Abstract
RNA-binding proteins (RBPs) form ribonucleoprotein (RNP) complexes that play crucial roles in RNA processing for gene regulation. The angiosperm sieve tube system contains a unique population of transcripts, some of which function as long-distance signaling agents involved in regulating organ development. These phloem-mobile mRNAs are translocated as RNP complexes. One such complex is based on a phloem RBP named Cucurbita maxima RNA-binding protein 50 (CmRBP50), a member of the polypyrimidine track binding protein family. The core of this RNP complex contains six additional phloem proteins. Here, requirements for assembly of this CmRBP50 RNP complex are reported. Phosphorylation sites on CmRBP50 were mapped, and then coimmunoprecipitation and protein overlay studies established that the phosphoserine residues, located at the C terminus of CmRBP50, are critical for RNP complex assembly. In vitro pull-down experiments revealed that three phloem proteins, C. maxima phloem protein 16, C. maxima GTP-binding protein, and C. maxima phosphoinositide-specific phospholipase-like protein, bind directly with CmRBP50. This interaction required CmRBP50 phosphorylation. Gel mobility-shift assays demonstrated that assembly of the CmRBP50-based protein complex results in a system having enhanced binding affinity for phloem-mobile mRNAs carrying polypyrimidine track binding motifs. This property would be essential for effective long-distance translocation of bound mRNA to the target tissues.
Keywords: Plant, Protein Phosphorylation, RNA Binding Protein, RNA-Protein Interaction, RNA Transport, Phloem transport, cucurbit, long-distance signaling, pumpkin, ribonucleoprotein complex formation
Introduction
It is well known that RNA-binding proteins (RBPs)3 form ribonucleoprotein (RNP) complexes that play crucial roles in RNA stability, processing, targeted delivery, and novel modes of mRNA protection (1–7). Generally, such RNP complexes function cell-autonomously. However, recent studies have established that in plants some RNP complexes can act non-cell-autonomously. In particular, studies on the angiosperm phloem have revealed the presence of a unique population of mRNA species present within the translocation stream that delivers nutrients and signaling molecules to developing regions of the plant (8, 9). The long-distance trafficking of some of these phloem-mobile RNA species has been shown to influence a range of processes, including organ development, systemic gene silencing and pathogen defense (10–17).
The sieve tube system of the angiosperm phloem is comprised of two cell types: the companion cells and their ontogenetically related sieve elements. The phloem conduit for translocation of nutrients and signaling molecules is established by files of sieve elements that, at maturity, are enucleate, have no vacuoles, and their cytoplasm has undergone a considerable reduction in complexity (8, 18). Mature sieve elements are interconnected by sieve plate pores, which establish a low-resistance pathway for pressure-driven flow of the translocation stream. Maintenance of this plasma membrane-lined sieve tube system is carried out by the companion cells that are connected to individual sieve elements by plasmodesmata that establish cytoplasmic continuity between these two cell types (18, 19).
The various mRNA, siRNA, and microRNA species, present in the phloem translocation stream (8, 9, 12, 20–23), are thought to be produced in the companion cells. Proteins within the companion cells then mediate the trafficking of these RNA species through the plasmodesmata into the sieve tube system. Long-distance translocation of these phloem-mobile RNAs appears to be mediated by phloem-specific RBPs (8, 24). A proteomics analysis of phloem proteins contained within exudates collected from pumpkin plants identified a diverse array of RBPs (25). The roles played by some of these proteins have been the subject of recent investigations. Pioneering studies were conducted on the pumpkin (Cucurbita maxima) phloem protein 16 (CmPP16), a phloem protein that binds RNA in a non-sequence-specific manner (26) and whose delivery into various target tissues appears to be regulated through an interaction with other phloem proteins (27). A pumpkin phloem small RNA binding protein 1 was shown to bind selectively to single-stranded siRNA species and to mediate their movement through plasmodesmata (12). Other phloem RBPs have been implicated in the control of systemic infection by plant viruses (28, 29).
Phloem-mobile mRNAs are considered to be translocated as RNP complexes. However, although some 82 phloem RBPs have been identified within pumpkin phloem exudates (25), only one RNP complex has been characterized thus far. This complex is based on a phloem RBP named C. maxima RNA-binding protein 50 (CmRBP50), a member of the polypyrimidine track binding (PTB) protein family. The core of this RNP complex was shown to contain six additional phloem proteins (30). Transcripts bound within CmRBP50-based RNP complexes were identified by purifying these complexes directly from pumpkin phloem exudates. All mRNA species extracted from these CmRBP50 complexes contained PTB motifs, indicating specificity in the formation of these phloem complexes. Of equal importance, heterografting studies performed between pumpkin, as the stock, and cucumber, as the scion, confirmed the presence of the pumpkin CmRBP50-based RNP complexes within the translocation stream of the grafted cucumber scions (30). The absence of cucumber orthologs, as components of the extracted CmRBP50-based RNP complexes, indicated that these specific phloem RNP complexes must be extremely stable in nature.
In this study, we report the requirements for assembly of this pumpkin phloem CmRBP50-based RNP complex. Phosphorylation sites on CmRBP50 were mapped and then coimmunoprecipitation, and protein overlay studies established that the phosophoserine residues, located at the C terminus of CmRBP50, are critical for RNP complex assembly. In vitro pull-down experiments revealed that three phloem proteins, CmPP16, CmGTPbP, and CmPSPL bind directly with CmRBP50 and that this interaction is mediated by phosphorylation. Gel mobility-shift assays demonstrated that assembly of the CmRBP50-based protein complex results in a system having enhanced binding affinity for phloem-mobile mRNAs carrying PTB motifs. This property is consistent with our earlier finding that these CmRBP-50-based RNP complexes are extremely stable and do not appear to dissociate during their translocation to sink tissues.
EXPERIMENTAL PROCEDURES
Plant Materials
Pumpkin (C. maxima cv. Big Max) plants were grown in a special insect- and pathogen-free greenhouse under natural daylight conditions. Pumpkin and Nicotiana benthamiana plants inoculated with the zucchini yellow mosaic virus (ZYMV) expression vector (31, 32) were grown in controlled environment chambers (Conviron, model PGR15). One-month-old N. benthamiana plants were also used for agroinfiltration experiments.
Bombardment, Agroinfiltration, and Protein Purification
Biolistic-mediated delivery of ZYMV-based constructs into pumpkin leaves was performed using a Helios gene gun system (Bio-Rad). ZYMV-based constructs were coupled on 1 μm of gold particles, and this gold/DNA mixture was bombarded (helium pressure of 650 p.s.i.) onto pumpkin seedlings at the two cotyledons and one true leaf stage. Agroinfiltration was performed using Agrobacterium strain GV2260 with pGWB2- or pGWB24-based constructs (33). Leaves from 20 plants per construct were harvested 5 days after infiltration. Recombinant proteins were purified from ZYMV-infected pumpkin leaves or agroinfiltrated N. benthamiana leaves using a two-step protocol. A HisTrap FF column (GE Healthcare) was used for the first step, and a c-Myc tagged protein mild purification kit (MBL International Corp.) was employed in the second step. Purification was performed according to the manufacturer's instructions, and purified proteins were stored at −80 °C for later use.
Protein Gel Blot Analysis, Coimmunoprecipitation, and Pull-down Assays
Protein gel blot analyses were performed as follows. Nitrocellulose membrane blots were blocked and then incubated with the following antibody preparation: anti-c-Myc monoclonal antibody (1:7000, Sigma-Aldrich), anti-phosphoserine monoclonal antibody (1:1000, EMD Biosciences), and anti-GST polyclonal antibody (1:5000, Covance). Secondary antibody incubation was performed with horseradish peroxidase-conjugated anti-rabbit (1:20,000, Sigma-Aldrich) or anti-mouse (1:80,000, Sigma-Aldrich). Blots were immunodetected using chemiluminescence reagent (PerkinElmer Life Sciences) and exposed to x-ray film (Research Products International).
For coimmunoprecipitation (co-IP) experiments, pumpkin phloem sap proteins (2 μg protein/μl) were mixed with 10 μg of purified recombinant protein for 1 h, and the co-IP was carried out using the ProFoundTM c-Myc tag IP/co-IP kit (Thermo Scientific). Elution fractions were separated on 12% SDS-PAGE gels and then stained with GelCode Blue reagent (Thermo Scientific). Pull-down experiments were performed as described previously (34) with some modification. Purified recombinant CmRBP50 or CmRBP50-TM (500 ng) was mixed with each purified GST-fused recombinant protein (500 ng) and incubated with c-Myc antibody-conjugated beads (MBL International Corp.) in 1× PBS buffer for 1 h at 4 °C. Beads were collected using a HandeeTM spin column (Thermo Scientific), washed three times with 1× PBS, and eluted using Myc elution peptide (MBL International Corp.).
Electrophoretic Mobility-shift Assays
Chemically synthesized 27-nucleotide RNA oligonucleotide probes containing PTB binding sites (PTBRS) were labeled with [γ-32P]ATP (20 μCi/μl) using a KinaseMax kit (Ambion) according to the manufacturer's instructions. The full-length CmGAIP RNA riboprobe was labeled in vitro with [α-32P]UTP (10 μCi/μl) using the MAXiscript system following the manufacturer's instructions (Ambion). All reactions were performed on ice in 20 μl of binding buffer (20 mm HEPES (pH 8.0), 50 mm KCl, 1 mm DTT, and 5% (v/v) glycerol). 32P-labeled PTBRS (10 nm) was used for gel mobility-shift assays with 250 ng of purified GST, CmRBP50, CmRBP50-TM, CmPP16, CmCPI, CmGTPbP, or CmPSPL. After each reaction, the mixture was incubated on ice for 30 min, and samples were separated on a 5% (v/v) nondenaturing polyacrylamide gel. For competition assays, a mixture of CmRBP50 and/or CmRBP50 interacting proteins was incubated with increasing amount of unlabeled 27-nucleotide RNA (PTBRS) for 15 min, followed by addition of [γ-32P]-radiolabeled PTBRS and further incubation for 15 min. For the Scatchard analysis, band intensities were quantified using the ImageQuant Tools software, version 5.2 (GE Healthcare). The Scatchard analysis and measurements of Kd, Bmax, and correlation coefficient (R2) were performed using Prism 4 software (GraphPad).
Phloem Sap Collection, Protein Fractionation, and Phosphoprotein Detection
Phloem sap was collected from well watered pumpkin plants as described previously (10). Protein fractionation of pumpkin phloem sap was performed as follows. Briefly, 20 ml of pumpkin phloem sap (5–20 mg protein/ml) was dialyzed at 4 °C against buffer A (50 mm Tris (pH 7.5), 1 mm EDTA, 1% (v/v) 2-mercaptomethanol) and centrifuged at 17,000 × g for 30 min at 4 °C. Clarified phloem sap proteins were loaded onto a HiTrap Q column (GE Healthcare), connected to an FPLC system (Amersham Biosciences). After washing the column with 20 column volumes of buffer A, phloem proteins were fractionated with a linear gradient of 0–500 mm NaCl in buffer A with 1 m NaCl. The phosphorylation status of fractionated phloem proteins was determined using Pro-Q Diamond reagent (Invitrogen), according to the manufacturer's instructions. Phloem proteins were visualized with SYPRO-Ruby reagent (Invitrogen).
Protein Expression Vectors
To engineer CmRBP50 mutants, the GeneTailor site-directed mutagenesis system (Invitrogen) was employed, according to the manufacturer's instructions. CmRBP50 was used as a template, and the primers used for mutagenesis are listed in supplemental Table S1. CmRBP5–0-QM was engineered using CmRBP50-TM as a template, and the primer set for CmRBP50-S223A mutagenesis was used. To clone CmGTPbP, CmCPI, CmPSPL, CmEP89, and CmHSP, the 5′ and 3′ SMARTTM RACE cDNA amplification kit (Clontech) was employed, according to the manufacturer's instructions, using the primers listed in supplemental Table S1.
To express the recombinant proteins, CmRBP50, CmRBP50-S206A, CmRBP50-S223A, CmRBP50-S429A, CmRBP50-TM, and CmRBP50-QM were subcloned into the SphI and KpnI sites on the ZYMV-based viral vector. For transient expression of recombinant proteins using agroinfiltration in N. benthamiana, the gateway binary vectors pGBW24 and pGBW2 were used (33). His-tagged CmPP16, CmGTPbP (GenBank accession no. JF326833), CmCPI (GenBank accession no. JF326832), CmPSPL (GenBank accession no. JF326829), CmEP89 (GenBank accession no. JF326830), CmHSP (GenBank accession no. JF326831), and GST were first amplified (using the primer sets listed in supplemental Table S1) for subcloning into the pENTRTM/D-TOPO vector (Invitrogen). All clones, except GST, were subcloned into pGWB24 by using the Gateway® LR ClonaseTM II enzyme mix (Invitrogen). GST was subcloned into pGWB2. All DNA sequences described above were confirmed by sequencing.
Protein Overlay Assays
Fractionated phloem sap proteins were separated on 13% SDS-PAGE gels and then transferred onto nitrocellulose membranes. Blots were overlaid with 2 μg of purified protein in BSA buffer (50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 5 mm EDTA, 0.1% (v/v) Triton X-100, 1 mg/ml BSA) for 1 h at 20 °C. Membranes were washed with 1 × TTBS (50 mm Tris-HCl (pH 7.5), 500 mm NaCl, 0.5% (v/v) Tween 20) three times for 5 min each and subjected to protein gel blot analysis procedures as described above with anti-c-Myc monoclonal antibody and anti-mouse horseradish peroxidase-conjugated secondary antibodies (Sigma-Aldrich).
RESULTS AND DISCUSSION
In Vivo Phosphorylation of CmRBP50
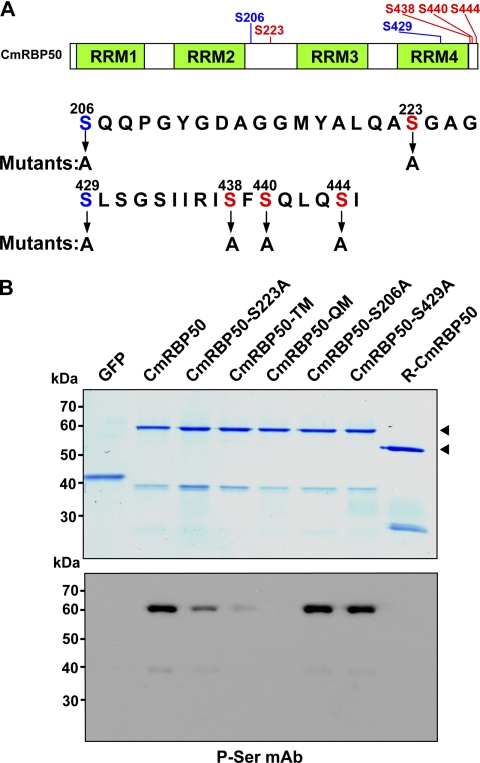
Our laboratory recently identified CmRBP50, a 50-kDa PTB protein that forms the core of a phloem-mobile RNP complex (30). Phosphorylation of CmRBP50 was shown to be required for RNP complex formation. To further explore this requirement, a web-based program was used to identify potential phosphorylation sites on CmRBP50. This analysis predicted five phosphoserine residues, Ser-206, Ser-223, Ser-429, Ser-438, Ser-440, and Ser-444 with a high probability (Fig. 1A). Preliminary mass spectroscopy-based phospho-peptide analysis indicated that the three C-terminal serine residues (Ser-438, Ser-440, and Ser-444) on phloem-purified CmRBP50 were phosphorylated.
FIGURE 1.
CmRBP50 C-terminal serine residues are phosphorylated in vivo. A, schematic of CmRBP50 indicating the location of the four conserved RNA recognition motifs (RRM1-RRM4) and the position of the putative phosphorylation sites (red and blue labeling) predicted using the bioinformatics software. Red-colored residues indicate sites whose phosphorylation appears to be essential for CmRBP50 RNP complex assembly. B, GelCode Blue staining of in planta-expressed and purified recombinant C-terminally c-Myc4-His6-tagged proteins (upper panel). Shown are CmRBP50 mutants in which Ser-206, Ser-223, or Ser-429 was replaced with Ala (CmRBP50-S206A, CmRBP50-S223A, and CmRBP50-S429A, respectively). CmRBP50-TM was generated by replacing Ser-438, Ser-440, and Ser-444 residues with Ala. CmRBP50-QM was engineered by replacing Ser-223 on CmRBP50-TM with Ala. R-CmRBP50 indicates recombinant His6-tagged protein purified from E. coli. Western blot analysis performed with an anti-phosphoserine monoclonal antibody (lower panel). The arrowheads indicate full-length CmRBP50. Note that the lower band in each lane represents a CmRBP50 autocleavage product as determined earlier by mass spectrometry (30).
Site-directed mutagenesis was employed to generate the following serine-to-alanine mutants; CmRBP50-S206A, CmRBP50-S223A, CmRBP50-S429A, CmRBP50-TM, and CmRBP50-QM (35). CmRBP50-TM was a triple mutant (TM) in which Ser-438, Ser-440, and Ser-444 were replaced by Ala, and CmRBP50-QM was a quadruple mutant (QM) derived from CmRBP50-TM in which Ser-223 was also replaced by Ala (Fig. 1A). For RNP complex assembly studies, wild-type and CmRBP50 mutants were first expressed in and purified from pumpkin leaves using a ZYMV-based vector system (31, 32, 36) (Fig, 1B, upper panel).
Western blot analysis, performed with a phosphoserine monoclonal antibody (35), confirmed that wild-type CmRBP50 was phosphorylated, whereas when expressed in and purified from Escherichia coli it was not, consistent with our earlier findings (30) (Fig. 1B, lower panel). With respect to the mutants, signals detected with CmRBP50-S206A and CmRBP50-S429A were similar in intensity to that obtained with CmRBP50, whereas CmRBP50-S223A was reduced relative to CmRBP50. Importantly, signal for CmRBP50-TM was greatly reduced, and CmRBP50-QM was not detected by the phosphoserine monoclonal antibody. Taken together, these experiments indicated that CmRBP50 residues Ser-223, Ser-438, Ser-440, and Ser-444 represent major sites phosphorylated in planta.
CmRBP50 C-terminal Phosphorylation Is Necessary for Interaction with Phloem Proteins
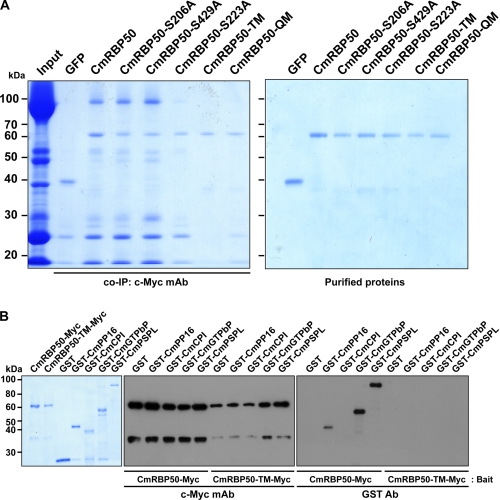
RNA-binding protein phosphorylation has recently been reported as a requirement for RNP complex formation (37, 38). To further investigate the role of CmRBP50 phosphorylation in protein-protein interaction, we next conducted co-IP and protein overlay assays using proteins contained within pumpkin phloem sap (30, 35, 39) (Fig. 2A and supplemental Fig. S1). Co-IP experiments using wild-type CmRBP50, CmRBP50-S206A, or CmBP50-S429A as bait yielded almost identical profiles for interacting phloem proteins (Fig. 2A, left panel). These studies indicated that CmRBP50 Ser-206 and Ser-429 are unlikely to be required for CmRBP50 complex formation.
FIGURE 2.
Phosphorylation of CmRBP50 C-terminal serine residues is essential for binding to proteins contained within the CmRBP50-based RNP complex. A, purified recombinant (c-Myc4-His6-tagged) GFP, CmRBP50, CmRBP50-S206A, CmRBP50-S223A, CmRBP50-S429A, CmRBP50-TM, or CmRBP50-QM was incubated with total pumpkin phloem sap (input, contained both proteins and mRNA) followed by co-IP using an anti-c-Myc monoclonal antibody (left panel). Note that CmRBP50 phosphorylation at its C terminus was necessary for RNP complex assembly. GFP was used as a negative control. Right panel, GelCode Blue staining of recombinant purified c-Myc4-His6-tagged proteins used in these pumpkin phloem co-IP experiments. B, Myc-tagged CmRBP50 or CmRBP50-TM was used to pull down the CmRBP50 interacting proteins. Bound proteins were detected by Western blot analysis using an anti-c-Myc monoclonal antibody (center panel) or GST polyclonal antibody (right panel). Left panel, recombinant purified proteins used in these pull-down assays as visualized by GelCode Blue staining.
Equivalent co-IP experiments conducted with CmRBP50-S223A indicated that this mutant form of CmRBP50 was compromised in its ability to interact with proteins within the pumpkin phloem sap to form a normal RNP complex. Similar co-IP experiments performed with CmRBP50-TM and CmRBP50-QM indicated that these proteins lacked the capacity to form RNP complexes (Fig. 2A). Equivalent results were obtained when these mutant proteins were tested in protein overlay assays. Here, CmRBP50-S223A retained the capacity to interact with phloem proteins in the lower molecular weight region (supplemental Fig. S1G), whereas the ability of both CmRBP50-TM and CmRBP50-QM to bind with phloem proteins was markedly reduced (supplemental Fig. S1, H and I, respectively).
CmRBP50 Binds Directly to CmPP16, CmGTPbP, and CmPSPL
We established earlier that the core for a CmRBP50 RNP complex contains six additional proteins, namely CmPP16, CmCPI, CmGTPbP, CmPSPL, CmEP89, and CmHSP (30). An Agrobacterium tumefaciens transient expression system, using N. benthamiana leaves, was employed to produce four of these proteins as GST fusions (Fig. 2B, left panel). Despite repeated attempts, analytical quantities of CmEP89 or CmHSP could not be obtained, as they appeared to be unstable during protein expression in N. benthamiana leaves.
In view of this situation, we proceeded to test the interaction between Myc-tagged CmRBP50 or CmRBP50-TM as bait and GST, GST-CmPP16, GST-CmCPI, GST-CmGTPbP, and GST-CmPSPL, respectively. These reaction mixtures were pulled down using a c-Myc affinity column and then tested with an anti-c-Myc mAb and a GST polyclonal Ab (Fig. 2B, center and right panel, respectively). Protein gel blot analysis, performed with anti-c-Myc mAb, confirmed the pull-down of the bait proteins CmRBP50 and CmRBP50-TM (Fig. 2B, center panel). Furthermore, these experiments established that CmRBP50 can directly interact with CmPP16, CmGTPbP, and CmPSPL but not with CmCPI. Consistent with our previous results, CmRBP50-TM failed to interact with any of these four interacting proteins (Fig. 2B, right panel). These data provided strong support for the hypothesis that, in contrast to Ser-223, the three phosphoserine residues located at the C-terminal region of CmRBP50 are essential for binding between CmRBP50 and CmPP16, and CmGTPbP and CmPSPL. These experiments also indicated that CmCPI likely binds indirectly through one of the protein components in the CmRBP50 core complex (30).
Assembly of the CmRBP50 RNP Complex
Numerous animal studies have proposed roles for RNP complexes in regulating target RNAs in mechanisms such as RNA splicing, biogenesis, and stability (40–43). The Arabidopsis genome encodes some 200 RNA-binding proteins (44), and our research group has provided evidence for the existence of numerous phloem-mobile RNA-binding proteins in the angiosperm phloem translocation stream (12, 25, 26, 30). Roles for such phloem-mobile RNP complexes have been proposed (8, 24), but detailed analyses regarding complex formation remain to be performed.
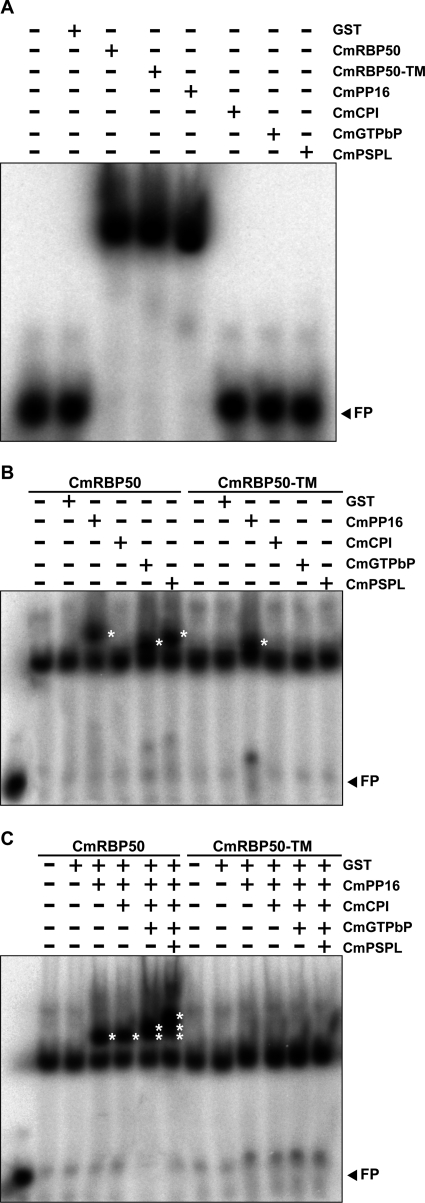
To investigate the assembly of the CmRBP50-based RNP complex, we next performed a series of gel mobility-shift assays using radiolabeled riboprobes containing PTB-binding motifs. Both wild-type CmRBP50 and CmRBP50-TM exhibited equivalent RNA binding activity (Fig. 3A). This finding is consistent with earlier studies showing that RNA binding-defective PTB proteins generally result from mutations located in the conserved RNA recognition motifs (45). We next tested whether any of the CmRBP50-interacting proteins had RNA-binding activity. With the exception of CmPP16, which we earlier showed binds RNA in a non-sequence-specific manner (26), none of the other proteins tested exhibited an RNA binding capacity (Fig. 3A). Parallel experiments performed with radiolabeled, full-length CmGAIP RNA, one of the PTB motif-containing transcripts shown to be transported through the phloem by the CmRBP50 complex (13, 30), yielded similar results (supplemental Fig. S2A). Taken together, these studies demonstrated that only CmRBP50 and CmPP16 within the CmRBP50 RNP complex have RNA-binding activity and that CmRBP50 phosphorylation specifically functions in protein-protein interaction, not RNA binding.
FIGURE 3.
Phosphorylation of CmRBP50 C-terminal serine residues potentiates assembly of a CmRBP50-based RNP complex. A, gel mobility-shift assays were performed using recombinant purified CmRBP50, CmRBP50-TM, CmPP16, CmCPI, CmGTPbP, or CmPSPL (250 ng each) and a 32P-labeled 27-nucleotide RNA probe containing PTB binding sites (PTBRS, 10 nm). Note that of the proteins contained in the CmRBP50 RNP complex, only CmRBP50 and CmPP16 had RNA-binding capacity. Equivalent binding by CmRBP50-TM and CmRBP50 to the PTBRS riboprobe indicated that binding is independent of phosphorylation. B, ability of individual CmRBP50 interacting proteins to bind CmRBP50 or CmRBP50-TM to form additional RNA complexes. CmRBP50 or CmRBP50-TM was mixed with GST, CmPP16, CmCPI, CmGTPbP, or CmPSPL for gel mobility-shift assays. The asterisks indicate the presence of additional bands compared with CmRBP50 or CmRBP50-TM alone. C, combinatorial analysis of CmRBP50-interacting proteins on the RNA binding properties of each assembled CmRBP50-based RNP complex. RNA-CmRBP50 complexes were assembled as indicated at the top of the panel. (Note that for B and C, 250-ng aliquots were added for each protein tested in a reaction mixture.) The asterisks indicate the presence of additional bands compared with CmRBP50/CmRBP50-TM alone. FP, free probe.
The influence of protein-protein interaction on CmRBP50 binding to the target PTB motif/GAIP RNA was next explored by mixing CmRBP50 or CmRBP50-TM with each interacting partner. Combinations of CmRBP50 plus CmPP16, CmRBP50 plus CmGTPbP, and CmRBP50 plus CmPSPL bound to both the PTB probe and GAIP transcript, and these RNP complexes exhibited more strongly shifted bands compared with the CmRBP50-RNA complex alone (Fig. 3B and supplemental Fig. S2B). As expected, parallel gel mobility-shift assays conducted with CmRBP50 plus CmCPI, CmRBP50-TM plus CmCPI, CmRBP50-TM plus CmGTPbP, and CmRBP50-TM plus CmPSPL exhibited similar patterns as CmRBP50 alone. These RNA binding experiments established that, in the presence of CmPP16, CmGTPbP, or CmPSPL, CmRBP50 can form protein-RNA complexes that display a greater shift in mobility as compared with CmRBP50 alone.
Our RNA binding experiments also revealed that the mobility shift observed with the CmRBP50 plus CmPP16 complex was greater than that for either CmRBP50 plus CmGTPbP or CmRBP50 plus CmPSPL (Fig. 3B). This could reflect a situation in which the molar ratio of CmPP16 binding to CmRBP50 may be higher than that for CmGTPbP or CmPSPL. Interestingly, our gel mobility-shift assays performed with radiolabeled full-length CmGAIP transcripts demonstrated that CmPP16, in the presence of either CmRBP50 or CmRBP50-TM, resulted in the formation of a second upper band (supplemental Fig. S2B). As CmPP16 does not bind to CmRBP50-TM in the absence of RNA (Fig. 2B), this result suggested that its interaction with the CmRBP50-TM-RNA complex may enhance the binding of CmPP16 to RNA (46).
Gel mobility-shift assays were next performed to examine the combinatorial effect of incubating CmRBP50 or CmRBP50-TM in the presence of either the PTB probe or GAIP transcript and combinations of CmPP16, CmCPI, CmGTPbP, and CmPSPL (Fig. 3C and supplemental Fig. S2C). As expected, CmRBP50 plus CmPP16 and CmRBP50 plus CmPP16 plus CmCPI showed similar mobility-shifted profiles. However, the combinations of CmRBP50, CmPP16, CmCPI, and CmGTPbP or CmRBP50, CmPP16, CmCPI, CmGTPbP, and CmPSPL resulted in a stepwise increase in the number of mobility-shifted bands (Fig. 3C and supplemental Fig. S2C). Taken together, these findings offer support for the hypothesis that CmPP16, CmGTPbP, and CmPSPL interact directly with CmRBP50 to assemble the core of the CmRBP50-based RNP complex.
CmRBP50-based Protein Complex Confers Enhanced RNA-binding Affinity
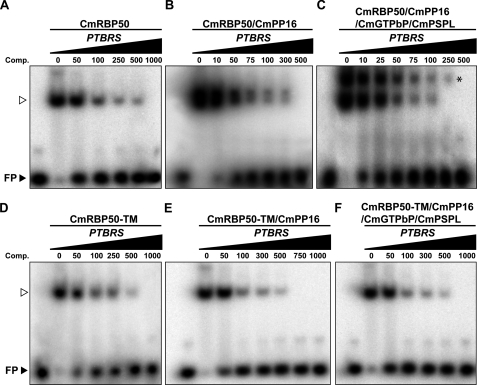
The influence of CmRBP50-protein complex assembly on RNA-binding affinity was next investigated using competition assays. Combinations of CmRBP50 alone, CmRBP50 with CmPP16, or CmRBP50 with CmPP16, CmGTPbP, and CmPSPL were preincubated with increasing concentrations of unlabeled PTBRS, followed by addition of a low concentration of radiolabeled PTBRS riboprobe (47). These experiments demonstrated that a complex comprised of CmRBP50, CmPP16, CmGTPbP, and CmPSPL was the most efficient at binding to the introduced PTB riboprobe (Fig. 4, A–C). Based on these competition assays, the RNP complex having the highest affinity for the PTBRS riboprobe is represented by the residual upper band in the presence of 250 femtomoles of unlabeled RNA (Fig. 4C). A parallel set of competition experiments was conducted using CmRBP50-TM (Fig. 4, D–F). Comparisons of the mobility-shifted bands in these CmRBP50-TM and CmRBP50 experiments further confirmed that the high-affinity RNP complex is likely composed of CmRBP50, CmPP16, CmGTPbP, and CmPSPL. Taken together, these experiments provided additional support for the hypothesis that assembly of a CmRBP50 protein complex results in an increase in RNA-binding affinity, and, thus, a more stable RNP complex.
FIGURE 4.
CmRBP50 RNP complexes have enhanced RNA binding affinity compared with CmRBP50 alone. Competition assays performed by preincubating CmRBP50 (A); CmRBP50 plus CmPP16 (B); CmRBP50, CmPP16, CmGTPbP, and CmPSPL (C); CmRBP50-TM (D); CmRBP50-TM plus CmPP16 (E); or CmRBP50-TM, CmPP16, CmGTPbP, and CmPSPL (F) with increasing concentrations of unlabeled 27-nucleotide RNA probe containing PTB binding sites (PTBRS, values in femtomoles), followed by competition with 32P-labeled PTBRS (10 femtomoles). The asterisk in C identifies the most stable RNP complex, likely composed of CmRBP50, CmPP16, CmGTPbP, and CmPSPL. The arrowheads indicate the position of the 32P-labeled 27-nt PTBRS bound by CmPBP50/CmRMP50-TM.
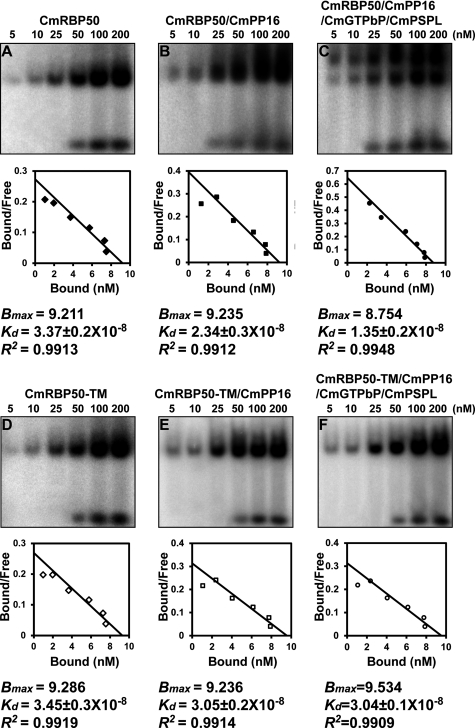
To measure the RNA binding constants associated with CmRBP50-based RNP complex assembly, gel mobility-shift assays were next conducted using reaction mixtures in which CmRBP50 or CmRBP50-TM was incubated with CmPP16 or CmPP16, CmGTPbP, and CmPSPL and increasing amounts of the PTBRS riboprobe. Band intensities were then measured, and a Scatchard plot analysis was performed to determine values for the dissociation constants (Kd) and Bmax (30). Based on these experiments, the calculated Kd values for CmRBP50, CmRBP50 plus CmPP16, and CmRBP50, CmPP16, CmGTPbP, and CmPSPL binding to the PTBRS riboprobe were 33.7 nm, 23.4 nm, and 13.5 nm, respectively (Fig. 5, A–C). As anticipated, the Kd values for CmRBP50-TM, CmRBP50-TM plus CmPP16 and CmRBP50-TM, CmPP16, CmGTPbP, and CmPSPL (Fig. 5, D–F) were higher than those measured with CmRBP50.
FIGURE 5.
Scatchard plot analyses establish that RNA binding by CmRBP50 increases in affinity in the presence of CmPP16, CmGTPbP, and CmPSPL. Purified recombinant CmRBP50 (A); CmRBP50 plus CmRBP16 (B); CmRBP50, CmPP16, CmGTPbP, and CmPSPL (C); CmRBP50-TM (D); CmRBP50-TM plus CmRBP16 (E); or CmRBP50-TM, CmPP16, CmGTPbP, and CmPSPL (F) was incubated with increasing amounts of a 32P-labeled 27-nt PTBRS riboprobe (concentrations from 5–200 nm and 250-ng aliquots were added for each protein tested in a reaction mixture). Autoradiograms (upper panels) were analyzed (ImageQuant Tools software, version 5.2, GE Healthcare) to quantify the bound and free forms of the PTBRS riboprobe, and measured values were employed for Scatchard plot analysis using the Prism 4 program (GraphPad) to determine the dissociation constants (Kd), Bmax, and correlation coefficients (R2) (lower panels).
CONCLUSIONS
Plant genomes contain a wide array of RNA-binding proteins that have been predicted to function in developmental processes, environmental adaptation, and genome organization (44). At least 82 non-redundant RNA-binding proteins were detected in the pumpkin phloem sap (25), but their functions mostly remain to be elucidated. In this study, we established the requirements for assembly of one such phloem RNP complex. The CmRBP50-based RNP complex has been shown to transport a specific set of mRNAs, including those encoding transcription factors, from mature leaves to developing tissues/organs (30).
CmRBP50 undergoes in vivo phosphorylation, and this posttranslational modification at its C terminus was shown to be necessary for assembly of the CmRBP50 RNP complex. It is also possible that CmRBP50 phosphorylation may be required for its cell-to-cell transport from the companion cell into the neighboring sieve element (35). Alternatively, phosphorylation may occur subsequent to CmRBP50 transport into the sieve element, followed by RNP complex assembly. In any event, assembly of the CmRBP50-based protein complex results in a system having enhanced binding affinity for phloem-mobile mRNAs carrying PTB motifs. This property would likely be essential to allow for effective long-distance translocation of bound mRNA. Such journeys could last for many hours to even days in large-vine species, such as the cucurbits. Disassembly of the CmRBP50 RNP complex within the target tissue(s) may be mediated by CmRBP50 dephosphorylation, involving a phosphatase located in the terminal phloem (25).
Supplementary Material
Acknowledgments
We thank Prof. Jingquan Yu, Zhejiang University, Hangzhou, China for supporting this cooperative research project. We also thank Dr. Zee-Yong Park, Gwangju Institute of Science and Technology, Gwangju, South Korea, for assistance with the initial phospho-peptide analysis of CmRBP50.
This work was supported by National Science Foundation grants IOS-0752997 and IOS-0918433 (to W. J. L.). P. L. was supported in part by a graduate fellowship from the Chinese Scholarship Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
- RBP
- RNA-binding protein
- RNP
- ribonucleoprotein
- CmPP16
- Cucurbita maxima phloem protein 16
- CmRBP50
- Cucurbita maxima RNA-Binding Protein 50
- PTB
- polypyrimidine tract binding
- ZYMV
- Zucchini yellow mosaic virus
- co-IP
- coimmunoprecipitation
- TM
- triple mutant
- QM
- quadruple mutant
- PTBRS
- 27-nucleotide RNA oligonucleotide probes containing PTB binding sites
- CmCPI
- Cucurbita maxima cysteine protease inhibitor
- CmGTPbP
- Cucurbita maxima GTP-binding protein
- CmPSPL
- Cucurbita maxima phosphoinositide-specific phospholipase-like protein
- CmEP89
- Cucurbita maxima-expressed protein 89
- CmHSP
- Cucurbita maxima heat shock protein-related protein
- CmGAIP
- Cucurbita maxima gibberellic acid-insensitive phloem.
REFERENCES
- 1. Guhaniyogi J., Brewer G. (2001) Gene 265, 11–23 [DOI] [PubMed] [Google Scholar]
- 2. Kloc M., Zearfoss N. R., Etkin L. D. (2002) Cell 108, 533–544 [DOI] [PubMed] [Google Scholar]
- 3. Shav-Tal Y., Singer R. H. (2005) J. Cell Sci. 118, 4077–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. St Johnston D. (2005) Nat. Rev. Mol. Cell Biol. 6, 363–375 [DOI] [PubMed] [Google Scholar]
- 5. Anderson P., Kedersha N. (2006) J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A., Slanchev K., le Sage C., Nagel R., Voorhoeve P. M., Van Duijse J., Ørom U. A., Lund A. H., Perrakis A., Raz E., Agami R. (2007) Cell 131, 1273–1286 [DOI] [PubMed] [Google Scholar]
- 7. Shyu A. B., Wilkinson M. F., van Hoof A. (2008) EMBO J. 27, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lough T. J., Lucas W. J. (2006) Annu. Rev. Plant Biol. 57, 203–232 [DOI] [PubMed] [Google Scholar]
- 9. Kehr J., Buhtz A. (2008) J. Exp. Bot. 59, 85–92 [DOI] [PubMed] [Google Scholar]
- 10. Ruiz-Medrano R., Xoconostle-Cázares B., Lucas W. J. (1999) Development 126, 4405–4419 [DOI] [PubMed] [Google Scholar]
- 11. Kim M., Canio W., Kessler S., Sinha N. (2001) Science 293, 287–289 [DOI] [PubMed] [Google Scholar]
- 12. Yoo B. C., Kragler F., Varkonyi-Gasic E., Haywood V., Archer-Evans S., Lee Y. M., Lough T. J., Lucas W. J. (2004) Plant Cell 16, 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haywood V., Yu T. S., Huang N. C., Lucas W. J. (2005) Plant J. 42, 49–68 [DOI] [PubMed] [Google Scholar]
- 14. Banerjee A. K., Chatterjee M., Yu Y., Suh S. G., Miller W. A., Hannapel D. J. (2006) Plant Cell 18, 3443–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aung K., Lin S. I., Wu C. C., Huang Y. T., Su C. L., Chiou T. J. (2006) Plant Physiol. 141, 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bari R., Datt Pant B., Stitt M., Scheible W. R. (2006) Plant Physiol. 141, 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baumberger N., Tsai C. H., Lie M., Havecker E., Baulcombe D. C. (2007) Curr. Biol. 17, 1609–1614 [DOI] [PubMed] [Google Scholar]
- 18. van Bel A. J. E. (2003) Plant Cell Environ. 26, 125–149 [Google Scholar]
- 19. van Bel A. J., Ehlers K., Knoblauch M. (2002) Trends Plant Sci. 7, 126–132 [DOI] [PubMed] [Google Scholar]
- 20. Zhang S., Sun L., Kragler F. (2009) Plant Physiol. 150, 378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dunoyer P., Brosnan C. A., Schott G., Wang Y., Jay F., Alioua A., Himber C., Voinnet O. (2010) EMBO J. 29, 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Molnar A., Melnyk C. W., Bassett A., Hardcastle T. J., Dunn R., Baulcombe D. C. (2010) Science 328, 872–875 [DOI] [PubMed] [Google Scholar]
- 23. Buhtz A., Pieritz J., Springer F., Kehr J. (2010) BMC Plant Biol. 10, Article No. 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lucas W. J., Yoo B. C., Kragler F. (2001) Nat. Rev. Mol. Cell Biol. 2, 849–857 [DOI] [PubMed] [Google Scholar]
- 25. Lin M. K., Lee Y. J., Lough T. J., Phinney B. S., Lucas W. J. (2009) Mol. Cell. Proteomics 8, 343–356 [DOI] [PubMed] [Google Scholar]
- 26. Xoconostle-Cázares B., Xiang Y., Ruiz-Medrano R., Wang H. L., Monzer J., Yoo B. C., McFarland K. C., Franceschi V. R., Lucas W. J. (1999) Science 283, 94–98 [DOI] [PubMed] [Google Scholar]
- 27. Aoki K., Suzui N., Fujimaki S., Dohmae N., Yonekura-Sakakibara K., Fujiwara T., Hayashi H., Yamaya T., Sakakibara H. (2005) Plant Cell 17, 1801–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gómez G., Pallás V. (2004) J. Virology 78, 10104–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gómez G., Torres H., Pallás V. (2005) Plant J. 41, 107–116 [DOI] [PubMed] [Google Scholar]
- 30. Ham B. K., Brandom J. L., Xoconostle-Cázares B., Ringgold V., Lough T. J., Lucas W. J. (2009) Plant Cell 21, 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin S. S., Hou R. F., Yeh S. D. (2002) Bot. Bull. Acad. Sinica 43, 261–269 [Google Scholar]
- 32. Hsu C. H., Lin S. S., Liu F. L., Su W. C., Yeh S. D. (2004) J. Allergy Clin. Immunol. 113, 1079–1085 [DOI] [PubMed] [Google Scholar]
- 33. Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007) J. Biosci. Bioeng. 104, 34–41 [DOI] [PubMed] [Google Scholar]
- 34. Patel M., Margaron Y., Fradet N., Yang Q., Wilkes B., Bouvier M., Hofmann K., Côté J. F. (2010) Curr. Biol. 20, 2021–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Taoka K., Ham B. K., Xoconostle-Cázares B., Rojas M. R., Lucas W. J. (2007) Plant Cell 19, 1866–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma Y., Miura E., Ham B. K., Cheng H. W., Lee Y. J., Lucas W. J. (2010) Plant J. 64, 536–550 [DOI] [PubMed] [Google Scholar]
- 37. Czudnochowski N., Vollmuth F., Baumann S., Vogel-Bachmayr K., Geyer M. (2010) J. Mol. Biol. 395, 28–41 [DOI] [PubMed] [Google Scholar]
- 38. Chen R., Yang Z., Zhou Q. (2004) J. Biol. Chem. 279, 4153–4160 [DOI] [PubMed] [Google Scholar]
- 39. Lee J. Y., Yoo B. C., Rojas M. R., Gomez-Ospina N., Staehelin L. A., Lucas W. J. (2003) Science 299, 392–396 [DOI] [PubMed] [Google Scholar]
- 40. Allemand E., Hastings M. L., Murray M. V., Myers M. P., Krainer A. R. (2007) Nat. Struct. Mol. Biol. 14, 630–638 [DOI] [PubMed] [Google Scholar]
- 41. Trabucchi M., Briata P., Garcia-Mayoral M., Haase A. D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M. G. (2009) Nature 459, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ishmael F. T., Fang X., Houser K. R., Pearce K., Abdelmohsen K., Zhan M., Gorospe M., Stellato C. (2011) J. Immunol. 186, 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. D'Orso I., Frasch A. C. (2001) J. Biol. Chem. 276, 15783–15793 [DOI] [PubMed] [Google Scholar]
- 44. Lorković Z. J. (2009) Trends Plant Sci. 14, 229–236 [DOI] [PubMed] [Google Scholar]
- 45. Auweter S. D., Allain F. H. (2008) Cell. Mol. Life Sci. 65, 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carroll K. L., Ghirlando R., Ames J. M., Corden J. L. (2007) RNA 13, 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang C., Meier U. T. (2004) EMBO J. 23, 1857–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.