Abstract
When deprived of anchorage to the extracellular matrix, fibroblasts arrest in G1 phase at least in part due to inactivation of G1 cyclin-dependent kinases. Despite great effort, how anchorage signals control the G1-S transition of fibroblasts remains highly elusive. We recently found that the mammalian target of rapamycin (mTOR) cascade might convey an anchorage signal that regulates S phase entry. Here, we show that Rho-associated kinase connects this signal to the TSC1/TSC2-RHEB-mTOR pathway. Expression of a constitutively active form of ROCK1 suppressed all of the anchorage deprivation effects suppressible by tsc2 mutation in rat embryonic fibroblasts. TSC2 contains one evolutionarily conserved ROCK target-like sequence, and an alanine substitution for Thr1203 in this sequence severely impaired the ability of ROCK1 to counteract the anchorage loss-imposed down-regulation of both G1 cell cycle factors and mTORC1 activity. Moreover, TSC2 Thr1203 underwent ROCK-dependent phosphorylation in vivo and could be phosphorylated by bacterially expressed active ROCK1 in vitro, providing biochemical evidence for a direct physical interaction between ROCK and TSC2.
Keywords: mTOR, Protein Kinases, Rat, Rho, Tuberous Sclerosis (Tsc), Cdk2, Cdk4, G1 Cyclin, Rho-associated Kinase, Tsc2
Introduction
Unless malignantly transformed, virtually all cells in adult mammals (other than those of hematopoietic origin) require anchorage (or adhesion) to the extracellular matrix (ECM)2 for their proliferation. The requirement for anchorage is absolute and cannot be overridden by simple growth factor stimulation (1–3). Anchorage to the ECM is sensed by heterodimeric integrins in the plasma membrane, and a signal is transmitted to various downstream effectors (4, 5). The major downstream effectors include the Rho-associated kinases (ROCK1 and ROCK2), which control actin-cytoskeleton organization and cell contractility and thereby contribute to cell spreading and migration (6–9). Stimulation of G protein-coupled receptors transforms GDP-bound RhoA GTPase into the active GTP-bound form. GTP-bound RhoA then activates ROCK effectively only when the cell is anchored to the ECM (10, 11). Activated ROCK phosphorylates LIM kinases, Adducin, and ERM (ezrin-radixin-moesin) proteins to control the assembly, stabilization, and membrane linkage of actin filaments. Activated ROCK also phosphorylates myosin light chain phosphatase and myosin light chain to induce cell contraction. Focal adhesion kinase (FAK) is another important mediator of integrin-derived signals that controls cell migration, survival, and cell cycle progression (12–14).
When deprived of an ECM anchorage, cells arrest in G1 phase with inactive Cdk4/6 and Cdk2 due largely to repression of the cyclin D1 and cyclin A genes and induction of p27KIP1 (15–18). Inactivation of CDK4 and CDK6 activates retinoblastoma protein (RB) and its cognates, which in turn bind and thereby inactivate the E2F-DP transcription factor complexes, shutting down a subset of genes essential or important for the onset of S phase, including Cdc6, cyclin A, E2F1, and Emi1 (19–23). Emi1 encodes an inhibitor of APC/CCDH1 ubiquitin ligase. Furthermore, CDC6 protein, the ATP-dependent remodeling factor that assembles prereplicative complexes required for S phase onset (24, 25) and activates p21WAF1/CIP1-bound CDK2 (26, 27), is completely eliminated by facilitated proteolysis exerted mainly by the APC/CCDH1 ubiquitin ligase at least in rat embryonic fibroblasts (28, 29).
The phosphatidylinositide 3-kinase (PI3K)-TSC-RHEB-mTOR pathway mediates a portion of the cell proliferation control signals associated with growth factors, energy availability, and amino acid availability (30–34). Growth factor-activated PI3K activates AKT/protein kinase B via protein kinase D, which in turn activates RHEB by inhibiting the TSC1/TSC2 complex (TSC1/2). Activated RHEB stimulates mTOR complex 1 (mTORC1) to enhance general translation by inactivating eIF4E-binding protein and activating S6 kinase 1 (S6K1), the latter via its Thr389 phosphorylation. Moreover, AMP-dependent kinase senses reduced ATP production and phosphorylates TSC2 to activate TSC1/2, which inactivates mTORC1. RAG small G proteins mediate an amino acid availability signal and activate mTORC1 in a RHEB-dependent but TSC1/2-independent manner (35–37). Additionally, several growth factor signals have been reported to negatively regulate TSC1/2 through different pathways. For instance, extracellular signal-regulated kinase (ERK)-mediated TSC2 phosphorylation dissociates TSC1/2 and thereby prevents it from inactivating mTORC1 (38). The Wnt signal activates mTORC1 by inhibiting GSK3-β, which phosphorylates TSC2 in an AMP-dependent kinase-primed phosphorylation-dependent manner (39). SRC kinase activates mTORC1 signaling (40). Notably, in relation to cell adhesion, FAK associates with TSC2 and apparently stimulates S6K1 activity by directly phosphorylating TSC2 (41). Germinal mutation of the TSC2 or TSC1 gene causes multiple benign tumors known as familial tuberous sclerosis (42), a rat version of which occurs in Eker rats in which one allele of Tsc2 is mutated by a retrotransposon insertion in exon 30 (43–45). Homozygous Eker rat cells (tsc2−/−) express a C-terminally truncated aberrant transcript but no TSC2-derived protein (46, 47).
We recently found that anchorage deprivation inactivates mTORC1, whereas mTORC1 activation by tsc2 mutation or activated mutant RHEB overexpression effectively suppresses virtually all of the effects of anchorage deprivation on the expression of major G1 cell cycle factors (29). When rat embryonic fibroblasts engineered to express CDC6 were deprived of an ECM anchorage, the exogenously expressed CDC6 as well as endogenous cyclin A and D-type cyclins disappeared or markedly diminished, and CDK4/6, CDK2, and mTORC1 became inactive. However, the effects of anchorage loss could be overridden by mutational inactivation of TSC2 or by activation of mTORC1 through overexpression of a constitutively active Rheb mutant. These interventions restored the expression of CDC6 and the cyclins as well as the activity of CDK4 and CDK6, although CDK2 remained largely inactive. These findings strongly suggest the possibility that the mTOR cascade mediates some of the anchorage signals that control cell proliferation. A thorough search for a molecule that connects the cell cycle-controlling anchorage signal to the mTOR pathway led us to identify ROCK.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
Anti-CDK4 and anti-β-actin antibodies were purchased from Sigma; anti-phospho-RB (Ser780) and anti-cyclin D3 were from MBL; anti-CDC6 was from NeoMarkers; and anti-TSC2, anti-phospho-RRX(S/T) (100G7E), anti-S6K1, anti-phospho-S6K1 (Thr389), anti-phospho-RB (Ser807/811), anti-ROCK1, anti-ROCK2, anti-LIM kinase 1 (LIMK1), and anti-phospho-LIMK1 (Thr508) were from Cell Signaling Technology. The rest of the antibodies used were obtained from Santa Cruz Biotechnology. Staurosporine, AICAR, and cytochalasin D were obtained from Sigma; rapamycin and Z-VAD-fluoromethyl ketone were from BIOMOL; and Y27632 was from Wako. The rat TSC2 isoform 4 (Tsc2i4) cDNA and homozygous Eker and wild-type rat embryonic fibroblasts (REFs) were gifts from O. Hino, Juntendo University. The cDNAs for human ROCK1 and its truncated constitutively active mutant were provided by S. Narumiya, Kyoto University. Mouse embryonic fibroblast was provided by K. Nakayama, Kyushu University.
Cell Culture and Construction
MEFs, REFs, and Eker REFs (in the latter of which both alleles of the Tsc2 gene are inactivated by mutation (43, 44)) were maintained in DMEM with 10% fetal calf serum (FCS). MEF, REF, and/or Eker cell clones constitutively expressing rat CDC6, human ROCK1, its constitutively active truncated mutant (amino acids 1–727) (48, 49), and/or rat TSC2i4 from the cytomegalovirus promoter were constructed using the Retroviral Gene Transfer and Expression System (Clontech) with appropriate drug-selectable marker genes. The drugs used for selection were G418, hygromycin, puromycin, blasticidin, and Zeocin.
Construction of Eker REF Cells with Inducible Wild-type and Mutant TSC2i4
The cDNA for the mutant TSC2i4 (TSC2i4A) in which Thr1203 was substituted with Ala was created by PCR mutagenesis and verified by DNA sequencing. The primers used were ‘5′-AATTCCCGGGACCGAGTCCGCTCCATGT-3′, 5′-GCTGGTGTTTCCGGCGGGCCTGCGGAC-3′, 5′-GTCCGCAGGCCCGCCGGAAACACCAGC-3′, and 5′-AATTAAGCTTGGGTTTGAATAGTGCACTTCTTCACGG-3′. The rat Tsc2i4 or Tsc2i4A cDNA was inserted into the pRevTRE response vector (Clontech). The pRevTet-Off vector and the pRevTRE response vector with the Tsc2i4 or Tsc2i4A cDNA were separately transfected into the EcoPack packaging cell line. The resulting pRevTet-Off virus-containing supernatant was used to infect Eker-Cdc6-aRK cells to produce a stable Tet-Off cell clone. The Tet-Off clone was then infected with pRevTRE-Tsc2i4 or -Tsc2i4A to obtain a pool of cell clones inducible for TSC2i4 or TSC2i4A. These were designated Eker-Cdc6-aRK-iTsc2i4 and Eker-Cdc6-aRK-iTsc2i4A, respectively, and used for analysis. Cells were maintained in DMEM with 10% FCS and 1 μg of doxycycline/ml.
Preparation of Whole Cell Extracts and Immunoblot Detection
Cells were lysed with an appropriate volume of 2× SDS sample buffer and heated at 95 °C for 5 min for protein denaturation. The cell lysates were electrophoresed and immunoblotted as described (28).
Methylcellulose Culture
Logarithmically proliferating REFs and Eker cells were harvested by trypsinization, embedded in semisolid 1.17% methylcellulose medium containing DMEM with 10% FCS, and cultured at 35 °C.
RNA Isolation and RT-PCR for Rat Tsc2
RNA was isolated from harvested cells with ISOGEN (Nippon Gene). First strand cDNAs were synthesized from 3 μg of total RNA by using oligo(dT) primers and PrimeScript II (Takara Bio). The PCR conditions were one cycle of 98 °C for 2 min for initial denaturation followed by 30 cycles of 98 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s for amplification and one cycle of 72 °C for 5 min for final extension. Individual PCR products were separated by 2% agarose gel electrophoresis. The following primers were used for amplification: for exon 25 detection: forward, TGGCCCATCACGTCATAGCC; reverse, CCCCAAGCTGGCACTGGTAAG; for exon 31 detection: forward, AGCGGCTGGCACAGCCAA; reverse, GCTGCCTCAAAGTCCTCCAATTCG.
Real Time RT-PCR
Semiquantification of the cyclin A, E2F1, and cyclin D1 transcripts was performed by real time RT-PCR as described (29).
Production of Histidine Oligomer-tagged Truncated Active ROCK1 and GST-fused TSC2i4 or -TSC2i4A in Escherichia coli
The truncated, active ROCK1 (amino acids 1–727) with an N-terminal histidine oligomer tag was expressed in E. coli with the pET28a vector (Merck) and purified with nickel beads. GST-fused TSC2i4 and TSC2i4A fragments (amino acids 1125–1809) were expressed in E. coli with the pGEX-5X-1 vector (GE Healthcare) and purified with glutathione beads.
In Vitro Phosphorylation of GST-TSC2i4 by Active ROCK1
The reaction was carried out at 30 °C for 30 min in 20 μl of 50 mm HEPES (pH 7.3) containing 10 mm MgCl2, 5 mm MnCl2, 25 μm ATP, 2 mm dithiothreitol, the active ROCK1, and the GST-TSC2i4 or -TSC2i4A fragment with or without 20 μm Y27632.
RESULTS
With the goal of understanding the anchorage signal cascades that control the G1-S transition, we pursued our recent finding of an epistatic relationship between anchorage loss and mTORC1 activation. We tentatively hypothesized that the TSC1/2-RHEB-mTORC1 cascade conveys a major anchorage signal that regulates the G1-S transition. If this hypothesis is correct, the enforced activation of TSC1/2 would have the same effects on G1 cell cycle factors as anchorage loss, and these effects would be suppressible by Tsc2 inactivation or active RHEB overexpression. Throughout this study, we used rat embryonic fibroblasts constitutively expressing CDC6 (REF-Cdc6) to monitor the effects of various treatments on CDC6 stability in addition to expression of other relevant G1 cell cycle factors because CDC6 stability is strictly regulated by anchorage signaling (28, 29). When rapidly proliferating REF-Cdc6 cells in tissue culture were treated with a chemical (AICAR) that activates AMP-dependent kinase and consequently TSC1/2, cyclin A and D-type cyclins (particularly D1 and D3) disappeared or were significantly diminished, there was a marked reduction in S6K1 phosphorylation at Thr389, and CDC6 was destabilized as observed in cells deprived of anchorage, although AICAR treatment yielded less severe effects (Fig. 1A). Consistent with the known effects of AICAR, Eker rat embryonic fibroblasts overexpressing CDC6 (Eker-Cdc6) in which the Tsc2 gene is homozygously inactivated (43, 44) resisted AICAR treatment with no apparent reduction in the levels of these cell cycle factors (Fig. 1B). Thus, with respect to G1 cell cycle effects and their Tsc2 dependence, AICAR treatment highly resembled anchorage deprivation. This resemblance was confirmed with similarly constructed CDC6-overexpressing mouse embryonic fibroblasts (supplemental Fig. S1A).
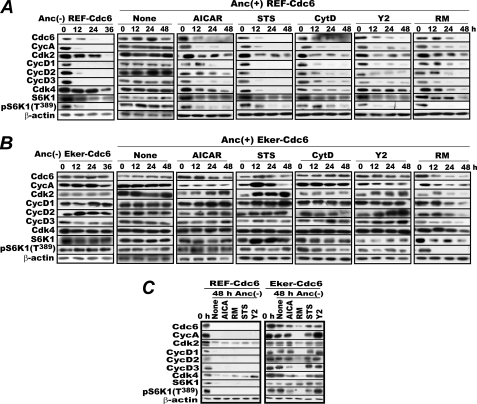
FIGURE 1.
Treatment with certain protein kinase inhibitors mimics anchorage deprivation. A, REF-Cdc6 cells logarithmically proliferating in anchorage-furnished (Anc(+)) culture dishes were treated with or without AICAR (1 mm), STS (1 μm), CytD (1 μm), Y2 (20 μm), or RM (50 nm) in the presence of Z-VAD-fluoromethyl ketone (25 μm), a pan-caspase inhibitor. In parallel, the cells were cultured in methylcellulose medium as an anchorage-deprivation control (Anc(−)). Cells were harvested at the indicated times and analyzed for their levels of CDC6, cyclin (Cyc) A, cyclin E, CDK2, D-type cyclins, CDK4, S6K1, and its Thr389 phosphorylated form by immunoblotting. B, the same experiments as in A were performed with Eker-Cdc6 cells. C, logarithmically proliferating REF-Cdc6 and Eker-Cdc6 cells were incubated in methylcellulose medium first for 24 h and then for an additional 24 h in the absence or presence of the same inhibitors as in A. Cells were harvested at 48 h and analyzed as above. As a control, cells of both types were similarly analyzed just before methylcellulose embedment (0 h).
Given this result, we began to search for inhibitors of anchorage-related protein kinases able to destabilize CDC6 despite the presence of anchorage with the hope that the target molecule of an effective inhibitor would be involved in anchorage signaling. The inhibitors examined were U0126 for MEK1/2; Su6656 for SRC kinase; lithium chloride for GSK-3β; staurosporine (STS) for protein kinase C, myosin light chain kinase, FAK, and others; and Y27632 (Y2) for ROCK. In addition, we tested cytochalasin D (CytD), which disrupts actin filaments and inhibits cell division, because proper actin filament assembly is controlled by cell-ECM adhesion (50). In the initial screen, only STS, Y2, and CytD were found to destabilize CDC6. We therefore proceeded to perform in-depth analyses of these compounds with respect to their effects on other G1 cell cycle factors, particularly cyclin A, D-type cyclins, and S6K1 Thr389 phosphorylation. REF-Cdc6 cells logarithmically proliferating in culture plates were treated with STS, Y2, or CytD for 48 h; harvested every 12 h; and lysed for immunoblot analysis. As positive controls, cells were treated with rapamycin (RM) or deprived of anchorage by culturing in methylcellulose medium, and they were processed similarly. Whenever the cells were treated with these compounds, Z-VAD-fluoromethyl ketone, a potent pan-caspase inhibitor, was added to prevent apoptosis, which is often co-induced by some of these compounds (see supplemental Fig. S2 for STS treatment) and can be misleading. As shown in Fig. 1A, treatment with any of these inhibitors reduced the levels of constitutively expressed CDC6, endogenous cyclins (A, D1, D2, and D3), and S6K1 Thr389 phosphorylation (a hallmark of active mTORC1). However, these reductions varied in extent and tended to be less severe than those caused by anchorage deprivation.
These effects were neither influenced by CDC6 overexpression nor limited to rat cells. The same results were obtained with original rat and mouse embryonic fibroblasts (supplemental Fig. S1, B and C). By contrast, when Eker-Cdc6 cells were treated similarly, none of these inhibitors (except for the positive control rapamycin) affected the levels of these factors (Fig. 1B). Moreover, when Eker-Cdc6 cells were deprived of anchorage and then treated similarly, only rapamycin affected the levels of these factors significantly (Fig. 1C). Thus, the effects of staurosporine, Y27632, and cytochalasin D absolutely depend on intact TSC2 both in the presence and absence of anchorage.
Expression of TSC2 Isoform 4 Restores Susceptibility of Eker-Cdc6 Cells to Anchorage Loss and Y2 but Not to Staurosporine or Cytochalasin D
TSC2 protein is not a single species but a collection of at least six variants generated by alternative splicing of its transcript (51–54). TSC2i4 is the product of an mRNA lacking both exon 25 and exon 31, and it consequently lacks 66 amino acids present in the full size isoform 1 (Fig. 2A). TSC2i4 is expressed in various tissues and is able to trans-genetically suppress the onset of tuberous sclerosis in Eker rats (47). We previously showed that complementation with this isoform restored the susceptibility of Eker-Cdc6 cells to anchorage loss and destabilized CDC6 (29). Herein, we first confirmed that all of the effects of anchorage deprivation on G1 cell cycle factors reappeared in the Tsc2i4-complemented Eker-Cdc6 cells (Eker-Cdc6-Tsc2i4). Logarithmically proliferating REF-Cdc6, Eker-Cdc6, and Eker-Cdc6-Tsc2i4 cells were cultured in methylcellulose medium and harvested at 12-h intervals for analysis of the levels of relevant G1 cell cycle factors and S6K1 Thr389 phosphorylation. As shown in Fig. 2B, unlike in Eker-Cdc6 cells but just like in REF-Cdc6 cells, CDC6, cyclin A, and the three D-type cyclins along with S6K1 Thr389 phosphorylation diminished or disappeared in Eker-Cdc6-Tsc2i4 cells upon deprivation of anchorage.
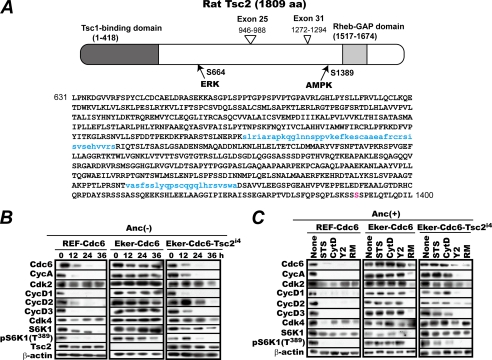
FIGURE 2.
A, schematic presentation of rat TSC2i4 generated by alternative splicing that removes exons 25 and 31. The two stretches of amino acids encoded by exons 25 and 31, which are absent in TSC2i4, are shown in lowercase blue letters. The amino acid (aa) numbers in the figure refer to full-size TSC2. The amino acid in red is the AMP-dependent kinase (AMPK) phosphorylation site (35). GAP, GTPase-activating protein. B, complementation with Tsc2i4 restores the susceptibility of Eker-Cdc6 cells to anchorage deprivation. Rapidly proliferating REF-Cdc6, Eker-Cdc6, and Eker-Cdc6 cells complemented with Tsc2i4 (Eker-Cdc6-Tsc2i4) were cultured in methylcellulose medium with cell sampling every 12 h for 36 h. The harvested cells were analyzed for the indicated factors by immunoblotting. C, complementation with Tsc2i4 restores the susceptibility of Eker-Cdc6 cells to Y2 but not to staurosporine or cytochalasin D. The same set of cells proliferating on culture dishes were treated with or without RM, STS, CytD, or Y2 for 24 h in the presence of Z-VAD as in Fig. 1A and analyzed for the indicated factors by immunoblotting. Anc, anchorage; Cyc, cyclin.
Given these results, we next examined whether or not complementation with Tsc2i4 would restore the susceptibility of Eker-Cdc6 to the inhibitors. Logarithmically proliferating REF-Cdc6, Eker-Cdc6, and Eker-Cdc6-Tsc2i4 cells were similarly treated with RM, STS, CytD, and Y2 for 24 h and analyzed for their levels of the G1 cell cycle factors and S6K1 Thr389 phosphorylation by immunoblotting. Confirming the Fig. 1 results, treatment with any one of these chemicals markedly decreased the levels of CDC6 and A- and D-type cyclins in REF-Cdc6 cells but not in Eker-Cdc6 cells (Fig. 2C). By contrast, Eker-Cdc6-Tsc2i4 cells showed divergent sensitivities to these chemicals. When the cells were treated with RM or Y2, these G1 factors diminished as expected. But when they were treated with STS or CytD, these factors suffered no obvious reduction. Thus, of the three, only the ROCK inhibitor retained the ability to induce the same effects as anchorage deprivation in Tsc2i4-complemented Eker cells, although the anchorage loss-induced down-regulation of G1 cell cycle factors was largely restored in these cells.
Rat Embryonic Fibroblasts Predominantly Express TSC2 Isoform 5 Containing Exon 25
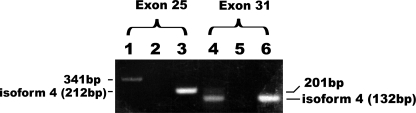
As described above, REFs were sensitive to staurosporine and cytochalasin D, but the Eker cells complemented with Tsc2i4 were not. This suggested that REFs must express other TSC2 isoforms. To test this, we prepared RNA from REF-Cdc6 cells and analyzed their Tsc2 transcripts for the presence of exons 25 and 31 by RT-PCR with appropriate primer sets (Fig. 3). As a size control for transcripts lacking both exon 25 and exon 31, the rat Tsc2i4 cDNA was amplified with the same primer sets. If the Tsc2 mRNA expressed in REFs contains exon 25, the size of the amplified fragment would be 341 bp, and if not, the size would be 212 bp. Similarly, if it contains exon 31, it would be 201 bp, and if not, it would be 132 bp. The product amplified for exon 25 was exclusively a 341-bp fragment, whereas for exon 31, both a 132-bp fragment and a weaker 201-bp fragment were amplified. Without reverse transcription, none of these fragments were detected. We therefore concluded that the Tsc2 transcripts expressed in REFs are predominantly isoform 5 containing exon 25 but not exon 31 with far fewer isoform 1 transcripts containing both.
FIGURE 3.
The major Tsc2 mRNA species expressed in rat embryonic fibroblasts is isoform 5. RNA was prepared from REF-Cdc6 cells, and the presence or absence of exon 25 and 31 sequences in the Tsc2 transcripts was determined by RT-PCR with specific primers followed by agarose gel electrophoresis. As a control for the absence of exons 25 and 31, the rat Tsc2i4 cDNA was similarly PCR-amplified (lanes 3 and 6). To confirm the absence of contamination by genomic DNA in the RNA preparation, the RNA preparation was amplified without reverse transcription (lanes 2 and 5). Lanes 1, 2, and 3 show the detection of exon 25, and lanes 4, 5, and 6 show the detection of exon 31. The PCR products from mRNAs with/without exon 25 were expected to be 341/212 bp, and those from mRNAs with/without exon 31 were expected to be 201/132 bp.
Expression of Truncated Constitutively Active ROCK1 Suppresses Effects of Anchorage Loss
The close phenotypic similarity between treatment with the ROCK inhibitor and anchorage deprivation implies that ROCK may mediate the anchorage signal to control the G1-S transition via the TSC1/2-RHEB-mTORC1 signaling cascade. We explored this possibility by examining the effects of enforced expression of a constitutively active form of ROCK1 on the G1 cell cycle factors that are destabilized or diminished upon anchorage deprivation. REF-Cdc6 cells that also expressed full-length, wild-type ROCK1 or C-terminally truncated, constitutively active ROCK1 (REF-Cdc6-RK and REF-Cdc6-aRK, respectively) (49) were constructed by retrovirus-mediated gene transfer and analyzed as in Fig. 2A with REF-Cdc6 and Eker-Cdc6 cells as controls. As shown in Fig. 4A, expression of the wild-type ROCK1 slightly delayed, but failed to prevent, the destabilization of CDC6 and the cyclins. By contrast, expression of the constitutively active ROCK1 up-regulated these factors and restored S6K1 Thr389 phosphorylation to an extent comparable with that caused by Tsc2 inactivation. The phenotypic similarity between active ROCK1 expression and Tsc2 inactivation was further extended. Just as in Eker-Cdc6 cells, cyclin D1 mRNA was up-regulated, and CDK4 remained active with the resultant induction of cyclin A and E2F1 mRNAs in anchorage-deprived RFE-Cdc6-aRK cells (Fig. 4, B and C) (29).
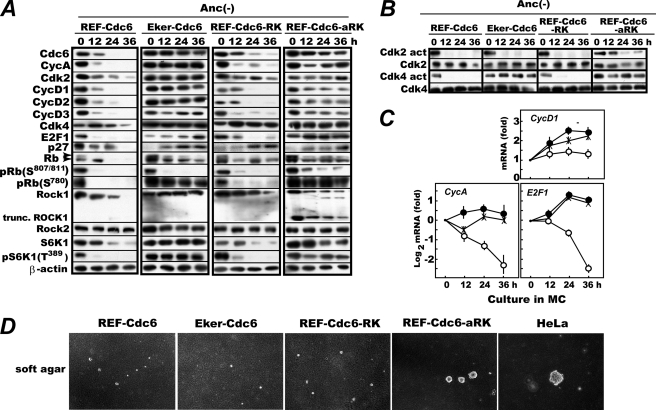
FIGURE 4.
Expression of a truncated, constitutively active ROCK1 overrides effects of anchorage deprivation; stabilizes CDC6, cyclin A, and D-type cyclins; and restores S6K1 Thr389 phosphorylation. A, logarithmically proliferating REF-Cdc6, Eker-Cdc6, REF-Cdc6-RK, and REF-Cdc6-aRK cells were cultured in methylcellulose medium, and cells were harvested every 12 h and analyzed for the indicated factors by immunoblotting. In parallel, the activities of Cdk2 and Cdk4 were assayed as described (29) (B), and the levels of cyclin D1, cyclin A, and E2F1 transcripts in REF-Cdc6 (open circles), Eker-Cdc6 (crosses), and REF-Cdc6-aRK (closed circles) cells were quantified by real time RT-PCR (C). Real time RT-PCR was performed on two or three independently isolated cell samples, and the data are shown as averages with S.D. bars. D, colony formation in soft agar. Logarithmically proliferating REF-Cdc6, Eker-Cdc6, REF-Cdc6-RK, and REF-Cdc6-aRK cells were cultured in DMEM containing 0.33% Noble agar layered on 0.5% bottom agar for 3 weeks with HeLa cells as a reference. Anc, anchorage; Cyc, cyclin.
There was, however, a slight dissimilarity between the two interventions. Upon anchorage deprivation, CDK2 was immediately inactivated in the Tsc2-inactive cells, whereas CDK2 activity persisted (albeit markedly attenuated) in the active ROCK1-expressing cells (Fig. 4B). Consistently, a low level of RB Ser807/811 phosphorylation was observed at 36 h in the latter cells (Fig. 4A). Perhaps as a combined consequence of the up-regulation of key G1 factors and the activation (albeit weak) of CDK2, REF-Cdc6-aRK cells, but not REF-Cdc6, Eker-Cdc6, or REF-Cdc6-RK cells, could form significant colonies in soft agar, although they were smaller than those of HeLa cells, a fully developed human cancer cell line used as a reference (Fig. 4D).
Notably, endogenous ROCK1 was unstable in REFs but stable in Eker or active ROCK1-expressing REFs during anchorage loss. In the REF-Cdc6 and REF-Cdc6-RK cells deprived of anchorage, endogenous and even exogenously expressed ROCK1 disappeared or significantly diminished at 36 h, whereas in Eker-Cdc6 and REF-Cdc6-aRK cells, endogenous ROCK1 was still present at this time point (Fig. 4A). By contrast, ROCK2 expression was virtually uninfluenced by anchorage deprivation or by the status of the mTOR cascade.
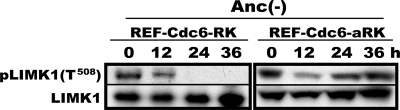
ROCK Activity in REFs Is Rapidly Lost upon Anchorage Deprivation
If ROCK indeed mediates an anchorage signal to regulate mTOR and control the expression and activation of G1 cell cycle factors, it must undergo inactivation upon anchorage loss. At least partial inactivation of ROCK upon anchorage loss was reported previously (10, 11). We examined whether or not ROCK strictly fulfilled this requirement in our experimental system. REF-Cdc6-RK and REF-Cdc6-aRK cells were deprived of anchorage and analyzed for the phosphorylation of Thr508 of LIM kinase 1, a specific in vivo substrate of ROCK1 and ROCK2 (56). When rapidly proliferating REF-Cdc6-RK cells were cultured in methylcellulose medium, the phosphorylation of LIM kinase 1 disappeared within 24 h, whereas in REF-Cdc6-aRK cells, it remained present after 36 h (Fig. 5). Thus, ROCK fulfilled this requirement.
FIGURE 5.
ROCK1 undergoes inactivation upon anchorage deprivation. In vivo ROCK1 activity was assayed by monitoring phosphorylation of LIM kinase 1 at Thr508. Logarithmically proliferating REF-Cdc6-RK and REF-Cdc6-aRK cells were cultured in methylcellulose medium for the indicated times, and LIMK1 and LIM kinase phosphorylated at Thr508 (pLIMK1(T508)) were immunodetected. Anc, anchorage.
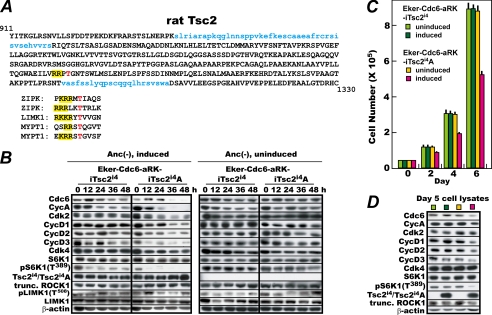
TSC2 Contains One ROCK Phosphorylation Target-like Sequence, and Alanine Substitution of It Attenuates Ability of TSC2 to Sensitize Eker-Cdc6 Cells to Active ROCK1
As shown above, expression of a constitutively active ROCK1 suppressed the effects of anchorage deprivation on the expression of CDC6 and the G1 cyclins similarly to the results obtained by mutational inactivation of Tsc2. Consequently, one obvious question is whether or not ROCK directly phosphorylates and thereby inactivates TSC2 or TSC1. A thorough search for a ROCK phosphorylation consensus sequence led us to identify one such sequence in TSC2 but not in TSC1 (Fig. 6A). This sequence (RRPT) (putative phosphorylation site and preceding basic amino acids in bold) at amino acids 1200–1203 contains the generally accepted minimal sequence ((R/K)X(S/T) or (R/K)XX(S/T)) for ROCK phosphorylation, which occurs at 32 locations in rat TSC2. More importantly, it resembles the actual in vivo ROCK phosphorylation sites of the well characterized target proteins zipper-interacting protein kinase (ZIPK), LIMK1, and the regulatory subunit of myosin light chain phosphatase (MYPT1) (7, 50, 56, 57). Furthermore, this sequence is present not only in full-sized TSC2 but also in the exon-lacking isoform 4, and it is conserved in humans, mice, cows, frogs, fish, and even fruit flies. By contrast, rat TSC1 contains three similar sequences (RRWKT, KKANS, and RKIT), but none of them are conserved even in human TSC1. We therefore focused on this highly conserved sequence in TSC2, constructed a mutant rat TSC2i4 (TSC2i4A) in which Thr1203 was substituted with unphosphorylatable alanine, and expressed it in Eker-Cdc6-aRK cells under the doxycycline-repressible system. As a control, Eker-Cdc6-aRK cells that could express wild-type TSC2i4 under the same repressible system were simultaneously constructed. These cells inducible for wild-type and mutant TSC2i4 (Eker-Cdc6-aRK-iTsc2i4 and Eker-Cdc6-aRK-iTsc2i4A, respectively) were then cultured in methylcellulose medium and analyzed as in Fig. 4. To induce TSC2i4 and TSC2i4A, doxycycline was removed 2 days before the start of the methylcellulose culture. As shown in Fig. 6B, in the Eker-Cdc6 cells expressing active ROCK1 and with wild-type TSC2i4 induced, CDC6 and the G1 cyclins were stably expressed with continued S6K1 Thr389 phosphorylation as expected. In contrast, in the Eker-Cdc6 cells expressing active ROCK1 and with mutant TSC2i4A induced, those factors diminished significantly within 36 h of anchorage deprivation together with S6K1 Thr389 phosphorylation. This was not caused by an unexpected interference of the ROCK1 activity by TSC2i4A because LIM kinase 1 was phosphorylated at Thr508 throughout anchorage deprivation. When expression of the mutant TSC2 was repressed by continuous addition of doxycycline, the G1 factors were stably expressed in the Eker-Cdc6-aRK-iTsc2i4A cells, and their levels were indistinguishable from those in the doxycycline-repressed Eker-Cdc6-aRK-iTsc2i4 cells. Thus, expression of TSC2i4A, but not TSC2i4, led to the down-regulation of CDC6, cyclin A, D-type cyclins, and S6K1 Thr389 phosphorylation in Eker-Cdc6-aRK cells. These results show that TSC2 Thr1203 is required for ROCK1 to activate mTORC1 and up-regulate the G1 factors in the absence of anchorage.
FIGURE 6.
Expression of a mutant form of TSC2i4 (TSC2i4A) in which Thr1203 is substituted with alanine fails to restore susceptibility of Eker-Cdc6 cells to active ROCK1 in absence of anchorage. A, the presence of a ROCK phosphorylation consensus sequence in rat TSC2. Upper panel, the amino acids in blue are those encoded by exons 25 and 31. The T in red is the putative phosphorylation site, and the preceding basic amino acids (RR) are highlighted in yellow. Lower panel, the ROCK phosphorylation sites and surrounding amino acid sequences of ZIPK1, LIMK1, and MYPT1. B, logarithmically proliferating Eker-Cdc6-aRK-iTsc2i4 and Eker-Cdc6-aRK-iTsc2i4A cells were induced for TSC2i4 or TSC2i4A by withdrawal of doxycycline or were uninduced. Two days later, the cells were cultured in methylcellulose medium, and samples were harvested every 12 h for 48 h and analyzed for the indicated factors by immunoblotting. C, expression of TSC2i4A retards cell proliferation in culture dishes. Logarithmically proliferating Eker-Cdc6-aRK-iTsc2i4 and Eker-Cdc6-aRK-iTsc2i4A cells were induced for TSC2i4 or TSC2i4A by withdrawal of doxycycline or were uninduced, cultured for 2 days, then replated at 4 × 104 cells each on three 10-cm plates for each sampling time point, and further cultured with sampling every other day until day 6. The cells were counted, and average cell numbers with S.D. values were calculated from the data of the three plates. Light and dark green, uninduced and induced Eker-Cdc6-aRK-iTsc2i4 cells, respectively; orange and dark red, uninduced and induced Eker-Cdc6-aRK-iTsc2i4A cells, respectively. D, on day 5 in C, the cells were harvested and examined for the indicated factors by immunoblotting. The same coloring as in C is used. Anc, anchorage; Cyc, cyclin.
The functional importance of phosphorylatable Thr1203 in TSC2 was further confirmed by the emergence of adverse effects on anchorage-furnished cell proliferation by its alanine mutation. When TSC2i4A was induced by withdrawal of doxycycline, Eker-Cdc6-aRK-iTsc2i4A cells cultured in dishes proliferated significantly less than uninduced Eker-Cdc6-aRK-iTsc2i4A cells or Eker-Cdc6-aRK-iTsc2i4 cells that were either induced or uninduced (Fig. 6C). The doubling time of the cells increased from roughly 35 to 40 h. The slow proliferation was accompanied by reduced mTORC1 signaling as indicated by lower levels of CDC6, cyclin A, D-type cyclins, and S6K1 Thr389 phosphorylation when cells were examined at day 5 (Fig. 6D).
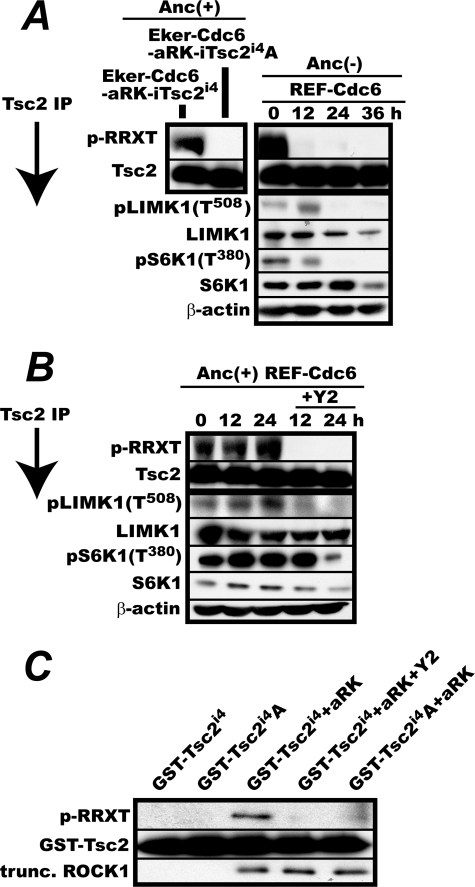
In Vivo and in Vitro Phosphorylation of TSC2 at Thr1203 by ROCK1
Given the data suggesting potential phosphorylation of TSC2 at Thr1203 by ROCK, we examined whether ROCK could phosphorylate TSC2 Thr1203 in vivo and in vitro. To detect phosphorylation of TSC2 Thr1203, we used a commercially available anti-phospho-RRX(S/T) antibody that recognizes a phosphorylated serine or threonine preceded by two arginine residues at positions −2 and −3, a sequence that is present at TSC2 Thr1203. TSC2 protein was immunoprecipitated from Eker-Cdc6-aRK-iTsc2i4 and Eker-Cdc6-aRK-iTsc2i4A cells logarithmically proliferating in doxycycline-free medium and immunoblotted with the anti-phospho-RRX(S/T) antibody. We found that only the TSC2 from the former cells reacted with the antibody (Fig. 7A, left panel). Furthermore, when logarithmically proliferating REF-Cdc6 cells were deprived of anchorage and similarly analyzed, the antibody reaction to TSC2 from REF-Cdc6 cells rapidly declined and was accompanied by a loss of both LIM kinase 1 Thr508 phosphorylation and S6K1 Thr389 phosphorylation (Fig. 7A, right panel). These results show that the antibody specifically detected phosphorylation of TSC2 at Thr1203 and that TSC2 Thr1203 was phosphorylated in anchorage-furnished proliferating REF cells in which ROCK was fully active.
FIGURE 7.
Phosphorylation of TSC2 Thr1203 by ROCK both in vivo and in vitro. A, phosphorylation of TSC2 but not TSC2T1203A during anchorage (Anc) deprivation in REFs expressing activated ROCK1. Left panel, validation of the specificity of the anti-phospho-RRX(S/T) antibody for the detection of TSC2 Thr1203 phosphorylation. Eker-Cdc6-aRK-iTsc2i4 and Eker-Cdc6-aRK-iTsc2i4A cells were cultured for 3 days in the absence of doxycycline and lysed. TSC2 was immunoprecipitated with the anti-TSC2 antibody and immunoblotted with the anti-phospho-RRX(S/T) (p-RRXT) antibody or the anti-TSC2 antibody. Right panel, logarithmically proliferating REF-Cdc6 cells were cultured in methylcellulose medium for 36 h with sampling every 12 h. Cells were lysed, immunoprecipitated for TSC2, and immunoblotted with the anti-phospho-RRX(S/T) or the anti-TSC2 antibody. In parallel, the levels of LIMK1, phosphorylated LIMK1 Thr508, S6K1, and phosphorylated S6K1 Thr389 in the cell lysates were determined by immunoblotting. B, treatment with the ROCK inhibitor Y2 abolishes TSC2 Thr1203 phosphorylation in vivo. Logarithmically proliferating REF-Cdc6 cells were cultured in anchorage-furnished culture dishes for 24 h with or without 20 μm Y2. Cells were harvested every 12 h and analyzed as in A. C, in vitro TSC2 Thr1203 phosphorylation by E. coli-expressed active ROCK1. An E. coli-expressed, truncated (trunc.) active ROCK1 (aRK) and GST-TSC2i4 or -TSC2i4A fragments were incubated in a reaction buffer containing ATP with or without Y2. Phosphorylation of GST-TSC2i4 at Thr1203 was detected with the anti-phospho-RRX(S/T) antibody.
To extend this finding, we proceeded to determine whether or not ROCK was responsible for TSC2 Thr1203 phosphorylation in vivo. Logarithmically proliferating REF-Cdc6 cells were treated with or without the specific ROCK inhibitor Y2 for 24 h. Cells were collected every 12 h, and TSC2 was immunoprecipitated and analyzed with the anti-phospho-RRX(S/T) antibody (Fig. 7B). When Y2 was added, Thr1203 phosphorylation vanished at the same time as LIM kinase 1 Thr508 phosphorylation, and S6K1 Thr1304 phosphorylation subsequently declined. Thus, TSC2 Thr1203 phosphorylation depended on ROCK activity.
Finally, we examined whether ROCK could phosphorylate TSC2 at Thr1203 in vitro. The truncated constitutively active ROCK1 used in the REF-Cdc6-aRK cells was N-terminally tagged with a histidine oligomer, expressed in E. coli, and purified with nickel beads. In parallel, we attempted to express the entire sequence of TSC2i4 in E. coli but found that, unlike ROCK1, this protein was extremely unstable. We therefore decided to express GST-fused fragments of TSC2i4 and TSC2i4A (amino acid 1125 to the C terminus). The expressed GST fusion proteins were purified with glutathione beads and used for an in vitro phosphorylation assay with ROCK1. To unequivocally identify the kinase, the ROCK inhibitor Y2 was included in some phosphorylation reactions. As shown in Fig. 7C, E. coli-expressed ROCK1 phosphorylated the GST-fused TSC2i4 but not the GST-fused TSC2i4A, and this phosphorylation was completely blocked by Y2. This result demonstrates that ROCK was able to phosphorylate TSC2 at Thr1203 in vitro.
DISCUSSION
How anchorage to the extracellular matrix is required for the G1-S transition of cells constituting solid organs is a fundamental question not only in cell cycle control but also in cancer research. Loss of anchorage invokes G1 arrest accompanied by CDC6 destabilization, the destruction of A- and D-type cyclins, and the inactivation of CDK2. Recently, we found that activation of mTORC1 by mutational inactivation of Tsc2 or overexpression of constitutively active RHEB suppresses most of the effects of anchorage deprivation on the expression of CDC6 and cyclins (29). This finding raised the possibility that the TSC1/2-RHEB-mTORC1 pathway might mediate an anchorage signal to control the expression of these cell cycle factors.
In this study, we show that ROCK connects a cell cycle-controlling anchorage signal to TSC2 via direct phosphorylation. There are three lines of evidence that led to this conclusion. First, expression of a constitutively active form of ROCK1 in anchorage-deprived REF-Cdc6 cells stabilized CDC6, cyclin A, and three D-type cyclins and activated mTORC1 as indicated by continued S6K1 Thr389 phosphorylation. Second, all the TSC2 isoforms of humans through fruit flies contain a sequence similar to the ROCK phosphorylation consensus sequence, and substitution of Thr1203 in that sequence with unphosphorylatable alanine largely abolished the ability of active ROCK1 to override the effects of anchorage loss. Third, TSC2 Thr1203 underwent phosphorylation in an active ROCK-dependent manner in vivo, and E. coli-expressed ROCK1 phosphorylated TSC2 Thr1203 in vitro.
How is the ROCK-mediated transduction of the anchorage signal to TSC2 or other targets regulated? Growth factor-activated RhoA can activate ROCK only when a cellular anchorage to the ECM is available as shown previously (10, 11) and in Fig. 5. This mechanism couples anchorage to ROCK activation, but it might not be the sole mechanism to control ROCK activity. As shown in Fig. 4, in REFs, both endogenous and constitutively expressed exogenous ROCK1 proteins disappeared or diminished during anchorage deprivation. By contrast, in Eker cells, endogenous ROCK1 was stably expressed. Thus, ROCK1 appeared to be stabilized in Eker cells despite the absence of anchorage. This observation suggests that a loss of anchorage led ROCK1 to become inactivated, which in turn might induce its own destabilization likely via the subsequent activation of TSC2, implying the presence of a positive feedback loop that involves the TSC-mTORC1 pathway and controls ROCK1 stability. Under this feedback control, ROCK1 signaling cannot immediately be restored when anchorage is resupplied once mTORC1 is inactivated. Notably, in sharp contrast, the expression of the ROCK2 protein was unaffected by anchorage loss and therefore the status of the mTORC1 pathway. The biological significance of this differential regulation is unknown, but it might be related to the specific role of ROCK2 suggested in mitosis (55).
Because FAK seems to directly phosphorylate TSC2 (41), FAK is a good candidate mediator of the anchorage signal that controls the TSC1/2-RHEB-mTOR pathway. However, as shown in Fig. 2C, treatment with staurosporine (a strong FAK inhibitor) destabilized CDC6 and the G1 cyclins in REF-Cdc6 cells but not in Tsc2i4-complemented Eker-Cdc6 cells despite the fact that the latter cells properly responded to anchorage deprivation by down-regulating these factors. Similarly, the depolymerization of actin fibers that occurs during anchorage deprivation could not be the mediator of the cell cycle-controlling anchorage loss signal because treatment with cytochalasin D, a strong inhibitor of actin polymerization, destabilized CDC6 and the G1 cyclins in REF-Cdc6 cells but not in Tsc2i4-complemented Eker-Cdc6 cells. Consistent with the differential sensitivity shown by the Tsc2i4-complemented Eker cells, none of the Tsc2 transcripts expressed in REF-Cdc6 cells that responded to staurosporine and cytochalasin were isoform 4; instead, most were isoform 5 with a much smaller number of isoform 1 transcripts. Therefore, it is possible that signals resulting from the inactivation of FAK or other staurosporine-sensitive kinases or from actin depolymerization require TSC2 isoform 5 (or 1) for their transmission to TSC2. Indeed, our recent experiments show that expression of TSC2 isoform 5 restores the susceptibility of Eker REFs to staurosporine and cytochalasin D.3 Because TSC2 isoform 5 (and 1) is abundantly expressed in brain, heart, and muscle (53), the connection between the TSC-Rheb-mTORC1 pathway and the signals from staurosporine-sensitive kinases or actin fibers might have specific biological functions associated with the cells in these tissues. In this regard, it is worth stressing that in cells expressing TSC2 isoform 1 or 5 not only anchorage signals but also signals from these targets would be needed for mTORC1 activation.
Supplementary Material
Acknowledgments
We thank Dr. O. Hino for the rat Tsc2i4 cDNA and homozygous Eker and wild-type rat embryonic fibroblasts, Dr. S. Narumiya for the human ROCK1 cDNAs, and Dr. K. Nakayama for MEF.
This work was supported by grants-in-aid for scientific research (S) and the Global Center of Excellence from the Ministry of Education, Science, and Culture of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
J. Park, S. Arakawa-Takeuchi, S. Jinno, and H. Okayama, unpublished data.
- ECM
- extracellular matrix
- ROCK
- Rho-associated kinase
- LIMK1
- LIM kinase 1
- FAK
- focal adhesion kinase
- RB
- retinoblastoma protein
- APC/C
- anaphase-promoting complex/cyclosome
- TSC1/2
- TSC1/TSC2 complex
- mTORC1
- mTOR complex 1
- S6K1
- S6 kinase 1
- Z
- benzyloxycarbonyl
- AICAR
- aminoimidazole carboxamide ribonucleotide
- REF
- rat embryonic fibroblast
- MEF
- mouse embryonic fibroblast
- STS
- staurosporine
- Y2
- Y27632
- CytD
- cytochalasin D
- RM
- rapamycin
- TSC2i4
- TSC2 isoform 4
- ZIPK
- zipper-interacting protein kinase.
REFERENCES
- 1. Otsuka H., Moskowitz M. (1975) J. Cell. Physiol. 87, 213–219 [DOI] [PubMed] [Google Scholar]
- 2. Rogne S., Rønning O. W., Myklebost O., Seglen P. O., Pettersen E. O. (1985) J. Cell. Physiol. 125, 528–532 [DOI] [PubMed] [Google Scholar]
- 3. Kume K., Jinno S., Miwatani H., Kizaka-Kondoh S., Terada Y., Nojima H., Okayama H. (1992) New Biologist 4, 504–511 [PubMed] [Google Scholar]
- 4. Boudreau N. J., Jones P. L. (1999) Biochem. J. 339, 481–488 [PMC free article] [PubMed] [Google Scholar]
- 5. Huveneers S., Truong H., Danen H. J. (2007) Int. J. Radiat. Biol. 83, 743–751 [DOI] [PubMed] [Google Scholar]
- 6. Ishizaki T., Maekawa M., Fujisawa K., Okawa K., Iwamatsu A., Fujita A., Watanabe N., Saito Y., Kakizuka A., Morii N., Narumiya S. (1996) EMBO J. 15, 1885–1893 [PMC free article] [PubMed] [Google Scholar]
- 7. Riento K., Ridley A. J. (2003) Nat. Rev. Mol. Cell Biol. 4, 446–456 [DOI] [PubMed] [Google Scholar]
- 8. Pellegrin S., Mellor H. (2007) J. Cell Sci. 120, 3491–3499 [DOI] [PubMed] [Google Scholar]
- 9. Narumiya S., Tanji M., Ishizaki T. (2009) Cancer Metastasis Rev. 28, 65–76 [DOI] [PubMed] [Google Scholar]
- 10. Ren X. D., Wang R., Li Q., Kahek L. A., Kaibuchi K., Clark R. A. (2004) J. Cell Sci. 117, 3511–3518 [DOI] [PubMed] [Google Scholar]
- 11. Bhadriraju K., Yang M., AlomRuiz S., Pirone D., Tan J., Chen C. S. (2007) Exp. Cell Res. 313, 3616–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schlaepfer D. D., Hauck C. R., Sieg D. J. (1999) Prog. Biophys. Mol. Biol. 71, 435–478 [DOI] [PubMed] [Google Scholar]
- 13. Mitra S. K., Hanson D. A., Schlaepfer D. D. (2005) Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 14. Zhao J., Bian Z. C., Yee K., Chen B. P., Chien S., Guan J. L. (2003) Mol. Cell 11, 1503–1515 [DOI] [PubMed] [Google Scholar]
- 15. Schulze A., Zerfass-Thome K., Bergès J., Middendorp S., Jansen-Dürr P., Henglein B. (1996) Mol. Cell. Biol. 16, 4632–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu X., Ohtsubo M., Böhmer R. M., Roberts J. M., Assoian R. K. (1996) J. Cell Biol. 133, 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang F., Orend G., Watanabe N., Hunter T., Ruoslahti E. (1996) Science 271, 499–502 [DOI] [PubMed] [Google Scholar]
- 18. Philips A., Huet X., Plet A., Rech J., Vié A., Blanchard J. M. (1999) Oncogene 18, 1819–1825 [DOI] [PubMed] [Google Scholar]
- 19. Hunter T., Pines J. (1994) Cell 79, 573–582 [DOI] [PubMed] [Google Scholar]
- 20. Sherr C. J. (1995) Trends Biochem. Sci. 20, 187–190 [DOI] [PubMed] [Google Scholar]
- 21. Sears R. C., Nevins J. R. (2002) J. Biol. Chem. 277, 11617–11620 [DOI] [PubMed] [Google Scholar]
- 22. Blais A., Dynlacht B. D. (2004) Curr. Opin. Genet. Dev. 14, 527–532 [DOI] [PubMed] [Google Scholar]
- 23. Hsu J. Y., Reimann J. D., Sørensen C. S., Lukas J., Jackson P. K. (2002) Nat. Cell Biol. 4, 358–366 [DOI] [PubMed] [Google Scholar]
- 24. Méndez J., Stillman B. (2000) Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelly T. J., Brown G. W. (2000) Annu. Rev. Biochem. 69, 829–880 [DOI] [PubMed] [Google Scholar]
- 26. Kan Q., Jinno S., Kobayashi K., Yamamoto H., Okayama H. (2008) J. Biol. Chem. 283, 17864–17872 [DOI] [PubMed] [Google Scholar]
- 27. Kan Q., Jinno S., Yamamoto H., Kobayashi K., Okayama H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4757–4762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jinno S., Yageta M., Nagata A., Okayama H. (2002) Oncogene 21, 1777–1784 [DOI] [PubMed] [Google Scholar]
- 29. Arakawa-Takeuchi S., Kobayashi K., Park J. H., Uranbileg B., Yamamoto H., Jinno S., Okayama H. (2010) FEBS Lett. 584, 2779–2785 [DOI] [PubMed] [Google Scholar]
- 30. Hay N., Sonenberg N. (2004) Genes Dev. 18, 1926–1945 [DOI] [PubMed] [Google Scholar]
- 31. Fingar D. C., Blenis J. (2004) Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 32. Manning B. D., Cantley L. C. (2003) Biochem. Soc. Trans. 31, 573–578 [DOI] [PubMed] [Google Scholar]
- 33. Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 34. Sarbassov D. D., Ali S. M., Sabatini D. M. (2005) Curr. Opin. Cell Biol. 17, 596–603 [DOI] [PubMed] [Google Scholar]
- 35. Inoki K., Zhu T., Guan K. L. (2003) Cell 115, 577–590 [DOI] [PubMed] [Google Scholar]
- 36. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. (2010) Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. (2005) Cell 121, 179–193 [DOI] [PubMed] [Google Scholar]
- 39. Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K., Wang C. Y., He X., MacDougald O. A., You M., Williams B. O., Guan K. L. (2006) Cell 126, 955–968 [DOI] [PubMed] [Google Scholar]
- 40. Vojtechová M., Turecková J., Kucerová D., Sloncová E., Vachtenheim J., Tuhácková Z. (2008) Neoplasia 10, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gan B., Yoo Y., Guan J. L. (2006) J. Biol. Chem. 281, 37321–37329 [DOI] [PubMed] [Google Scholar]
- 42. Cheadle J. P., Reeve M. P., Sampson J. R., Kwiatkowski D. J. (2000) Hum. Genet. 107, 97–114 [DOI] [PubMed] [Google Scholar]
- 43. Yeung R. S., Xiao G. H., Jin F., Lee W. C., Testa J. R., Knudson A. G. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 11413–11416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kobayashi T., Hirayama Y., Kobayashi E., Kubo Y., Hino O. (1995) Nat. Genet. 9, 70–74 [DOI] [PubMed] [Google Scholar]
- 45. Xiao G. H., Jin F., Yeung R. S. (1995) Oncogene 11, 81–87 [PubMed] [Google Scholar]
- 46. Soucek T., Yeung R. S., Hengstschläger M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15653–15658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Momose S., Kobayashi T., Mitani H., Hirabayashi M., Ito K., Ueda M., Nabeshima Y., Hino O. (2002) Hum. Mol. Genet. 11, 2997–3006 [DOI] [PubMed] [Google Scholar]
- 48. Ishizaki T., Naito M., Fujisawa K., Maekawa M., Watanabe N., Saito Y., Narumiya S. (1997) FEBS Lett. 404, 118–124 [DOI] [PubMed] [Google Scholar]
- 49. Itoh K., Yoshioka K., Akedo H., Uehata M., Ishizaki T., Narumiya S. (1999) Nat. Med. 5, 221–225 [DOI] [PubMed] [Google Scholar]
- 50. Woodsome T. P., Polzin A., Kitazawa K., Eto M., Kitazawa T. (2006) J. Cell Sci. 119, 1769–1780 [DOI] [PubMed] [Google Scholar]
- 51. Kobayashi T., Nishizawa M., Hirayama Y., Kobayashi E., Hino O. (1995) Nucleic Acids Res. 23, 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu L., Sterner C., Maheshwar M. M., Wilson P. J., Nellist M., Short P. M., Haines J. L., Sampson J. R., Ramesh V. (1995) Genomics 27, 475–480 [DOI] [PubMed] [Google Scholar]
- 53. Xiao G. H., Jin F., Yeung R. S. (1995) Cell Growth Differ. 6, 1185–1191 [PubMed] [Google Scholar]
- 54. Olsson P. G., Schofield J. N., Edwards Y. H., Frischauf A. M. (1996) Mamm. Genome 7, 212–215 [DOI] [PubMed] [Google Scholar]
- 55. Lowery D. M., Clauser K. R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S. E., Gammeltoft S., Carr S. A., Yaffe M. B. (2007) EMBO J. 26, 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ohashi K., Nagata K., Maekawa M., Ishizaki T., Narumiya S., Mizuno K. (2000) J. Biol. Chem. 275, 3577–3582 [DOI] [PubMed] [Google Scholar]
- 57. Hagerty L., Weitzel D. H., Chambers J., Fortner C. N., Brush M. H., Loiselle D., Hosoya H., Haystead T. A. (2007) J. Biol. Chem. 282, 4884–4893 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.