Abstract
The RNA polymerase II (pol II) initiation and elongation factor elongation factor TFIIF can be extensively phosphorylated in vivo, although the significance of this modification has not been clear. We now show that phosphorylation of recombinant TFIIF by casein kinase 2 (CK2) reduces or eliminates some of the functions of TFIIF while paradoxically leaving others intact. Phospho-IIF is fully functional in binding to free pol II and is able to support the initiation of transcription. However, the phosphorylated factor does not bind to stalled elongation complexes as measured in a gel mobility shift assay. Significantly, phosphorylation strongly reduces (or for some truncated versions of RAP74, eliminates) stimulation of transcript elongation by TFIIF. Thus, although TFIIF must participate at the initiation of transcription, its ability to continue its association with pol II and stimulate transcript elongation can be specifically regulated by CK2. This is particularly interesting because CK2 is required for initiation at a subset of pol II promoters. Modulation of TFIIF function could be important in controlling promoter-proximal pausing by pol II during the early stage of transcript elongation.
Keywords: Gene Regulation, RNA Polymerase II, RNA Synthesis, Transcription Elongation Factors, Transcription Initiation Factors, TFIIF
Introduction
TFIIF is a general pol II3 transcription factor (1) that in metazoans consists of two subunits designated RAP30 and RAP74. It was originally purified on the basis of its association with pol II (2). TFIIF assists pol II in binding to promoter sequences as part of preinitiation complex (PIC) formation (3). TFIIF is also involved, along with the general transcription factor TFIIB, in selecting the exact site at which transcription begins (4–8). After initiation, TFIIF is less tightly associated with the transcription complex, but it can bind to both stalled and freely elongating complexes (3, 9, 10). During transcript elongation in metazoan systems, TFIIF considerably stimulates the overall rate of RNA synthesis (11–14). It apparently functions to maintain the appropriate location of the transcript 3′ end with the active site and thus reduces transient pausing (15). TFIIF synergizes with TFIIS to facilitate transcript elongation on nucleosomal templates in vitro (16). In accordance with its in vitro properties during both initiation and elongation, TFIIF was shown to associate with both promoter and coding regions of transcriptionally active metazoan genes in vivo (17–19). TFIIF may also be involved in recycling of pol II by stimulating the phosphatase activity of Fcp1 for the C-terminal domain of the largest subunit of pol II (3).
Given the broad range of the involvement of TFIIF in transcription by pol II, it is important to understand to what extent TFIIF activity can be regulated. It was previously reported that both subunits of TFIIF can be phosphorylated in vivo (2, 20–22). Earlier in vitro studies reached different conclusions on the effects of these modifications on TFIIF activity. Kitajima et al. (23) compared the properties of TFIIF purified from HeLa cells before and after treatment with phosphatase. They found that the dephosphorylated protein was less active in all assays and showed a lower affinity for pol II. These effects were attributed specifically to the RAP74 subunit. Consistent with that report, Cabrejos et al. (24) observed that phosphorylation by CK2 increased the apparent activity of TFIIF in supporting transcription in vitro. However, Rossignol et al. (25) showed that some phosphorylation events can down-regulate TFIIF activity in transcript elongation assays. These authors also showed that treatment of TFIIF with HeLa extracts resulted in phosphorylation of three peptides within the central so-called charged domain of RAP74. All of these peptides contain consensus sites for CK2 (25). CK2 is involved in many aspects of gene expression. It plays a major role in controlling transcription by RNA polymerases I and III (26–28). CK2 also functions during transcription by pol II. It was shown to phosphorylate TFIIF and the TFIIF-associated C-terminal domain phosphatase Fcp1 (25, 29, 30). Phosphorylation of Fcp1 by CK2 stimulated its phosphatase activity and enhanced its binding to RAP74 (29). CK2 is essential for the activity of some pol II promoters in vitro, and it associates with pol II promoter regions in vivo as judged by ChIP analysis (31). CK2 colocalizes with productively transcribing RNA pol II and RAP74 on polytene chromosomes of Chironomus salivary gland cells (18). In that study it was concluded that both CK2 and TFIIF travel with the elongating form of pol II.
On the basis of the established roles of CK2 in transcriptional regulation and preliminary experiments showing that phosphorylation of TFIIF can modulate its function, we have investigated the effects of CK2 modification on TFIIF activity. We now show that the in vitro activity of TFIIF can be selectively affected by CK2. CK2 modification completely eliminates factor binding to stalled transcript elongation complexes in a mobility shift assay and at least strongly reduces stimulation of transcript elongation by TFIIF. However, the ability of TFIIF to support transcription complex assembly is only slightly impaired. Remarkably, CK2 phosphorylation has no effect on the interaction of TFIIF and free pol II, although CK2 modification eliminates factor interaction with transcriptionally engaged pol II. These results are significant in light of the requirement for CK2 for the activity of some pol II promoters (31), the obligatory participation of TFIIF in effective transcript elongation complexes in vitro (32), and the regulated pausing of pol II immediately downstream of the start of transcription at many metazoan genes (10, 33–35).
EXPERIMENTAL PROCEDURES
Reagents
NTPs were obtained from GE Healthcare, 32P-labeled NTPs from New England Nuclear, streptavidin-coated magnetic beads from Invitrogen, Ni-nitrilotriacetic acid (Ni-NTA) beads from Qiagen, and RNasin Plus from Promega. CpA dinucleotide was custom-synthesized by Dharmacon. Casein kinase 2 was purchased from New England Biolabs, anti-RAP74 antibody (N-16, sc-234) was obtained from Santa Cruz Biotechnology, and anti-Rpb3 (POLR2C, ab14252) was from Abcam.
Plasmids and Templates
All templates contained the adenovirus major late promoter. Plasmids pML20–40(6G) and pML20–40(31G) were described previously (36). The pML20–40(31G)m template was obtained from pML20–40(31G) by eliminating a downstream PvuII site. Working templates were generated by PCR amplification from one of these plasmids. The biotinylated upstream primer contained a PvuII site. Its 5′ end was located 100 bp upstream of the transcription start site. Except for the pML20–40(6G) template, downstream template ends were generated by restriction cleavage.
Proteins and Factors
Human TBP, TFIIB, and TFIIE were recombinant proteins prepared as described (37, 38). Recombinant human RAP30, RAP74, and truncated and mutated versions of RAP74 were expressed individually in Escherichia coli, purified on Ni-NTA-agarose in the presence of 8 m urea, and dialyzed against buffer BC500 (20 mm Tris-HCl (pH 7.9), 0.2 mm EDTA, 20% glycerol, 500 mm KCl) containing 5 mm β-mercaptoethanol and 0.25 mm PMSF. (Numbers for the BC series buffers indicate the millimolar KCl concentration.) TFIIF was generated by mixing purified RAP30 and RAP74 at a 1:1 molar ratio under denaturing conditions (4 m urea in BC500) followed by dialysis against BC500 and further purification by gel filtration. Serine-to-alanine variants of RAP74 were generated using the QuikChange site-directed mutagenesis kit (Agilent Technologies) and verified by sequencing.
TFIIH was purified from HeLa nuclear extract using a modification of the procedure of Maldonado et al. (37, 39). Nuclear extract was applied to a phosphocellulose column in BC100, washed extensively in BC300, and developed with a linear gradient from BC300 to BC1000 with the buffers supplemented with 1 mm DTT and 0.25 mm PMSF. TFIIH activity, which eluted around 0.5 m KCl, was further purified by step elution from a DE52 column (Whatman) at 0.35 m KCl followed by binding and elution from a mono Q column (GE Healthcare) developed with a linear gradient of BC100 to BC800 (buffers supplemented with 1 mm DTT and 0.25 mm PMSF). TFIIH activity eluted from mono Q between 0.25 and 0.35 m KCl. Human pol II was purified from HeLa cell nuclear pellets by the protocol of Maldonado et al. (39) with the modifications described by Újvári and Luse (38).
Phosphorylation and Purification of TFIIF
24 pmol TFIIF in a volume of 800 μl was incubated at 30 °C for 60 min in 20 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 75 mm KCl, 35 mm NaCl, 0.2 mm ATP, 80 μg/ml BSA, 0.25 mm PMSF, 0.01% Triton X-100, and 5 units/μl CK2. The reactions in Fig. 1 also contained a trace amount of [γ-32P]ATP. Mock reactions were performed identically, except that ATP and CK2 were omitted. Kinased or mock-treated proteins were mixed with 2.1 ml of BB500 (20 mm Tris-HCl (pH 7.9), 0.05% Nonidet P-40, 1.5 mm β-mercaptoethanol, 0.2 mm PMSF, 20% glycerol, 500 mm KCl) containing 10 mm imidazole and 20 μl of Ni-NTA beads. Tubes were gently mixed by rotation at 4 °C for 1 h. Beads were washed four times with the binding buffer to remove the kinase. Proteins were eluted from the beads with 50 μl of BB200 (same as BB500 but with 200 mm KCl) containing 200 mm imidazole and 0.25 mg/ml BSA, followed by dialysis against BB100 containing 10 mm NaF, 2 mm Na β-glycerophosphate, and 2 mm Na pyrophosphate. Protein integrity and concentrations were determined by Western blot analysis.
FIGURE 1.
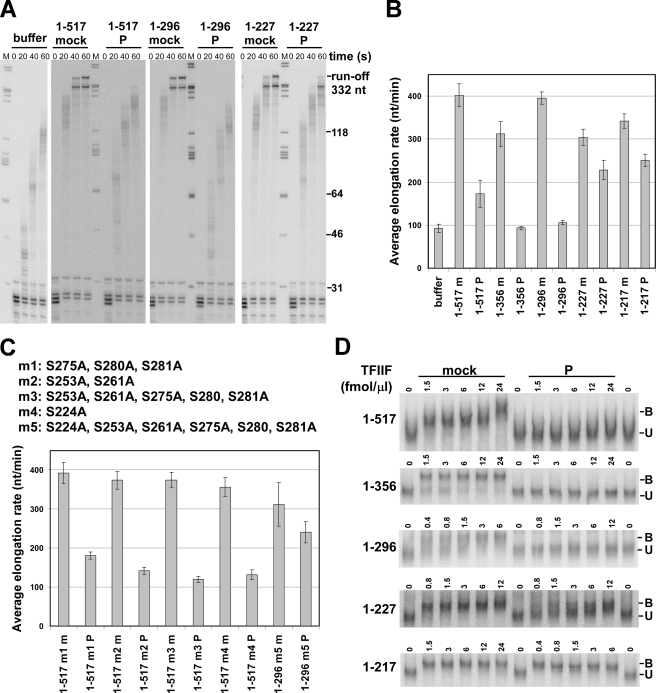
Phosphorylation of TFIIF by CK2. A, RAP74, RAP30 and C-terminal truncations of RAP74 are shown diagrammatically, and the length of each protein is given. Vertical black bars with numbers below the diagram indicate consensus CK2 phosphorylation sites (S/TXXE/D). The numbers give the position of the Ser or Thr residue within that site. Two SD sites, which could also be modified by CK2, are labeled in gray above the diagram and marked by gray bars. Sites labeled with an asterisk were shown to be phosphorylated by HeLa whole cell extract (25). Domain designations are based on Lei et al. (14) and Kamada et al. (51) for RAP74 and Gaiser et al. (52) for RAP30. B, TFIIF containing full-length or truncated RAP74 was phosphorylated by recombinant CK2 in the presence of [γ-32P]ATP. Proteins were separated by SDS-PAGE. Signal intensity for the gel on the left was adjusted to be darker below the dashed line to reveal labeling of the RAP30 subunit. The positions of the different labeled subunits are indicated on the right.
Transcription Initiation Assay
Preinitiation complexes were assembled as described (38) using 133 ng of bead-attached pML20–40(6G) template per 100 μl and varying amounts of TFIIF as indicated in Fig. 3. Transcription was initiated with 0.25 mm CpA (initiating at position −1), 50 μm UTP and GTP, 1 μm [α-32P]CTP, and 50 μm dATP at 30 °C for 5 min followed by incubation with 50 μm CTP for 5 min. Complexes were washed, and the 21-mer RNAs were then recovered by phenol-chloroform extraction and resolved on 15% denaturing acrylamide gels.
FIGURE 3.
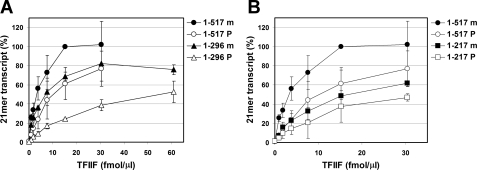
CK2 phosphorylated TFIIF is active in assembly of preinitiation complexes. PICs were formed in the presence of varying amounts of mock-treated or phosphorylated TFIIF, and transcription was initiated by limited NTP addition to yield 21-mer transcripts. A, comparison of RNA yields when TFIIF(1–517) versus TFIIF(1–296) was used. B, comparison of RNA yields when TFIIF(1–517) versus TFIIF(1–217) was used. For both panels, 100% corresponds to the level of RNA made with mock-treated TFIIF(1–517) at saturating concentrations (15 fmol/μl). The error bars indicate mean ± S.D. based on three to five repeats.
Transcript Elongation Assay
PCR-amplified pML20–40(31G)m templates were cut with NdeI so that run-off transcription resulted in a 332-nt RNA. Preinitiation complexes were assembled using ∼600 ng of template per 100 μl and were rinsed with BC100 containing 1 mm DTT and 0.5 mg/ml BSA, followed by a rinse with M65 (20 mm Tris-HCl (pH 7.9), 0.25 mm EDTA, 5% glycerol, 10 mm Na β-glycerophosphate, 10 mm MgCl2, 1 mm DTT, 65 mm KCl) containing 0.25 mg/ml BSA to remove free factors. Complexes were resuspended in M65, and transcription was initiated with 0.5 mm ATP (initiating at position +1), 50 μm UTP, and 2.5 μm [α32-P]CTP at 30 °C for 1.5 min, followed by incubation with 50 μm CTP for 0.5 min. The transcription complexes, which now contained primarily 30-nt transcripts, were washed once with high salt buffer (20 mm Tris-HCl (pH 7.9), 1 mm DTT, 1.6 m KCl, 0.25 mg/ml BSA) and twice with M40 (same as M65 but with 40 mm KCl) containing 0.25 mg/ml BSA. Complexes were released from magnetic beads by treatment with 4 units/μl PvuII for 15 min at 30 °C in the M40 plus BSA buffer. Following the addition of 1.5 μg of herring sperm DNA per μg of template, the supernatants were separated from the beads. Reactions were diluted with M40 plus BSA, supplemented with RNAsin Plus to a final concentration of 0.4 units/μl, mixed with mock-treated or phosphorylated TFIIF, brought to final concentrations of 56 mm KCl and 10 mm MgCl2, and kept on ice until needed. Before the addition of nucleotides, reactions were brought to 30 °C, and an aliquot was removed for the zero time point. Transcription was restarted by addition of all four NTPs (preincubated at 30 °C) to a final concentration of 0.25 mm. Aliquots were removed at 20, 40, and 60 s and pipetted into phenol-chloroform. Transcripts were resolved on 8% denaturing acrylamide gels. Average elongation rates were calculated by determining the average transcript lengths (in comparison to DNA markers) in lanes where run-off transcripts had not yet accumulated significantly.
Analysis of Elongation Complexes by Native Gel Electrophoresis
High salt-washed complexes containing labeled 30-mer transcripts were generated as described above for the elongation assay. Complexes were released from the beads by cleavage with Cac8 I for 15 min at 30 °C (which cuts both upstream and downstream of the complexes), followed by addition of 1.5 μg of herring sperm DNA per μg of template. The supernates were diluted with M40 plus BSA, followed by the addition of mock-treated TFIIF, phosphorylated TFIIF, or buffer, as indicated in Fig. 2. After 10 min at room temperature, glycerol was added to 18%, and the samples were resolved on 4% native acrylamide gels (40) at 7 Watt for 3 h at 4 °C.
FIGURE 2.
Phosphorylation of TFIIF by CK2 impairs its activity during elongation. A, high salt-washed transcription complexes containing labeled 30-nt RNAs were chased with 0.25 mm NTPs at 30 °C for the indicated times with saturating amounts of the indicated TFIIF proteins (48 to 96 fmol/μl) or with buffer only. The positions of selected DNA molecular weight markers (M) are indicated to the right of the panel. B, average elongation rates were calculated from results like those in A by determining the average length of transcripts in lanes where the polymerase had not yet reached the end of the template. The lengths of the RAP74 subunit and the modification states of TFIIF (m, mock; P, phosphorylated) are shown. The error bars indicate mean ± S.D. based on three to nine repeats. C, RAP74 variants with the indicated Ser-to-Ala substitutions were made either with full-length RAP74 (m1-m4) or the 1–296 truncation (m5). Assays identical to those in A were performed with CK2-modified (P) or mock-phosphorylated (m) TFIIF preparations containing the indicated variants. Average elongation rates were determined as in B. The error bars indicate mean ± S.D. for three determinations. D, electrophoretic mobility shift assay with high salt-washed ternary complexes containing 30-nt labeled transcripts. Mock-treated or phosphorylated TFIIF was added to complexes at the indicated concentrations, and samples were run on 4% native acrylamide gels. The composition of the RAP74 subunit of TFIIF is indicated to the left of each gel. Unbound (U) and bound (B) complexes are labeled.
RNA Polymerase II Binding Assay
12 pmol mock-treated or phosphorylated TFIIF was mixed with 3 or 6 pmol pol II in BB100 containing 10 mm imidazole and 0.5 mg/ml BSA in a final volume of 50 μl for 1 h at 4 °C. 50 μl of BB100 was added containing 5 μl of Ni-NTA beads, and the reactions were rotated at 4 °C for 1 h. Beads were washed four times with 500 μl of the same buffer, and bound proteins were eluted with BB100 containing 200 mm imidazole and 0.25 mg/ml BSA. The eluted proteins were resolved on SDS-PAGE. RAP74 and the Rpb3 subunit of pol II were detected by Western blot analysis. About 10–20% of the input pol II was retained on the nickel resin through binding to TFIIF.
RESULTS
Phosphorylation of TFIIF by CK2
Our laboratory has been studying the retention of transcription initiation factors during transcript elongation by pol II in vitro. During the course of these experiments, we observed that addition of small amounts of HeLa nuclear extract to PICs resulted in lower rates of subsequent transcript elongation. Given the known effects of TFIIF on elongation and the report indicating that CK2 in HeLa whole cell extracts is an efficient kinase of TFIIF (25), we decided to investigate the effects of phosphorylation of TFIIF by CK2 during transcript elongation.
In Fig. 1A, the locations of consensus CK2 modification sites within RAP74 (S/TXXE/D) are indicated by the black numbers below the diagram of the full-length RAP74 protein. The gray numbers above the diagram indicate two sites (with sequence SD) that could also be modified by CK2. Sites previously reported to be phosphorylated by HeLa whole cell extract (25) are marked by asterisks. We incubated TFIIF reconstituted from recombinant RAP30 and RAP74 with recombinant CK2 in the presence of [γ-32P]ATP and continued the reactions until incorporation of radioactivity reached its maximal level (Fig. 1). The results in Fig. 1B show that both RAP74 and RAP30 are phosphorylated under these conditions, with most of the radioactive signal going into RAP74 (lane 1). As demonstrated below, we eventually determined that phosphorylation of RAP74 is responsible for the functional effects of CK2 on TFIIF. We therefore also tested a series of C-terminal truncation mutants of RAP74, generously provided to us by Z. Burton (14, 41). The locations of the C termini of each of these mutants are listed in Fig. 1A. All TFIIF proteins containing RAP74 truncations were phosphorylated by CK2 on both TFIIF subunits, as shown in Fig. 1B, lanes 2-5. The observed differences in modification level among these proteins may have resulted from different accessibility of the modification sites in the various truncated forms of RAP74. RAP30 was phosphorylated to the same limited extent in all constructs tested.
CK2 Phosphorylation Impairs the Activity of TFIIF during Transcript Elongation
We first tested the effect of phosphorylation by CK2 on the ability of TFIIF to stimulate the rate of elongation. We assembled transcription complexes on bead-bound templates containing the adenovirus major late promoter with purified pol II and purified or recombinant general transcription factors. Complexes containing radiolabeled transcripts were stalled at position 30 and washed successively with high salt (1.6 m KCl) and low salt (40 mm KCl) buffers. This procedure removes all transcription factors and leaves only engaged polymerase in the transcription complex (32). Complexes were released from the beads by restriction enzyme cleavage, incubated with buffer or saturating amounts of TFIIF, and chased at 30 °C to the end of the templates with 0.25 mm NTPs (which is roughly the NTP concentration needed to support half-maximal transcription rates (42)). Reactions were stopped at different times, and the resulting transcripts were resolved on denaturing acrylamide gels. Calculated average elongation rates ± S.D. are plotted in Fig. 2B.
Because TFIIF is removed from the stalled complexes during the high salt wash, complexes supplemented only with buffer displayed a slow elongation rate, 93 ± 10 nt/min, as expected from previous studies (43). Addition of native, mock-treated TFIIF increased the elongation rate 4.3-fold (402 ± 10 nt/min), also as expected (43). However, if TFIIF was phosphorylated by CK2, it was able to stimulate the elongation rate by only 1.9-fold (173 ± 31 nt/min). Truncation of RAP74 from the C terminus resulted in at most a 25% reduction in the ability of TFIIF to stimulate transcript elongation (Fig. 2, A and B). The effects on CK2 phosphorylation of RAP74-truncated versions of TFIIF, however, varied strongly with the extent of truncation. (For the sake of brevity we will refer to TFIIF complexes bearing RAP74 truncations by the length of RAP74 remaining. Full-length RAP74 is 517 amino acids.) Phosphorylated TFIIF(1–356) and TFIIF(1–296) were unable to stimulate the elongation rate of pol II. However, when phosphorylated TFIIF(1–227) and TFIIF(1–217) were tested, elongation rates dropped only slightly compared with mock-treated counterparts. Modification of RAP30 apparently does not affect TFIIF activity because RAP30 was phosphorylated to about the same extent in all TFIIF preparations tested.
Results with the truncated versions of RAP74 suggested that the functional target of CK2 modification in transcript elongation is the central, so-called charged domain of RAP74. We therefore prepared and tested a series of point mutants of RAP74 in which serine residues within the charged domain were changed to alanines. Within this region, Rossignol et al. (25) identified a peptide including serines 280 and 281 as a major site of RAP74 phosphorylation in HeLa extracts. These serines are located within a CK2 consensus site. There is also a third serine (Ser-275) in a less-favored context (SD) for CK2 modification. Changing these three residues to alanine within the context of full-length RAP74 generated the m1 variant (Fig. 2C). Wang and Burton (41) suggested that serines at positions 253 and 261 are important sites for phosphorylation of RAP74 by CK2. We therefore created variant m2, in which these residues were converted to alanine. In the m3 variant, we combined all five alterations from m1 and m2 (Fig. 2C). TFIIF(1–227) contains a CK2 consensus site at position 224 and is strongly modified by CK2 (Fig. 1B). A global proteomic study demonstrated that Ser-224 is also modified in HeLa cells (44). On this basis, we generated a fourth RAP74 variant, m4, with the single Ser-to-Ala change at 224 (Fig. 2C).
The four TFIIF serine substitution variants were phosphorylated with CK2 and tested for their ability to stimulate transcript elongation using the salt-washed 30-mer complexes as in Fig. 2A. The mock-phosphorylated variants served as controls. We were surprised to find that all of the variants were essentially as sensitive to CK2 modification as TFIIF containing wild-type RAP74 (Fig. 2C). In an attempt to produce a variant that was not inhibited by phosphorylation, we began with the RAP74 1–296 construct, the shortest version of RAP74 that is significantly responsive to CK2 modification. Within RAP74 1–296, we changed all six serines altered in m3 and m4 to alanine. TFIIF containing the resulting m5 variant was only slightly inhibited in the transcript elongation assay by CK2 modification (Fig. 2C). Thus, there is no single serine or small set of serines within the charged domain of RAP74 whose phosphorylation by CK2 is solely responsible for the reduction in TFIIF function. Loss of inhibition by CK2 was only observed when the Ser-to-Ala change at 224 was added to the other five alterations. However, CK2 modification of Ser-224 alone is clearly not sufficient to cause inhibition of TFIIF function (m4 variant). Also, it is important to note that the m3 variant (all serine changes except 224) was generated in full-length RAP74, whereas m5 (with the additional change at 224) was based on the 1–296 truncation.
TFIIF Phosphorylated by CK2 Does Not Bind to Stalled Elongation Complexes
What is the basis for the loss of function by modified TFIIF in the elongation assay? Phosphorylation by CK2 does not simply destroy TFIIF activity because all of the modified proteins function in an initiation assay (see Fig. 3, below). We have also demonstrated that dephosphorylation of CK2-phosphorylated TFIIF with shrimp alkaline phosphatase restored full activity in our elongation assay (data not shown). Loss of function by TFIIF during transcript elongation could reflect a failure to bind to the elongation complex. To test this, we used a previously described protocol involving native gel electrophoresis (40). High salt-washed complexes containing radiolabeled 30-mer RNAs were incubated with mock-treated or CK2-modified TFIIF and resolved on low-percentage polyacrylamide gels (Fig. 2D). All mock-treated TFIIF preparations bound to the stalled elongation complexes in a concentration-dependent manner, as indicated by the appearance of slower-migrating complexes. Significantly, phosphorylated TFIIF(1–517), TFIIF(1–356), and TFIIF(1–296) all failed to bind the 30-mer complexes (Fig. 2D). Phosphorylated TFIIF(1–227) did bind to the elongation complexes but only with intermediate affinity because binding was detected only at high concentrations of the factor (Fig. 2D). CK2-modified TFIIF(1–217) was able to bind to elongation complexes even at very low factor concentrations. These results are generally consistent with the deficits displayed by the various phosphorylated forms of TFIIF in the elongation assay, except that TFIIF(1–517) retains some stimulatory effect on transcript elongation (Fig. 2B).
CK2 Phosphorylated TFIIF Is Able to Support Preinitiation Complex Assembly
We also tested the effect of CK2 modification on the ability of TFIIF to support pol II PIC assembly. PICs were formed using purified pol II, purified or recombinant general transcription factors, and varying concentrations of mock-treated or CK2-phosphorylated TFIIF. Transcription was initiated with a limited set of NTPs to synthesize 21-mer RNAs. The results of these studies are summarized in Fig. 3. Truncation of RAP74 led to a reduction in the ability of (nonmodified) TFIIF to support transcription complex assembly, as expected from earlier studies (41, 43). Note that our purified pol II and TFIIH contain no detectable TFIIF because synthesis of the 21-mer was completely dependent on the addition of TFIIF. Phosphorylation by CK2 reduced the activity of both TFIIF(1–517) and TFIIF(1–296) in this assay, but this deficit was much less than that seen in the transcript elongation or elongation complex binding assays (Fig. 2). The ability of phosphorylated TFIIF(1–217) to support 21-mer synthesis was only slightly reduced, in this case consistent with the performance of this modified factor in the assays shown in Fig. 2. These results indicate that phosphorylation of RAP74 can also modulate the ability of TFIIF to function in transcription complex assembly.
CK2-phosphorylated TFIIF Binds Efficiently to Free pol II
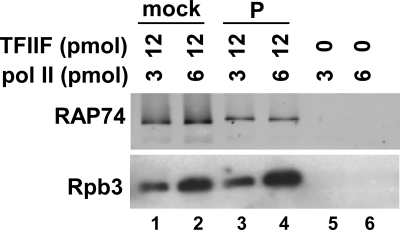
Although phosphorylated TFIIF cannot bind to stalled elongation complexes, it can support the assembly of PICs. Given that TFIIF is believed to recruit free pol II to the PIC, our results could be explained if phosphorylation selectively interferes with the interaction of TFIIF with transcriptionally engaged polymerase. Alternatively, phosphorylated TFIIF might function in the PIC only through interactions with components other than pol II. We therefore tested the ability of CK2-phosphorylated TFIIF to bind to free polymerase (Fig. 4). A fixed amount of mock-treated or phosphorylated TFIIF was incubated with different amounts of pol II in solution. Complexes were then bound to Ni-NTA beads via the C-terminal histidine tag on the subunits of TFIIF. Following washing to remove nonspecifically interacting proteins, bound proteins were eluted with imidazole. Western blotting indicates that in the absence of TFIIF, polymerase did not bind nonspecifically to the beads (Fig. 4, lanes 5 and 6). However, about the same amount of polymerase could be pulled down in the presence of mock-treated or phosphorylated TFIIF (lanes 1–4). Furthermore, the amount of polymerase recovered was proportional to the starting amount (compare lane 1 to lane 2 and lane 3 to lane 4), consistent with specific binding. These results indicate that phosphorylation of TFIIF by CK2 does not change the ability of the factor to interact with free polymerase.
FIGURE 4.
CK2 phosphorylated TFIIF binds to free pol II. The indicated amounts of pol II and phosphorylated or mock-phosphorylated TFIIF were mixed and incubated with Ni-NTA beads. Proteins eluted from the beads were detected by Western blot analysis using antibodies to RAP74 or the Rpb3 subunit of pol II.
DISCUSSION
TFIIF is both a transcription initiation factor and, in metazoans, an elongation factor. It can be phosphorylated in vivo, but the functional consequences of this modification are not well understood. CK2 has been suggested to have a role in pol II transcription, and TFIIF is a known substrate for CK2. We demonstrate here that phosphorylation by CK2 can affect a specific subset of the multiple functions of TFIIF. Binding to the stalled elongation complex is eliminated as judged by mobility shift, and stimulation of elongation is strongly reduced or eliminated, whereas interaction with free pol II is unaffected, and support for initiation is only partially affected. Our observations also indicate that there are significant differences in the interaction of TFIIF with free and transcriptionally engaged pol II, which is particularly interesting in light of recent structural studies describing the interaction of yeast TFIIF with yeast pol II (45) and the yeast PIC (46). It should be noted that a previous study of CK2 modification of TFIIF reached a different conclusion from ours (24). Those authors reported that phosphorylation of TFIIF with CK2 leads to modest increases in TFIIF function. Although we cannot fully explain the discrepancy between our findings and the earlier report, it should be noted that the experimental approaches in the earlier study were significantly different from those used here. The transcription assay in the earlier study used calf thymus pol II to synthesize a 400 nt RNA in 45 min reactions with one NTP very limiting. Polymerase and transcription factors were present throughout these reactions. In such an experiment it is not possible to distinguish separate effects on transcription complex assembly, initiation, reinitiation of transcription, and transcript elongation.
TFIIF presumably has different roles as a part of the pre-initiation complex and the elongation complex. In assembly of the PIC, TFIIF is thought to function as a chaperone for pol II (3). Because phosphorylation by CK2 does not inhibit the association of free pol II with TFIIF, the pol II chaperone activity should not be affected by modification. TFIIF also plays a role in conjunction with TFIIB in locating the start site of transcription (4–8), probably by positioning TFIIB near the pol II active site (47). Yeast Tfg1 and Tfg2 are homologues of human RAP74 and RAP30. Tfg1 and Tfg2 dimerize through their N-terminal domains. These dimerized regions in turn interact with the lobe domain of the Rpb2 subunit of pol II, well downstream of the polymerase active site and outside of the central cleft that binds template (45, 46). The central charged domain of Tfg1 is largely unstructured in the PIC. At least one contact between yeast pol II and residue 480 of Tfg1 places the charged domain of Tfg1 close to the active site cleft and TFIIB (46). It is thus reasonable to suppose that modification of RAP74 could disturb pol II/TFIIB interactions in the human PIC, leading to somewhat reduced RNA synthesis activity as we observed.
The failure (or reduction) in elongation stimulation by CK2-phosphorylated TFIIF can be directly attributed to the lack of interaction with pol II in the elongation complex. However, it then seems paradoxical that phosphorylation of TFIIF does not affect its interaction with free pol II because, as just noted, the Tfg1/Tfg2 dimerization domain (and by inference, the corresponding RAP74/RAP30 domain) has extensive interactions with the Rpb2 lobe domain in both the binary complex with free pol II (45) and in the PIC (46). This dimerization domain/Rpb2 interaction should not be affected by CK2 phosphorylation because the activity of TFIIF(1–217), which retains the full RAP74/RAP30 interface, is not significantly inhibited by CK2 modification. In this context it is interesting that residues from the N- and C-terminal ends of the Tfg1 charged domain were reported to interact with the Rpb1 jaw domain at the downstream end of the cleft in yeast pol II-TFIIF binary complexes (45), distant from proposed charged domain interactions in the PIC. It was proposed that the generally unstructured charged domain of Tfg1 might occupy the cleft in the binary complex and thereby prevent entry of nonspecific DNA (45), consistent with the known ability of TFIIF to reduce pol II interactions with non-promoter DNA (48). In elongation complexes, however, template DNA would displace the charged domain of RAP74 from the cleft. Phosphorylation within the charged domain would not be expected to lessen interaction with the positively charged pol II cleft, consistent with our results on binding of phospho-TFIIF and pol II (Fig. 4). Our results suggest that the proposed relocation of the charged domain in the elongation complex renders the RAP74-pol II interaction very sensitive to phosphorylation by CK2 because significant reduction in the effect of CK2 modification was only obtained with six serine-to-alanine changes within the charged domain of the truncated 1–296 version of RAP74 (Fig. 2C).
Regardless of the exact mechanism of CK2 action, we would stress the potential significance of a transcription-associated kinase that can selectively control critical aspects of TFIIF function. It is important to recall that, in contrast to the case with yeast (49, 50), metazoan TFIIF is found within the transcribed regions of active genes (17, 18). This is consistent with the well documented ability of metazoan TFIIF to stimulate transcript elongation in vitro (11, 12, 14). It is now appreciated that most metazoan genes are controlled at least in part through the regulated pausing of pol II immediately downstream of the start of transcription (10, 33–35). The detailed mechanism through which such pausing is regulated is not yet known, but results from in vitro studies implicate the controlled association of TFIIF with transcribing pol II as a central aspect of this process (32). Our results suggest that CK2 modification of TFIIF could participate in this regulatory mechanism because phosphorylation by CK2 would not affect the function of TFIIF as a chaperone for pol II and a participant in PIC assembly but would interfere with the ability of TFIIF to support effective transcript elongation. This model seems particularly attractive because CK2 was shown to be required for pol II activity at some promoters in vitro and is associated with promoters in vivo (31).
In summary, phosphorylation of human TFIIF by CK2 affects an interesting subset of the roles TFIIF plays in transcription. The availability of a form of TFIIF that discriminates between free and engaged pol II should provide a useful tool to further our understanding of the functional interactions of TFIIF with polymerase. It should be informative to explore further the possibility that regulation of the efficiency of transcript elongation by pol II involves the selective control of TFIIF activity through phosphorylation.
Acknowledgment
We thank Zachary Burton for the expression constructs for the truncated versions of RAP74.
This work was supported by Public Health Service grant GM 29487 from the National Institute of General Medical Sciences (to D. S. L.).
- pol II
- RNA polymerase II
- CK2
- casein kinase 2
- PIC
- preinitiation complex
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Hahn S. (2004) Nat. Struct. Mol. Biol. 11, 394–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sopta M., Carthew R. W., Greenblatt J. (1985) J. Biol. Chem. 260, 10353–10360 [PubMed] [Google Scholar]
- 3. Sims R. J., 3rd, Belotserkovskaya R., Reinberg D. (2004) Genes Dev. 18, 2437–2468 [DOI] [PubMed] [Google Scholar]
- 4. Sun Z. W., Hampsey M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 3127–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghazy M. A., Brodie S. A., Ammerman M. L., Ziegler L. M., Ponticelli A. S. (2004) Mol. Cell. Biol. 24, 10975–10985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freire-Picos M. A., Krishnamurthy S., Sun Z. W., Hampsey M. (2005) Nucleic Acids Res. 33, 5045–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Majovski R. C., Khaperskyy D. A., Ghazy M. A., Ponticelli A. S. (2005) J. Biol. Chem. 280, 34917–34923 [DOI] [PubMed] [Google Scholar]
- 8. Khaperskyy D. A., Ammerman M. L., Majovski R. C., Ponticelli A. S. (2008) Mol. Cell. Biol. 28, 3757–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zawel L., Kumar K. P., Reinberg D. (1995) Genes Dev. 9, 1479–1490 [DOI] [PubMed] [Google Scholar]
- 10. Price D. H. (2008) Mol. Cell 30, 7–10 [DOI] [PubMed] [Google Scholar]
- 11. Price D. H., Sluder A. E., Greenleaf A. L. (1989) Mol. Cell. Biol. 9, 1465–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Izban M. G., Luse D. S. (1992) J. Biol. Chem. 267, 13647–13655 [PubMed] [Google Scholar]
- 13. Kephart D. D., Wang B. Q., Burton Z. F., Price D. H. (1994) J. Biol. Chem. 269, 13536–13543 [PubMed] [Google Scholar]
- 14. Lei L., Ren D., Finkelstein A., Burton Z. F. (1998) Mol. Cell. Biol. 18, 2130–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C., Zobeck K. L., Burton Z. F. (2005) Mol. Cell. Biol. 25, 3583–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luse D. S., Spangler L. C., Újvári A. (2011) J. Biol. Chem. 286, 6040–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cojocaru M., Jeronimo C., Forget D., Bouchard A., Bergeron D., Côte P., Poirier G. G., Greenblatt J., Coulombe B. (2008) Biochem. J. 409, 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Egyházi E., Ossoinak A., Filhol-Cochet O., Cochet C., Pigon A. (1999) Mol. Cell. Biochem. 191, 149–159 [PubMed] [Google Scholar]
- 19. Wang Y., Fairley J. A., Roberts S. G. E. (2010) Curr. Biol. 20, 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burton Z. F., Killeen M., Sopta M., Ortolan L. G., Greenblatt J. (1988) Mol. Cell. Biol. 8, 1602–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flores O., Maldonado E., Reinberg D. (1989) J. Biol. Chem. 264, 8913–8921 [PubMed] [Google Scholar]
- 22. Yonaha M., Aso T., Kobayashi Y., Vasavada H., Yasukochi Y., Weissman S. M., Kitajima S. (1993) Nucleic Acids Res. 21, 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitajima S., Chibazakura T., Yonaha M., Yasukochi Y. (1994) J. Biol. Chem. 269, 29970–29977 [PubMed] [Google Scholar]
- 24. Cabrejos M. E., Allende C. C., Maldonado E. (2004) J. Cell. Biochem. 93, 2–10 [DOI] [PubMed] [Google Scholar]
- 25. Rossignol M., Keriel A., Staub A., Egly J. M. (1999) J. Biol. Chem. 274, 22387–22392 [DOI] [PubMed] [Google Scholar]
- 26. Johnston I. M., Allison S. J., Morton J. P., Schramm L., Scott P. H., White R. J. (2002) Mol. Cell. Biol. 22, 3757–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu P., Wu S., Hernandez N. (2003) Mol. Cell 12, 699–709 [DOI] [PubMed] [Google Scholar]
- 28. Bierhoff H., Dundr M., Michels A. A., Grummt I. (2008) Mol. Cell. Biol. 28, 4988–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palancade B., Dubois M. F., Bensaude O. (2002) J. Biol. Chem. 277, 36061–36067 [DOI] [PubMed] [Google Scholar]
- 30. Friedl E. M., Lane W. S., Erdjument-Bromage H., Tempst P., Reinberg D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2328–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lewis B. A., Sims R. J., 3rd, Lane W. S., Reinberg D. (2005) Mol. Cell 18, 471–481 [DOI] [PubMed] [Google Scholar]
- 32. Cheng B., Price D. H. (2007) J. Biol. Chem. 282, 21901–21912 [DOI] [PubMed] [Google Scholar]
- 33. Nechaev S., Adelman K. (2008) Cell Cycle 7, 1539–1544 [DOI] [PubMed] [Google Scholar]
- 34. Margaritis T., Holstege F. C. P. (2008) Cell 133, 581–584 [DOI] [PubMed] [Google Scholar]
- 35. Gilmour D. S. (2009) Chromosoma 118, 1–10 [DOI] [PubMed] [Google Scholar]
- 36. Pal M., Luse D. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5700–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pal M., Ponticelli A. S., Luse D. S. (2005) Mol. Cell 19, 101–110 [DOI] [PubMed] [Google Scholar]
- 38. Újvári A., Luse D. S. (2006) Nat. Struct. Mol. Biol. 13, 49–54 [DOI] [PubMed] [Google Scholar]
- 39. Maldonado E., Drapkin R., Reinberg D. (1996) Methods Enzymol. 274, 72–100 [DOI] [PubMed] [Google Scholar]
- 40. Cheng B., Price D. H. (2008) Nucleic. Acids Res. 36, e135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang B. Q., Burton Z. F. (1995) J. Biol. Chem. 270, 27035–27044 [DOI] [PubMed] [Google Scholar]
- 42. Kadesch T. R., Chamberlin M. J. (1982) J. Biol. Chem. 257, 5286–5295 [PubMed] [Google Scholar]
- 43. Lei L., Ren D., Burton Z. F. (1999) Mol. Cell. Biol. 19, 8372–8382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 45. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eichner J., Chen H. T., Warfield L., Hahn S. (2010) EMBO J. 29, 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen H. T., Hahn S. (2004) Cell 119, 169–180 [DOI] [PubMed] [Google Scholar]
- 48. Conaway J. W., Conaway R. C. (1990) Science 248, 1550–1553 [DOI] [PubMed] [Google Scholar]
- 49. Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J. F. (2002) Mol. Cell. Biol. 22, 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mayer A., Lidschreiber M., Siebert M., Leike K., Söding J., Cramer P. (2010) Nat. Struct. Mol. Biol. 17, 1272–1278 [DOI] [PubMed] [Google Scholar]
- 51. Kamada K., De Angelis J., Roeder R. G., Burley S. K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3115–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gaiser F., Tan S., Richmond T. J. (2000) J. Mol. Biol. 302, 1119–1127 [DOI] [PubMed] [Google Scholar]