 118 mutants were sequenced within the YdgR open reading frame to identify mutations, and consisted of 27 single point mutants (22 unique) and 17 double point mutants.
The mutants resulting in single amino acid changes were re-assayed to confirm their phenotype, resulting in the identification of 8 false positives. The remaining 35 single mutants and the 29 isolated double mutants are shown in Table 1. Comparison of A600 values of the native and mutant clones 4–5 h post-induction was used to calculate a numerical growth advantage for each of the 35 single mutants (Table 2).
TABLE 1.
Mutants isolated in the primary screen
| Single mutants, library H2–4 | Single mutants, library H6–8 | Double mutants, library H2–4 | Double mutants, library H6–8 |
|---|---|---|---|
| E56G | M154Ka | L52P, K130E | L221R, V316L |
| S59P | L190V | S64Y, A99E | Q242H, F288L |
| I60N | N196K | S67T, A68P | G255D, N306K |
| A68P | F197I | V70I, L80F | I256F, I259F |
| G78C | V252E | Y71C, A75D | E263D, E317G |
| G86R | A264P | W79R, I91N | M275L, F279I |
| L98R | K274I | G86S, P133T | E284G, A285V |
| I100V | A285V | G89R, K130T | F289I, E317G |
| G101D | F289L | A95T, P133L | V290A, Q320H |
| I122N | F289S | A99V, K130I | Y292S, I331T |
| A123V | M295K | Y116C, M119K | T297R, A315T |
| G127D | T297A | M117I, S134P | L299P, R305H |
| L136R | N300I | L299R, L312M | |
| L137H | N300Y | F302C, R305P | |
| F301I | P308R, A371V | ||
| A303G | L312S, A405T | ||
| R305C | Q322L, A323P | ||
| N306I | |||
| Q320L | |||
| L324V | |||
| P326Q |
a Mutation occurs outside of the mutagenized region.
TABLE 2.
Membrane expression and transport characteristics of mutants isolated in the primary screen
| Mutant | Growth advantagea | Uptake of β-Ala-Lys(AMCA)b | Membrane expressionc | Corrected uptaked | Predicted location |
|---|---|---|---|---|---|
| % | % | % | |||
| Empty vector | 2.8 ± 0.5 | 0 ± 1 | 0 | 0 | |
| Wild-type | 1.0 ± 0.2 | 100 ± 5 | 100 | 100 | |
| Y156A | 2.5 ± 0.5 | 2 ± 1 | 96 | 2 | Helix 5 |
| No membrane localization | |||||
| I100V | 2.6 ± 0.5 | 3 ± 1 | 0 | 0 | Helix 3 |
| L137H | 2.4 ± 0.6 | 1 ± 4 | 0 | 0 | Helix 4 |
| LOF mutants | |||||
| E56G | 2.3 ± 0.1 | 10 ± 4 | 89 | 11 | Helix 2 |
| S59P | 2.5 ± 0.3 | -1 ± 2 | 100 | -1 | Helix 2 |
| I60N | 3.3 ± 0.5 | 1 ± 2 | 106 | 1 | Helix 2 |
| A68P | 2.6 ± 0.3 | 1 ± 1 | 105 | 1 | Helix 2 |
| G78C | 2.3 ± 0.4 | 1 ± 3 | 91 | 1 | Helix 2 |
| L98R | 2.4 ± 0.4 | 1 ± 1 | 114 | 1 | Helix 3 |
| L136R | 2.5 ± 0.5 | 0 ± 1 | 79 | 0 | Helix 4 |
| A285V | 2.3 ± 0.2 | 11 ± 6 | 105 | 10 | Helix 7 |
| F289L | 2.4 ± 0.2 | 1 ± 1 | 81 | 1 | Helix 7 |
| F289S | 2.1 ± 0.3 | -1 ± 4 | 67 | -1 | Helix 7 |
| M295K | 2.2 ± 0.2 | -1 ± 2 | 82 | -1 | Helix 7 |
| T297A | 2.4 ± 0.1 | 3 ± 1 | 103 | 3 | Helix 7 |
| N300I | 2.7 ± 0.5 | 7 ± 1 | 116 | 6 | Helix 7 |
| F301I | 2.7 ± 0.2 | 2 ± 1 | 75 | 2 | Helix 7 |
| Q320L | 2.2 ± 0.5 | 7 ± 3 | 127 | 6 | Helix 8 |
| Partial LOFs | |||||
| G86R | 1.7 ± 0.3 | 0 ± 3 | 94 | 0 | Loop 2–3 |
| G101D | 1.2 ± 0.3 | 35 ± 12 | 113 | 31 | Helix 3 |
| I122N | 1.5 ± 0.1 | 17 ± 4 | 117 | 15 | Helix 4 |
| A123V | 1.3 ± 0.5 | 23 ± 1 | 61 | 38 | Helix 4 |
| G127D | 1.3 ± 0.2 | 60 ± 8 | 24 | 240 | Helix 4 |
| N196K | 1.9 ± 0.2 | 35 ± 4 | 120 | 29 | Helix 6 |
| A264P | 1.7 ± 0.3 | 39 ± 2 | 88 | 44 | Helix HB |
| N300Y | 3.3 ± 0.4 | 25 ± 8 | 62 | 40 | Helix 7 |
| A303G | 1.3 ± 0.2 | 42 ± 9 | 94 | 45 | Helix 7 |
| R305C | 1.7 ± 0.1 | 3 ± 1 | 62 | 5 | Helix 7 |
| N306I | 1.4 ± 0.2 | 21 ± 1 | 100 | 21 | Helix 7 |
| P326Q | 1.6 ± 0.3 | 2 ± 2 | 57 | 4 | Helix 8 |
| Selectivity mutants | |||||
| M154K | 2.4 ± 0.2 | 257 ± 70 | 93 | 276 | Helix 5 |
| L190V | 2.6 ± 0.5 | 105 ± 25 | 89 | 118 | Helix 6 |
| F197I | 2.7 ± 0.4 | 142 ± 26 | 86 | 165 | Helix 6 |
| V252E | 2.2 ± 0.2 | 87 ± 7 | 84 | 104 | Helix HB |
| K274I | 2.5 ± 0.3 | 59 ± 21 | 55 | 107 | Helix 7 |
| L324V | 1.8 ± 0.5 | 242 ± 11 | 80 | 303 | Helix 8 |
a Data are represented as the fold difference in A600 of mutant versus native transporter in the presence and absence of alafosfalin 5 h post-induction: (Amutant + Alafos./AWT + Alafos)/(Amutant − Alafos/AWT − Alafos). Error values represent S.D. of triplicate measurements from at least two different experiments.
b Fluorescence values are normalized after setting the value of the overexpressed native transporter to 100%.
c % Membrane expression was determined by quantitation of the band intensity on the Western blots shown in Fig. 2 using ImageJ software (22) and normalized by setting the value of the overexpressed native transporter to 100%.
d Corrected uptake represents: (% uptake of β-Ala-Lys(AMCA)/% Membrane expression) × 100.
Functional Analysis Reveals Four Different Phenotypes
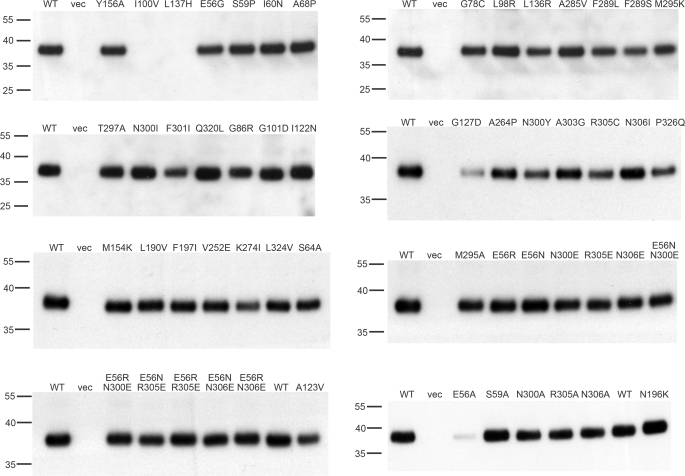
The 35 single mutants were then further characterized. To determine whether their phenotype is simply due to loss of protein expression or membrane insertion, membrane localization was determined by isolation of the membrane fraction from small-scale cultures and verification of the presence of protein in the membrane fraction by Western blot using an α-His antibody to detect the C-terminal His6 tag of the protein. This analysis revealed that two mutants, I100V and L137H, show no expression in the membrane fraction, whereas G127D is present in the membrane at very low levels, and K274I is expressed at only 55% as compared with native YdgR (Table 2, Fig. 2). All other mutants show expression levels of 60% or more, and interpretation of their transport phenotype is not affected by their expression level.
FIGURE 2.
Membrane localization of all mutants. The localization of mutants to the plasma membrane was tested by preparation of E. coli membranes, analysis by SDS-PAGE and Western blot, and quantitation of the band intensity. Molecular mass markers (kDa) are indicated to the left of each Western blot. The quantitation of the expression level of each mutant is recorded in Tables 2 or 4. WT, membranes from cells overexpressing native YdgR; vec, membranes from cells expressing the empty vector.
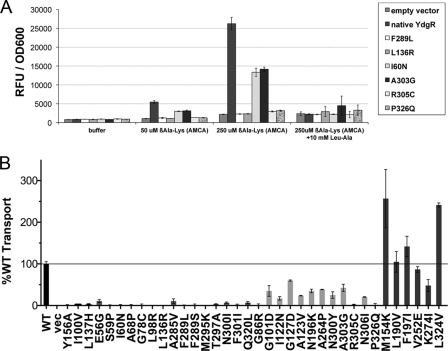
To further analyze the transport activity of the individual mutants, their ability to take up the fluorescent substrate β-Ala-Lys(AMCA), a known substrate of YdgR (14), was assessed and compared with that of the native transporter (Fig. 3). By compiling the data from the alafosfalin growth assay and the β-Ala-Lys(AMCA) uptake assay, the 35 mutants can be divided into four phenotypic categories (Table 2). The first category includes the two mutants that fail to localize to the membrane. The second and largest category comprises 15 mutants that show a 2-fold or greater growth advantage in the presence of alafosfalin, and less than 10% uptake activity as compared with the native transporter. Most of these mutants represent amino acid substitutions that dramatically alter the properties of the side chain, such as hydrophobic to charged or vice versa, or mutations to glycine or proline, which may significantly alter the protein structure. However, we have also identified more conservative mutations that lead to the same phenotype, including F289L, T297A, and F301I.
FIGURE 3.
Uptake of β-Ala-Lys(AMCA) reveals distinct phenotypes. The ability of each mutant to take up the fluorescent dipeptide substrate β-Ala-Lys(AMCA) was measured and compared with that of the native transporter. A, representative data for uptake by native YdgR, empty vector, and 6 mutants are shown under conditions including either buffer alone, β-Ala-Lys(AMCA) alone, or β-Ala-Lys(AMCA) plus 10 mm of a dipeptide competitor (Leu-Ala, leucyl-alanine). Data are shown as raw fluorescence units (RFU) normalized to A600. Error bars represent standard deviations from triplicate measurements. B, after subtraction of nonspecific transport, as measured by residual transport by cells expressing the empty vector, the transport ability of each mutant was normalized with respect to the native transporter, which was set to 100%. Error bars represent standard deviations of triplicate measurements from at least two different experiments. Wild-type (WT), vector (vec), and Y156A controls are shown as black bars, mutants are grouped and shaded according to the phenotypic categories shown in Table 2. A black line is used to indicate 100% transport.
The third category represents 12 mutants with a slight growth advantage in the presence of alafosfalin, and uptake activity ranging from ∼20 to 60% of the native transporter. The mutants G86R, R305C, and P326Q show very little transport in the β-Ala-Lys(AMCA) uptake assay, however, do retain some ability to take up alafosfalin, and have therefore been included in this category. Similarly, N300Y seems unable to transport alafosfalin, but still retains significant ability to transport β-Ala-Lys(AMCA), particularly when correcting for its reduced expression level. Again, many of these mutations represent substitution of one type of amino acid for another, however, they show milder phenotypes than the complete loss-of-function mutants. Most interesting, the G127D mutant maintains significant transport activity despite its low expression level (Table 2).
A fourth intriguing group of mutants, which we refer to as selectivity mutants, are those that exhibit a loss-of-function phenotype in the alafosfalin growth assay, yet show uptake of β-Ala-Lys(AMCA) equivalent to or, in some cases, even greater than that of the native transporter (Table 2). This would suggest that these mutations do not affect the ability of the protein to transport substrates, but rather may affect substrate selectivity or affinity. Although the uptake assay is performed in minimal media with only one potential substrate present, the alafosfalin growth assay is performed in complex media (LB), which contains other potential di- and tripeptide substrates. Although the presence of competitors in complex media does not inhibit transport of alafosfalin by the native transporter, higher affinity interaction with competitors, and/or lower affinity interaction with alafosfalin, may prevent alafosfalin uptake in these mutants.
Selectivity Mutants Show Lower Apparent Affinity for Alafosfalin
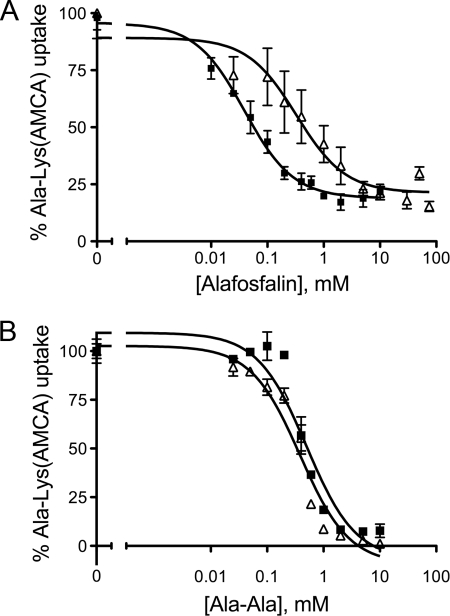
To test the hypothesis that these mutants have a lower apparent affinity for alafosfalin than the native transporter, we determined the IC50 and Ki values for alafosfalin for these six mutants by performing the β-Ala-Lys(AMCA) uptake assay in the presence of increasing concentrations of alafosfalin (Fig. 4A, Table 3). This experiment reveals that mutants M154K, L190V, F197I, V252E, and L324V have Ki values 3.5–13.5-fold higher than the native transporter, indicating weaker interaction with alafosfalin. In contrast, the mutant K274I has a Ki value equal to that of the native transporter.
FIGURE 4.
Competitive binding data for native YdgR and the V252E mutant. A, inhibition of β-Ala-Lys(AMCA) uptake by alafosfalin. Representative data for the native transporter (black squares) and the V252E mutant (white triangles) from one experiment are shown. B, inhibition of β-Ala-Lys(AMCA) uptake by alanyl-alanine (Ala-Ala). Representative data for the native transporter (black squares) and the V252E mutant (white triangles) are shown. Data for all mutants are summarized in Table 3.
TABLE 3.
Apparent affinities of the selectivity mutants for alafosfalin and alanyl-alanine (Ala-Ala)
| YdgR mutant | Alafosfalin |
Ala-Ala |
||
|---|---|---|---|---|
| IC50a | Kib | IC50a | Kib | |
| mm | ||||
| Wild-type | 0.11 ± 0.07 | 0.07 ± 0.08 | 0.33 ± 0.16 | 0.21 ± 0.2 |
| M154K | 0.39 ± 0.12 | 0.25 ± 0.16 | 0.12 ± 0.01 | 0.08 ± 0.01 |
| L190V | 0.59 ± 0.03 | 0.38 ± 0.04 | 0.32 ± 0.06 | 0.20 ± 0.08 |
| F197I | 0.71 ± 0.02 | 0.45 ± 0.03 | 0.48 ± 0.01 | 0.31 ± 0.01 |
| V252E | 0.44 ± 0.21 | 0.28 ± 0.27 | 0.33 ± 0.05 | 0.21 ± 0.06 |
| K274I | 0.08 ± 0.04 | 0.05 ± 0.05 | 0.31 ± 0.07 | 0.20 ± 0.09 |
| L324V | 1.5 ± 0.07 | 0.96 ± 0.09 | 0.56 ± 0.10 | 0.36 ± 0.13 |
a IC50 values represent averages from at least two experiments with error calculated as S.D.
b Ki values were determined using the equation: Ki = IC50/(1 + [substrate]/KD. The KD value used was 0.44 ± 0.05 mm, which is the apparent affinity of YdgR for β-Ala-Lys(AMCA) as determined in Ref. 14.
The structure of alafosfalin mimics that of the dipeptide alanyl-alanine, however, in alafosfalin (l-alanyl-l-1-aminoethylphosphonic acid) the carboxyl group on the C-terminal moiety is replaced by phosphonic acid. To determine whether these mutants show lower apparent affinity for dipeptide substrates with an unmodified carboxyl terminus, the IC50 and Ki values for alanyl-alanine were also determined (Fig. 4B, Table 3). For alanyl-alanine, the Ki values for five mutants are within ±2-fold of that of the native transporter, and for one mutant, M154K, the apparent affinity is higher than that of the native transporter. This suggests that the lower apparent affinity for alafosfalin for 5 of these mutants is not a general property of substrate binding, but rather may be specific for the substrate alafosfalin. We conclude that the loss of apparent affinity for alafosfalin is related to the presence of the additional C-terminal charged hydroxyl, either through direct steric hindrance of binding or unfavorable charge-charge interactions. As the K274I mutant shows apparent affinities similar to those of the native transporter for both alafosfalin and alanyl-alanine, it is not clear from these experiments why this mutant shows a LOF phenotype in the alafosfalin growth assay.
Localizing Mutations Based on Homology Modeling
To attempt to correlate the phenotype of individual mutations with their location in the transporter, we generated a homology model of YdgR based on the structure of PepTso (17). PepTso shares 26% identity and 45% similarity with YdgR (supplemental Fig. S1). Almost all of the residues in the peptide binding site of PepTso are conserved in YdgR, and also localize to the substrate cavity in our model (supplemental Fig. S1, Fig. 5, A and B).
The majority of LOF mutants isolated in our screen are located in helices 2, 4, and 7 of our homology model, with the majority in helix 7 (Table 2, supplemental Fig. S1). Significantly fewer mutants were found in helices 3, 6, and 8. This correlates well with data from other MFS family members, as based on the structures of LacY, GlpT, EmrD, and PepTso, helices 2, 4, and 7 are located in the hydrophilic cavity representing the substrate translocation pathway and contain many critical functional residues (15, 16–18). Residues that when mutated result in a partial LOF phenotype are located primarily in helix 4 (Ile122, Ala123, Gly127) or close to the end of helix 7 (Asn300, Ala303, Arg305, Asn306), and appear to be more distant from the substrate binding site (17) (Fig. 5, supplemental Fig. S1).
Most of the LOF or partial LOF mutations isolated in our screen are located in the transmembrane helices, and, as mentioned previously, represent quite severe mutations. Therefore, many of these mutations most probably lead to dramatic changes in helix packing (I60N, L98R, I122N, L136R) or introduce kinks or bends in the helices (S59P, A68P, G78C, G86R, G101D, G127D, A264P, P326Q). However, several mutations are more conservative (A285V, F289L, F301I) yet also result in complete loss of function. Phe289 is of particular interest as in our model this residue is in a position equivalent to that of Trp312 in PepTso, one of the few residues lining the substrate cavity that is not strictly conserved in YdgR (Fig. 5, A and B, supplemental Fig. S1).
Another residue of interest is Met295, as it is located in the substrate binding cavity of our homology model, and the mutant M295K shows a complete loss of transport function (Table 2, Fig. 5, A and B). To determine whether the side chain of Met295 plays a role in substrate binding, we created mutant M295A and measured its ability to transport substrates in both the alafosfalin growth assay and the β-Ala-Lys(AMCA) uptake assay. M295A is unable to transport alafosfalin, with a growth advantage of 2.3 ± 0.1 over native YdgR (Table 4). However, M295A shows 70% β-Ala-Lys(AMCA) uptake activity as compared with native YdgR, which increases to 89% when accounting for the reduced expression level of the mutant (Fig. 7A, Table 4). Therefore we conclude that this residue does not play a significant role in mediating substrate interaction in general, but may modulate affinity toward certain substrates, as it is selective against alafosfalin.
TABLE 4.
Membrane expression and transport characteristics of alanine and charge-swap mutants
| Mutant | Growth advantagea | Uptake of β-Ala-Lys(AMCA)b | Membrane expressionc | Corrected uptaked |
|---|---|---|---|---|
| % | % | % | ||
| E56A | 2.6 ± 0.5 | 9 ± 1 | 7 | 129 |
| E56N | 15 ± 5 | 106 | 14 | |
| E56R | 4 ± 3 | 95 | 4 | |
| S59A | 0.8 ± 0.2 | 149 ± 36 | 111 | 134 |
| S64A | 95 ± 19 | 72 | 132 | |
| M295A | 2.3 ± 0.1 | 72 ± 8 | 81 | 89 |
| N300A | 2.6 ± 0.1 | 24 ± 13 | 88 | 27 |
| N300E | 9 ± 4 | 85 | 11 | |
| R305A | 2.4 ± 0.6 | 7 ± 3 | 85 | 8 |
| R305E | 9 ± 4 | 79 | 11 | |
| N306A | 1.2 ± 0.1 | 72 ± 8 | 87 | 8 |
| N306E | 8 ± 3 | 87 | 9 | |
| E56N, N300E | −2 ± 2 | 77 | −3 | |
| E56R, N300E | −2 ± 1 | 92 | −2 | |
| E56N, R305E | 35 ± 15 | 81 | 43 | |
| E56R, R305E | 220 ± 66 | 101 | 218 | |
| E56N, N306E | −2 ± 2 | 93 | −2 | |
| E56R, N306E | −2 ± 2 | 96 | −2 |
a Data are represented as the fold-difference in A600 of mutant versus native transporter in the presence and absence of alafosfalin 5 h post-induction: (Amutant + Alafos/AWT + Alafos.)/(Amutant–Alafos/AWT − Alafos). Error values represent S.D. of triplicate measurements from at least two different experiments.
b Fluorescence values are normalized after setting the value of the overexpressed native transporter to 100%.
c % Membrane expression was determined by quantitation of the band intensity on the Western blots shown in Fig. 2 using ImageJ software (22) and normalized by setting the value of the overexpressed native transporter to 100%.
d Corrected uptake represents: (% uptake of β-Ala-Lys(AMCA)/% membrane expression) × 100.
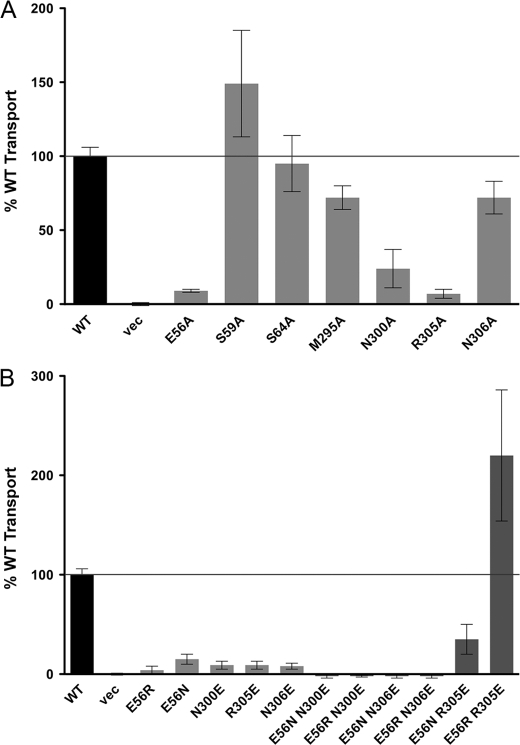
FIGURE 7.
Glu56 and Arg305 are potential periplasmic gating residues. The ability of each mutant to take up the fluorescent dipeptide substrate β-Ala-Lys(AMCA) was measured and compared with that of the native transporter. Error bars represent standard deviations of triplicate measurements from at least two different experiments. Wild-type (WT) and vector (vec) controls are shown as black bars, single mutants are shaded in light gray, and double mutants in dark gray. A black line is used to indicate 100% transport. A, the ability of E56A, S59A, S64A, M295A, N300A, R305A, and N306A to take up β-Ala-Lys(AMCA) is compared with wild-type (WT) and vector (vec) controls. B, the ability of single and double charge-swap mutations of Glu56, Ans300, Arg305, and Asn306 to take up β-Ala-Lys(AMCA) is compared with wild-type (WT) and vector (vec) controls.
The residues that when mutated show selectivity against alafosfalin are, perhaps surprisingly, located distant from the substrate binding cavity (Fig. 5, C and D). Residues Leu190 and Phe197 are located in helix 6 and most likely affect packing with the adjacent helices 1, 3, and 4. Val252 is located in helix HB of the membrane hairpin, which is not thought to play a role in transport, but may also affect packing of adjacent helices. Met154 is located at the cytoplasmic end of helix 5, whereas Lys274 is located at the cytoplasmic end of helix 7. Leu324 is located in helix 8, within the strictly conserved “LNP” motif, and its mutation may affect the interface with helices 10 and 7 (Fig. 5, C and D, supplemental Fig. S1).
Several polar or charged residues that were isolated as LOF mutants (Glu56, Ser59, Asn300, Arg305, Asn306) are located in the periplasmic halves of helices 2 and 7, and may form part of a hydrogen bonding network critical for closing of the periplasmic cavity, as seen in LacY and GlpT (27, 28). As the putative gating residues of the peptide transporter PepTso, His61 and Glu316, are not conserved in YdgR and many other bacterial PTRs (17) (supplemental Fig. S1), we hypothesized that one or more of these residues are potential candidates for the periplasmic gating residues of YdgR. To compare the positions of these residues to the putative gating residues of PepTso and the previously characterized gating residues of LacY, we aligned the structures of PepTso, LacY, and the YdgR homology model. Glu56 and Arg305 are within close proximity in our homology model, and are located in positions similar to the LacY gating residues Ile40 and Asn245 (28) (Fig. 6).
Glu56 and Arg305 Are Potential Gating Residues
To determine whether any of the charged residues near the periplasmic ends of helix 2 and 7 isolated in our screen may be responsible for periplasmic gating we first mutated Glu56, Ser59, Asn300, Arg305, and Asn306 to alanine. If the side chains of any of these residues are involved in periplasmic gating, we would expect mutation to alanine to result in a LOF phenotype similar to that of the original mutation.
We tested these new mutants in the alafosfalin growth assay and β-Ala-Lys(AMCA) uptake assay. This analysis reveals that mutants N300A and R305A have phenotypes identical to those of the original mutation isolated in the screen, suggesting a functional role for the side chains of these amino acids (Fig. 7A, Table 4). E56A also shows a loss-of-function phenotype, however, this is due to a dramatic loss in membrane expression (Table 4, Fig. 2). However, because E56G also displays a LOF phenotype and localizes to the membrane, we hypothesized that the Glu56 side chain is also critical for function. In contrast, as Ser59 and Asn306 show an increase in transport activity as compared with the original mutation, we conclude that these residues are unlikely to be critical gating residues. Additionally, we mutated Ser64 to alanine, as this residue is in a position equivalent to that of His61 in PepTso (supplemental Fig. S1) and tested this mutant in the β-Ala-Lys(AMCA) uptake assay, where it displays activity equal to that of the native transporter, making this side chain unlikely to play a role in gating (Fig. 7A, Table 4).
To test whether E56 interacts with Asn300 or Arg305 we created several “charge-swapped” transporters, in which Glu56 is mutated to either asparagine or arginine, and Asn300 or Arg305 is mutated to glutamate. We also included Asn306 in this study due to its proximity to Arg305. We expected that transporters containing the single mutations would be inactive, whereas if the side chains of these residues form a hydrogen bond or charge-based interaction, a double mutant in which the residues are swapped may rescue activity. When tested, the single mutants all show no activity in the β-Ala-Lys(AMCA) uptake assay (Fig. 7B, Table 4). The double mutants tested are also inactive, with the exception of the E56N/R305E double mutant, in which transport activity is restored to 35% of the native transporter, and the E56R/R305E double mutant, in which transport activity is dramatically increased to 220% of the native transporter (Fig. 7B, Table 4). We therefore propose that Glu56 and Arg305 are the periplasmic gating residues in YdgR, fulfilling the roles of His61 and Glu316 in PepTso.
DISCUSSION
We have identified residues in the E. coli YdgR peptide transporter that compromise transport ability when mutated, based on random mutagenesis and selection for mutants with a loss-of-function phenotype. By generating a homology model of YdgR based on the structure of PepTso, we have localized these mutations to specific transmembrane helices with proximity to the substrate translocation pathway. Based on our homology model, our functional data, and studies of PepTso, LacY, GlpT, and PepT1 several interesting insights emerge regarding individual residues as well as YdgR function in general.
The phenotypes of several mutants correlate well with the results of mutagenesis of other PTR or MFS members. Two of the mutants isolated in our study, Gly78 and Gly86, confirm the importance of the conserved MFS motif in loop 2–3, and are both conserved in prokaryotic homologues of YdgR (supplemental Fig. S1). Gly78 is equivalent to Gly64 in LacY, which seems critical for maintaining conformational flexibility, as only mutation to alanine is tolerated at this position (29). In agreement with studies on LacY, mutation of Gly78 shows a more severe phenotype than mutation of Gly86 (6).
We were also able to identify Phe289 as the likely equivalent of Trp312 of PepTso, one of the few residues in the substrate binding pocket that is not strictly conserved between YdgR and PepTso. Both phenotypes of the Phe289 mutants, two of which were isolated in our screen, and our homology model point to a role for Phe289 equivalent to that of Trp312. In PepTso, Trp312 is forming hydrophobic interactions with Trp446 (Trp423 in YdgR) in the substrate binding cavity. In human PEPT1, mutation of the Trp312 equivalent Trp294 to alanine also results in reduced transport activity (26), providing additional support for the role of this residue in substrate transport.
We also propose Met295 as an additional residue in the substrate binding pocket that may mediate substrate affinity. This residue is conserved in many bacterial species, although not in PepTso. As mutation of Met295 to alanine creates a selectivity mutant, it is clear that even changes to less conserved residues in the substrate binding pocket or in residues influencing packing of the helices lining the substrate cavity may affect substrate affinity and selectivity.
Because even conservative mutations distant from the substrate translocation pathway, such as L190V or L324V, lead to changes in the apparent affinity for selected substrates, we propose that YdgR undergoes large and dynamic conformational transitions, as has been shown for LacY and GlpT (27, 30). Due to the large number of mutations we isolated in helix 7, it seems that in particular the interhelical interface of this helix is critical for proper protein function. This is in agreement with structural work on PepTso. Comparison between LacY in the open-in conformation and PepTso in the occluded conformation shows the greatest displacement in helix 7 (17). This underscores the similarity between YdgR and MFS transporters.
Although the crystal structure of PepTso identified the potential periplasmic gating residues His61 and Glu316, these residues are not conserved in most other prokaryotic PTR family members. Through our mutagenesis screen, we identified several candidate amino acids that may play a role in periplasmic gating, and we were able to confirm that two of these residues, Glu56 and Arg305, are critical for transport function, most likely through formation of a charge-based interaction that may regulate gating. As these residues are located on the periplasmic termini of helices 2 and 7 in our homology model, they would also be positioned close to the small extracellular cavity seen in the PepTso structure, formed by the periplasmic halves of helices 2, 7, 11, and 12 (17). As this cavity has been proposed as the entry site for peptide substrates (17), these residues are likely to be properly positioned for coupling of gating and substrate binding.
Comparison of the locations of the putative periplasmic gating residues of PepTso and YdgR with those of LacY indicates that the position of the periplasmic gate of YdgR seems to be much closer to that of LacY than that of PepTso (Fig. 7B). The putative gating residues of YdgR show higher conservation within bacterial PTR family members than His61 and Glu316 of PepTso, however, they are also not conserved in more distant homologues (supplemental Fig. S1). This suggests that periplasmic gating may not be strictly conserved within the PTR family, and more than one periplasmic gating mechanism may have evolved. Taken together, the results of our analysis suggest that YdgR shares many functional similarities with both PepTso and LacY, thus adding support to the hypothesis that members of the MFS superfamily, and perhaps other proton-coupled transporter families, share a common fold and mechanism, despite transporting a wide range of substrates.
Supplementary Material
Acknowledgments
We thank Stefan Westermann and Tim Clausen for critical reading of the manuscript, and Subhodeep Sarker for assistance with GraphPad Prism 5.0. We are grateful to Daniel Harder and Hannelore Daniel for helpful discussions at the start of this project and for providing us with initial amounts of β-Ala-Lys(AMCA).
This work was supported by Austrian Science Fund, SFB35 projects F3507 and F3502, the Federal Ministry of Economy, Family and Youth through “Laura Bassi Centre of Expertise” initiative project Number 253275, and Boehringer Ingelheim.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PTR
- peptide transporter
- MFS
- major facilitator superfamily
- AMCA
- 7-amino-4-methylcoumarin-3-acetic acid
- LOF
- loss-of-function
- PDB
- Protein Data Bank.
REFERENCES
- 1. Paulsen I. T., Skurray R. A. (1994) Trends Biochem. Sci. 19, 404 [DOI] [PubMed] [Google Scholar]
- 2. Steiner H. Y., Naider F., Becker J. M. (1995) Mol. Microbiol. 16, 825–834 [DOI] [PubMed] [Google Scholar]
- 3. Chang A. B., Lin R., Keith Studley W., Tran C. V., Saier M. H., Jr. (2004) Mol. Membr. Biol. 21, 171–181 [DOI] [PubMed] [Google Scholar]
- 4. Daniel H., Spanier B., Kottra G., Weitz D. (2006) Physiology 21, 93–102 [DOI] [PubMed] [Google Scholar]
- 5. Henderson P. J., Maiden M. C. (1990) Philos. Trans. R. Soc. Lond. B Biol. Sci. 326, 391–410 [DOI] [PubMed] [Google Scholar]
- 6. Jessen-Marshall A. E., Paul N. J., Brooker R. J. (1995) J. Biol. Chem. 270, 16251–16257 [DOI] [PubMed] [Google Scholar]
- 7. Hauser M., Kauffman S., Naider F., Becker J. M. (2005) Mol. Membr. Biol. 22, 215–227 [DOI] [PubMed] [Google Scholar]
- 8. Kulkarni A. A., Haworth I. S., Lee V. H. (2003) Biochem. Biophys. Res. Commun. 306, 177–185 [DOI] [PubMed] [Google Scholar]
- 9. Fei Y. J., Liu J. C., Fujita T., Liang R., Ganapathy V., Leibach F. H. (1998) Biochem. Biophys. Res. Commun. 246, 39–44 [DOI] [PubMed] [Google Scholar]
- 10. Kulkarni A. A., Haworth I. S., Uchiyama T., Lee V. H. (2003) J. Biol. Chem. 278, 51833–51840 [DOI] [PubMed] [Google Scholar]
- 11. Daniel H., Morse E. L., Adibi S. A. (1992) J. Biol. Chem. 267, 9565–9573 [PubMed] [Google Scholar]
- 12. Brandsch M., Knütter I., Bosse-Doenecke E. (2008) J. Pharm. Pharmacol. 60, 543–585 [DOI] [PubMed] [Google Scholar]
- 13. Harder D., Stolz J., Casagrande F., Obrdlik P., Weitz D., Fotiadis D., Daniel H. (2008) FEBS J. 275, 3290–3298 [DOI] [PubMed] [Google Scholar]
- 14. Weitz D., Harder D., Casagrande F., Fotiadis D., Obrdlik P., Kelety B., Daniel H. (2007) J. Biol. Chem. 282, 2832–2839 [DOI] [PubMed] [Google Scholar]
- 15. Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 16. Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 17. Newstead S., Drew D., Cameron A. D., Postis V. L., Xia X., Fowler P. W., Ingram J. C., Carpenter E. P., Sansom M. S., McPherson M. J., Baldwin S. A., Iwata S. (2011) EMBO J. 30, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yin Y., He X., Szewczyk P., Nguyen T., Chang G. (2006) Science 312, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Spiess C., Beil A., Ehrmann M. (1999) Cell 97, 339–347 [DOI] [PubMed] [Google Scholar]
- 20. Liu H., Naismith J. H. (2008) BMC Biotechnol. 8, 91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Surade S., Klein M., Stolt-Bergner P. C., Muenke C., Roy A., Michel H. (2006) Protein Sci. 15, 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abramoff M. D., Magelhaes P. J., Ram S. J. (2004) Biophotonics Int. 11, 36–42 [Google Scholar]
- 23. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 24. Laskowski R., MacArthur M., Moss D., Thornton J. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 25. Gibson M. M., Price M., Higgins C. F. (1984) J. Bacteriol. 160, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bolger M. B., Haworth I. S., Yeung A. K., Ann D., von Grafenstein H., Hamm-Alvarez S., Okamoto C. T., Kim K. J., Basu S. K., Wu S., Lee V. H. (1998) J. Pharm. Sci. 87, 1286–1291 [DOI] [PubMed] [Google Scholar]
- 27. Law C. J., Almqvist J., Bernstein A., Goetz R. M., Huang Y., Soudant C., Laaksonen A., Hovmöller S., Wang D. N. (2008) J. Mol. Biol. 378, 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Y., Nie Y., Kaback H. R. (2009) J. Mol. Biol. 394, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jessen-Marshall A. E., Parker N. J., Brooker R. J. (1997) J. Bacteriol. 179, 2616–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guan L., Kaback H. R. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 67–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Söding J., Biegert A., Lupas A. N. (2005) Nucleic Acids Res. 33, W244–W248 (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.