Abstract
The cell wall of mycobacteria consists of an outer membrane, analogous to that of Gram-negative bacteria, attached to the peptidoglycan (PG) via a connecting polysaccharide arabinogalactan (AG). Although the primary structure of these components is fairly well deciphered, issues such as the coverage of the PG layer by covalently attached mycolates in the outer membrane and the spatial details of the mycolic acid attachment to the arabinan have remained unknown. It is also not understood how these components work together to lead to the classical acid-fast staining of mycobacteria. Because the majority of Mycobacterium tuberculosis bacteria in established experimental animal infections are acid-fast negative, clearly cell wall changes are occurring. To address both the spatial properties of mycobacterial cell walls and to begin to study the differences between bacteria grown in animals and cultures, the cell walls of Mycobacterium leprae grown in armadillos was characterized and compared with that of M. tuberculosis grown in culture. Most fundamentally, it was determined that the cell wall of M. leprae contained significantly more mycolic acids attached to PG than that of in vitro grown M. tuberculosis (mycolate:PG ratios of 21:10 versus 16:10, respectively). In keeping with this difference, more arabinogalactan (AG) molecules, linking the mycolic acids to PG, were found. Differences in the structures of the AG were also found; the AG of M. leprae is smaller than that of M. tuberculosis, although the same basic structural motifs are retained.
Keywords: Carbohydrate Structure, Cell Wall, Fatty Acid, Mass Spectrometry (MS), Membrane Bilayer, Leprosy, Mycolic Acids, Tuberculosis
Introduction
The hallmark of mycobacteria is their cell wall consisting of a peptidoglycan layer attached to a mycolic acid containing outer membrane via the polysaccharide arabinogalactan. Profound and fundamental questions remain about this architecture, including how division occurs and how molecules, both nutrients and drugs, enter the cell. Although its impermeability is stressed (1), anomalies exist such as the susceptibility of in vitro grown Mycobacterium tuberculosis to lysozyme at concentrations between 0.1 and 3 mg/ml.2 Also, the acid fastness of in vivo bacteria varies, in a manner dependent upon their growth state (2). Considered together, these phenomena point to the need to understand the cell wall physical spatial organization and how the cell wall changes during in vivo growth.
Mycobacterium leprae cannot be cultured in vitro and is propagated in nine-banded armadillos (Dasypus novemcinctus) (3). In this animal, as in the mouse foot pad model and human lepromatous leprosy, the bacteria grow logarithmically but with a very slow generation time of 12–14 days (4). Although there is a robust humoral response to M. leprae in armadillo, it would appear that the immune system does little to slow the growth of the bacteria and that the slow growth rate is an intrinsic property of the highly attenuated M. leprae, marked by less than 50% genomic coding capacity (5). This is in contrast to M. tuberculosis that presents an initial doubling time in animal models of about 2.4 days (5) until the adaptive immune response is fully activated and stops M. tuberculosis bacilli replication (6), although the bacteria are still viable. The nongrowing M. tuberculosis is resistant to chemotherapy and to clearing by the immune system. Thus, it is important to study the cell wall barrier of mycobacteria under all of these conditions, and here we begin with a study of in vivo M. leprae.
Previous cell wall analyses of in vivo grown leprosy bacilli reported the presence of the characteristic mycobacterial cell wall sugars arabinose and galactose and the amino acids diaminopimelic acid (DAP),3 alanine, and glutamic acid (7, 8). Further analysis of the arabinogalactan and peptidoglycan from M. leprae revealed structural similarities with related mycobacterial species (9). Torrelles et al. (10) reported that the arabinan architecture of M. leprae lipoarabinomannan, an important cell envelope component, is simpler than that of M. tuberculosis with a high degree of exposed nonmannosylated capped arabinan termini (10). More recently, it was found that in the case of M. leprae peptidoglycan, the muramic acid residues are exclusively N-acetylated, unlike M. tuberculosis where both N-acetylated and N-glycosylated versions of muramic acid exist (11). However, a comprehensive and quantitative comparison of the cell wall core of in vivo grown M. leprae versus in vitro grown M. tuberculosis is lacking.
The cell wall core of in vitro grown M. tuberculosis has been studied in great detail. Analyses of the O-alkylated alditols of AG-derived oligosaccharides revealed the following: (i) the attachment of arabinan side chains to a linear galactan chain, with a specific hexaarabinoside at its nonreducing end (12); (ii) the linker disaccharide (Rha-GlcNAc)-P at the reducing end (13) of the entire AG molecule; and (iii) the location of the outer membrane-located mycolyl esters at the nonreducing end of the arabinan (14). The use of a partially purified endo-arabinase from Mycobacterium smegmatis that specifically cleaves the α-1,5 linkages allowed released oligoarabinans to be characterized by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF) (15, 16) and the realization that two hexaarabinosides are connected to yield an Ara17 unit (see Fig. 2). Additionally, the nonessentiality of the arabinogalactan biosynthetic genes in the closely related Corynebacterium glutamicum allowed definition of the nature of the attachment of the arabinan to the galactan (17). Finally, a recent report from our group has defined the entire primary structure of the cell wall mycolyl arabinogalactan identifying the location of the attached succinyl residues and re-defining the structure of the interior arabinan chain of M. tuberculosis and M. smegmatis (16).
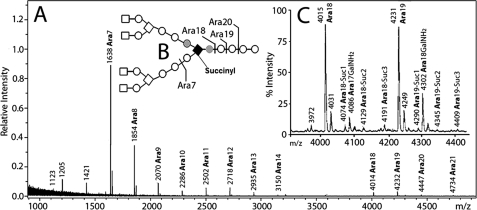
FIGURE 2.
A, MALDI-TOF mass spectrum of predominantly nongalactosaminylated arabinans (after acetylation) released by endogenous arabinase from the cell wall core (mAGP) of M. leprae. Both M + Na+ and, at 16 atomic mass units higher, M + K+ ions are seen. The major species released are Ara7, Ara18, and Ara19 that are formed as illustrated in B. C, expansion of the spectrum from m/z 3900 to m/z 4500 showing the presence of 1–3 succinyl groups on Ara18 and Ara19. Also seen are the galactosaminylated arabinans for Ara18 and Ara19 as the galactosaminylated and nongalactosaminylated arabinans were not perfectly separated from each other.
Recent cryotomography EM studies (18, 19) unequivocally showed that covalently attached mycolic acids are present as part of a true outer membrane. This observation gives rise to the question of what percentage of the inner leaflet of this outer membrane is composed of mycolic acid esters covalently attached to peptidoglycan via the AG as compared with noncovalently attached lipids, such as the trehalose mycolates that are also located in the outer membrane.
In this study, endo-arabinase digestion coupled with carefully designed methylation and compositional analyses were applied to the cell wall core of M. leprae and to the cell wall of in vitro grown M. tuberculosis. The results lead to a model focusing on the spatial arrangement of the mycolic acids of the two cell wall cores and provide new perspectives on the distinct outer membrane of Mycobacterium spp.
EXPERIMENTAL PROCEDURES
Bacterial Cultures
M. leprae was obtained from armadillo livers and spleens as described previously (20). M. tuberculosis (H37Rv) and M. smegmatis mc2 155 were grown and harvested as described previously (16).
Preparation of Mycolyl Arabinogalactan Peptidoglycan Cell Wall Core (mAGP)
Mycobacterial cells were disrupted mechanically in 10 mm PBS buffer (pH 8) using a French press (SIM-AMINCO systems) at 1500 p.s.i. followed by centrifuging at 21,000 × g for 30 min. The pellet obtained was suspended in 30 ml of Milli-Q water containing 2% SDS and stirred gently at room temperature overnight. This was followed by centrifugation (25,000 × g) to remove the supernatant. The pellet was suspended in 30 ml of Milli-Q water containing 2% SDS and stirred gently at 100 °C for 1 h, followed by centrifuging at 25,000 × g for 30 min, and the supernatant was removed. The pellet was washed with water (three times) followed by centrifugations as above. After the water washes, the pellet was suspended in 30 ml of 80% acetone/Milli-Q water and centrifuged at 25,000 × g for 20 min to remove SDS. This was again followed by two more water washes. Finally, the material was treated with organic solvents (CHCl3:CH3OH, 2:1; overnight extraction) to remove any remaining noncovalently attached mycolic acids. The resulting insoluble final pellet was considered as mAGP and is a cell wall with only the covalently attached inner leaflet of the outer membrane.
Analysis of Cell Wall Components
The resulting mAGP (1 mg in the case of both that from M. leprae and M. tuberculosis) was treated with 2 m TFA (500 μl) for 2 h at 120 °C after adding the internal standards 3-O-methyl glucose and α-aminoadipic acid (for sugar and amino acid quantitation, respectively). After cooling down to room temperature, it was extracted twice with 500 μl of CHCl3. The organic phases were pooled, dried under N2, and processed further for mycolate analysis. The aqueous phase was divided to two equal parts, for sugar and DAP analyses.
Derivatization of Cell Wall Sugars to Alditol Acetates
The aqueous extract obtained after TFA hydrolysis was dried and reduced with NaBD4 (10 mg/ml in 1 m NH4OH:C2H5OH) for 4 h at room temperature. Excess NaBD4 was removed by adding 20 μl of CH3COOH (glacial acetic acid), and the tubes were dried under N2, followed by two more CH3OH washes and subsequent drying steps. The samples were per-acetylated using 100 μl of acetic anhydride and heated for 1 h at 100 °C. After acetylation, samples were cooled to room temperature and extracted with CHCl3:H2O (1:1), and the CHCl3 layer was dried and analyzed by GC/MS.
GC/MS Analysis of Cell Wall Sugars
GC/MS analyses were carried out using a CP3800 gas chromatograph (Varian Inc., Palo Alto, CA) equipped with an MS320 mass spectrometer. The samples were run on a DB 5 column (30 m × 0.20 mm inner diameter). The oven temperature was held at 50 °C for 1 min and programmed at 30 °C/min to 150 °C and then programmed at 5 °C/min to 275 °C.
Per-O-methylation of Cell Wall Glycosyl Residues
The mycobacterial cell wall core (mAGP) was subjected to methylation as per Ciucanu and Karek (21). Typically, five pellets of NaOH and 3 ml of anhydrous DMSO were ground using a mortar and a pestle, and the slurry was added to a sample of mAGP, followed by an addition of 500 μl of CH3I. This mixture was vigorously shaken for 1 h at room temperature. The reaction was quenched with a dropwise addition of 1 ml of water and was extracted three times with 2 ml of CHCl3. The CHCl3 extracts were pooled, again extracted with distilled water, and finally dried under N2. An aliquot of the methylated sample was subjected to hydrolysis, reduction, and acetylation by the alditol acetate procedure as described above for the identification and quantitation of linked sugars.
Quantitation of Per-O-methylated Sugars
GC/MS analysis of the partially methylated and partially acetylated alditols was used to identify them. The GC/MS conditions were as described earlier for neutral sugar analysis. Quantitation of the different components was achieved using the effective carbon response theory (22). For this, the partially methylated cell wall sugars were run on a GC (Shimazu, Columbia, MD) equipped with a flame ionization detector. The molar response factors of partially methylated sugars from the effective carbon response theory (22) were then used to calculate the ratios of the variously linked glycosyl residues of the cell wall core.
Analysis of Diaminopimelic Acid
The remaining 50% of the aqueous phase from TFA hydrolysis of mAGP was dried and treated with 6 n HCl at 120 °C for 16 h to release all amino acids, including DAP. The products were derivatized with n-propyl chloroformate (“EZ:faast” kit; Phenomenex, Torrance CA) and analyzed by GC/MS. In this procedure, the amino acids are purified after 6 m HCl hydrolysis by cation exchange before derivatization with n-propyl chloroformate in the presence of n-propyl alcohol. A standard of DAP was prepared in the same fashion; it and the unknown samples contained α-aminoadipic acid as an internal standard.
Analysis of Mycolic Acids
The organic layer of the 2 m TFA-hydrolyzed mAGP contains mycolic acids covalently attached to arabinosyl residues (presumably the α-branched and β-hydroxylated components of the mycolate structure account for the acid stability of the ester (23)). Although the arabinosyl mycolate can be analyzed directly by LC/MS, better sensitivity results after de-esterification. Thus, arabinosylated mycolic acids released by 2 m TFA from the mAGP were subjected to alkaline hydrolysis (24), acidified with HCl, and extracted twice with diethyl ether. The pooled ether fractions were dried under a stream of nitrogen and dissolved in 200 μl of CHCl3:CH3OH (2:1) for LC/MS analysis. The compound, 1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine-N-nonadecanoyl, was used as an internal standard for quantitation of mycolates (Avanti Polar Lipids, Alabaster, AL). For a standard, large amounts of pure mycolic acids were prepared from M. tuberculosis cells so that they could be weighed.
The ESI/APCI-MS (negative mode) was performed on an Agilent 6220 TOF mass spectrometer equipped with a MultiMode source. The HPLC was equipped with an Agilent 1200 binary pump. Two tandemly attached Waters XBridge (C18 2.1 × 150 mm, 5 μm particle size) columns were used for the separation of the mycolic acids. The column temperature was 45 °C. The separation was done using a gradient of 100% solvent A (99% CH3OH + 1% 500 mm ammonium acetate) to 100% solvent B (79% n-propyl alcohol + 20% hexane + 1% 500 mm ammonium acetate) (25). The column eluent was introduced into the multimode source operated in the negative ion mode. The drying gas temperature was 300 °C, and the vaporizer temperature was set at 200 °C. Typically, 5 μl of sample was injected for analysis, with a flow rate of 0.32 ml/min, and the total run time was 45 min. The fragmentor voltage was set to 120 V. The mass spectrum was acquired from m/z 250 to 3200 Da with a frequency of 1 scan/s. MS data were analyzed using Mass Hunter software (Agilent) and a custom data base of M. tuberculosis lipids (40).
To analyze the size of the α chain, the mycolates were subjected to LC/MS/MS (25). An Agilent 6520 qTOF was used for MS/MS analyses of the mycolic acids. The chromatographic setup was similar to the one described above for mycolic acid analysis. Mass spectra were acquired in Auto MS/MS mode with N2 as a collision gas, and collision energy of 75 eV was used for fragmentation.
Treatment of the Cell Wall Core with Endogenous Arabinase and Purification of Arabinans
mAGP was incubated with a partially purified (15) M. smegmatis arabinase at 37 °C for 12 h, followed by extraction with CHCl3:CH3OH:H2O (10:10:3) in which, surprisingly, the arabinan is soluble (16). The 10:10:3 extract was dried and extracted with CHCl3:CH3OH:H2O (8:4:3); and the aqueous fraction was collected. This fraction was passed through a P-2 column (BioGel, 1 × 25 cm) and eluted with 50 mm sodium acetate buffer (pH 5). The collected fractions were hydrolyzed and derivatized as alditol acetates and further analyzed by GC/MS to identify which fraction contained galactosamine. As in our earlier work, it was observed that the earlier fractions of the P-2 column contained the majority of the galactosamine residues (16). The dried samples from the P-2 column (showing higher arabinan content) were per-O-acetylated with 100 μl of acetic anhydride in the presence of 50 μl of pyridine (room temperature, overnight) and were extracted in CHCl3:H2O (1:1). The CHCl3 layer upon concentration was analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) in the positive ion reflector mode on a Bruker Ultraflex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA). The sample (1 μl) was mixed with 1 μl of matrix (dihydroxybenzoic acid) at 10 mg/ml in 50% acetonitrile in 0.1% TFA) and allowed to air-dry on the MALDI target plate.
Analysis of Succinates
Succinates from the cell wall core (mAGP) were obtained through octanolysis using 3 n HCl in 1-octanol (99%, 100 μl) at 120 °C for 30 min (16). The resultant octyl succinate derivatives were extracted with 2 ml each of hexane and a supersaturated solution of sodium bicarbonate. The hexane layer was further analyzed by GC/MS with a DB-5 column. The initial temperature of the column was at 60 °C, held for 1 min. The temperature was increased to 330 °C at a rate of 30 °C/min. The retention times were compared with that of standard succinic acid and were further quantified by comparing the area of internal standard glutaric acid (as octyl ester), which was also subjected to octanolysis as per the procedure (16).
RESULTS
Comparative Compositional Analysis of the mAGP of in Vivo Grown M. leprae and in Vitro Grown M. tuberculosis
Crude cell walls (27,000 × g pellets after centrifugation of the bacteria broken by a French press) were prepared from M. leprae isolated from armadillos and from M. tuberculosis grown in culture. These included the entire outer membrane but unfortunately were contaminated with fragments of plasma membrane. Hence the crude cell walls were extracted with chloroform to yield the cell wall core composed only of the covalently attached mycolic acids, arabinogalactan and peptidoglycan (mAGP). The only aspects of the outer membrane remaining were the mycolates covalently attached to the AG·PG complex because all of the free mycolate-containing neutral lipids had been removed.
To obtain compositional data on mAGP from the two mycobacterial species, the glycosyl residues were analyzed as alditol acetates after release by 2 m TFA. DAP, released by 6 m HCl hydrolysis, was analyzed after derivatization with n-propyl chloroformate, and the mycolic acids were measured as free mycolate anions after base hydrolysis (Table 1). All three analyses were done from the same sample rather than using separate aliquots for each analysis to increase the accuracy of the ratios of the different components.
TABLE 1.
The composition of the cell wall core (mAGP) from M. tuberculosis and M. leprae
| Origin of mAGP | mol % (standard deviation in parenthesesa) normalized to 1 Rha residue (boldface type) |
Macromolecular component ratiosb (propagated standard deviation in parentheses) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rha | Arac | Gal | GalNAcd | Total glycosyl residues in AG | GlcNAcd | MurNAcd | DAP | Myc | PGe | AG | Myc | % substitution of AG with Mycf | |
| M. tuberculosis | 0.95 (0.05) | 53.6 (0.92) | 21.3 (0.69) | 0.81 (0.05) | 2.2 (0.46) | 1.91 (0.1) | 7.3 (0.37) | 12.0 (1.0) | 10 | 1.3 (0.03) | 16.4 (2.2) | 79 (9.6) | |
| 1 | 54 | 23 | 0.9 | 79 | 2.3 | 2.0 | 7.7 | 12.7 | |||||
| M. leprae | 1.39 (0.08) | 62 (1.3) | 18 (2.4) | 0.7 (0.2) | 2.0 (0.6) | 1.7 (0.6) | 4.6 (0.4) | 9.4 (1) | 10 | 3.0 (0.23) | 20.8 (4.4) | 42 (2.7) | |
| 1 | 45 | 13 | 0.5 | 59.5 | 1.4 | 1.2 | 3.3 | 6.7 | |||||
a Four separate samples of mAGP from M. tuberculosis and M. leprae were analyzed. The ratios are the average from these four analyses, and the standard deviation is calculated from them. The amount of mAGP from both sources used for analyses was approximately 1 mg.
b These ratios are calculated from the DAP, Rha, and Myc values; see text for justification.
c The total number of Ara residues includes those attached to the mycolic acids and not liberated by acid hydrolysis.
d GlcNAc, GalNH2, and MurNAc cannot be accurately quantitated due to the fact that acid conditions either fail to fully hydrolyze them or stronger conditions degrade them after liberation. Thus, these values are not used for further calculations.
e The PG is chosen to include 10 disaccharide/peptide repeating units for clarity of ratios. There is always one DAP per PG repeating unit.
f Data were calculated based on the ratio of Myc to AG recognizing that 16 mycolic acids per AG is fully mycolylated AG.
The ratios of DAP, Rha, and mycolate can be used to calculate the ratios of PG:AG:mycolate as shown in Table 1. DAP is used as a marker for PG because it is quantitatively released by the 6 m HCl and not significantly degraded by the acid. It is not possible to accurately quantitate the PG amino sugars due to the fact they are not completely released by 2 m TFA, and the stronger acid conditions required for their hydrolysis also degrades them. Thus, their ratio to Rha is underestimated in Table 1. The component used for AG quantitation is the rhamnosyl residue because one rhamnosyl unit is present per AG chain, and it is released and not degraded by the 2 m TFA. The mycolates are accurately measured due to their efficient release, without degradation, by base.
The data (Table 1) show that compared with M. tuberculosis, M. leprae mAGP has a 1.3 (20.8/16.4)-fold increase in the amount of mycolic acids and a 2.3 (3.0/1.3)-fold increase in arabinogalactan for a given amount of peptidoglycan. This requires that more mycolic acids be covalently attached to PG and more AG be present to make that attachment. These changes are modeled below.
Comparison of the Mycolates of M. leprae and M. tuberculosis
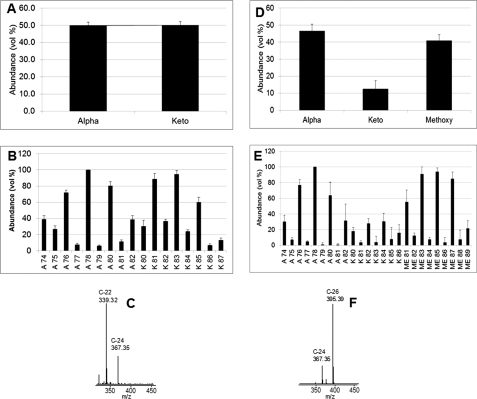
The mycolic acid amounts shown in Table 1 represent all the different classes obtained by integrating the entire complex mycolate peak in the LC/MS total ion chromatogram and comparing that area with that of an internal standard. Further information on the mycolate species present is also available from the LC/MS analysis. Thus, because the LC/MS was run with high mass resolution, the number of oxygen atoms present per mycolate molecule is readily determined from the exact molecular weights of each mycolate. This allowed the mycolates to be readily assigned to their appropriate class of α, keto, and methoxy. In keeping with the data of others (26), the M. leprae mycolates are present as α- and keto-mycolate classes, but the methoxy-mycolates are lacking (Fig. 1A). The α- and keto-mycolates were equally distributed (Fig. 1A). The α class mycolates with total carbon numbers of 78 and 80 and the keto class of C-83 were most abundant in M. leprae (Fig. 1B). Tandem mass spectrometry (LC/MS-MS) analysis of the α-mycolate from M. leprae (α-78) (Fig. 1C) showed that the predominant α chain was C-22; C-24 α chains are also present as originally shown (27). For purposes of comparison, the same data are shown for the mycolates of in vitro grown M. tuberculosis (Fig. 1, D–F); these data are consistent with analyses of others (28, 29). The most notable differences between the mycolates of the two species are the lack of methoxy mycolates in M. leprae and the presence of C-22 (with some C-24) α chains in M. leprae as compared with C-26 (with some C-24) α chains in in vitro grown M. tuberculosis.
FIGURE 1.
A, relative abundance of the two major classes of mycolic acids, α and keto, present in M. leprae bacteria. B, chain length distribution of these M. leprae mycolic acids. C, LC/MS-MS analysis showing that the C-22 α chain is predominant in the mycolic acids of M. leprae (spectrum shown is from the α-78 where m/z 1136 was targeted). D, relative abundance of the three major classes of mycolic acids, α, methoxy, and keto, present in M. tuberculosis bacteria. E, chain length distribution of these M. tuberculosis mycolic acids. F, LC/MS-MS analysis showing that the C-26 α chain is predominant in the mycolic acids of M. tuberculosis (spectrum shown is from the α-78 where m/z 1136 was targeted). Four separate samples were analyzed, and the values presented here are the average of those; the standard deviation is shown on the graphs.
Analysis of AG
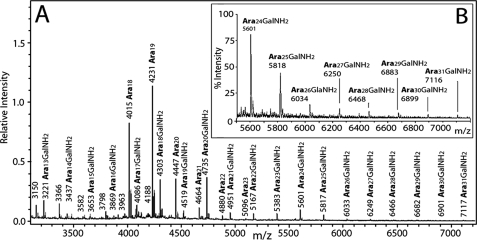
Glycosyl linkage analysis of the AG of M. leprae and M. tuberculosis as determined by per-O-methylation (Table 2) as well as the neutral sugar analysis (Table 1) reflects the lower amounts of galactofuranose and to some extent the arabinofuranose in the M. leprae AG compared with M. tuberculosis AG (M. leprae with 56 and 83% the amount of M. tuberculosis for galactofuranose and arabinofuranose, respectively). Otherwise, the polymers are quite similar by linkage composition, which suggests a similar branching structure. To confirm this, the arabinan chains present on the M. leprae cell wall core were treated with a partially purified endo-arabinase enzyme, from M. smegmatis (15). The released oligo-arabinans were purified on a P-2 sizing column (16) into fractions poor and rich in galactosamine, which were analyzed by MALDI-TOF MS (Figs. 2 and 3 respectively). Although the endogenous arabinase prefers the α-1,5 cleavage of the residue in front of a branching point (16), it also cuts at various positions on the α-5-linked arabinan, resulting in multiple cleavages forming different sizes of arabinans as evident by MALDI-TOF spectra (Figs. 2 and 3). MALDI-TOF MS of the nongalactosaminylated per-O-acetylated arabinans identified a pattern of arabinans ranging from Ara5 to Ara21, with Ara7 being the most predominant (Fig. 2A) and Ara18 being the dominant high molecular weight oligo-arabinan. The lack of Ara4 suggests that in the M. leprae arabinan the nonreducing terminus is fully branched (i.e. the pentaarabinoside shown in Figs. 2 and 3) like that of M. tuberculosis (16). Also very small amounts of arabinosyl oligomers from Ara14–17 are found in M. leprae, as is the case for M. tuberculosis (16), because of the enzyme's preference to only rarely cleave in long stretches of α-1,5 Araf residues (16). For the same reason, oligomers of arabinose greater than Ara21 are also only rarely formed; however, these arabinosyl oligomers, if detected, give information on how long are the internal arabinan chains linking the branched region to the galactan. Unlike the case with M. tuberculosis, we could not find nongalactosaminylated arabinans in M. leprae with a degree of polymerization greater than Ara21; in M. tuberculosis up to Ara30 was found. However, such fragments are weak in M. tuberculosis, and one cannot be sure if this reflects an actual change in structure, especially because the more easily ionized Ara31GalNH2 was found in M. leprae (Fig. 3 and see below).
TABLE 2.
Glycosyl linkage composition of M. tuberculosis and M. leprae cell walls
Four separate samples of mAGP from M. tuberculosis and M. leprae were analyzed. The ratios are the average from these four analyses, and the standard deviation is calculated from them.
| Glycosyl residue | M. lepraea (mol/59 mol) | Theoretical 2 Ara22 units and 1 Gal14 unit | M. tuberculosisb (mol/79 mol) | Theoretical 2 Ara26 units and 1 Gal22 unit |
|---|---|---|---|---|
| T-Araf | 5 (0.09) | 8.0 | 7 (0.25) | 8 |
| 2-Araf | 5 (0.27) | 8.0 | 7 (0.38) | 8 |
| 5-Araf | 25 (0.37) | 22 | 29 (0.40) | 28 |
| 3,5-Araf | 9 (0.22) | 6.0 | 8 (0.20) | 6 |
| 4-Rhap | 1 (0.07) | 1.0 | 1 (0.07) | 1 |
| T-Galf | 1 (0.10) | 1.0 | 2 (0.27) | 1 |
| 5-Galf | 5 (0.17) | 5.5 | 13 (0.06) | 9.5 |
| 6-Galf | 3 (0.8) | 5.5 | 10 (0.04) | 9.5 |
| 5,6-Galf | 4 (0.76) | 2.0 | 3 (0.01) | 2 |
FIGURE 3.
A, MALDI-TOF mass spectrum of the fraction enriched in galactosaminylated arabinans (after acetylation) released by endogenous arabinase from the cell wall core (mAGP) of M. leprae. Both M + Na+ and, at 16 atomic mass units higher, M + K+ ions are seen. The galactosaminylated species released are a series running from Ara17GalNH2 through Ara31GalNH2. B, expansion of the spectrum from m/z 5500 to m/z 7200 showing more clearly Ara24GalNH2 through Ara31GalNH2. The fragments are formed as in the illustration shown in Fig. 2B except succinyl is replaced by GalNH2.
Careful examination of the expanded spectra of Fig. 2B revealed the presence of succinyl groups on the arabinan chains of M. leprae. The succinyl groups are 58 mass units heavier than the respective nonsuccinylated arabinans (M + 58) and, as in the cell wall of M. tuberculosis (16), multiple succinylation was observed. However, all ions resulting from succinylation were present at a much less intensity in the case of M. leprae (Fig. 2B). Succinylation was not observed in the galactosaminylated arabinans, a feature that was first observed in M. tuberculosis (16).
A direct analysis for succinyl groups in mAGP prepared from M. leprae was performed by GC/MS analysis after octanolysis of the cell wall to form succinyl dioctyl esters (10, 16). GC/MS analysis led to the identification of a peak that co-eluted with authentic dioctyl succinate and gave an identical mass spectrum ([M + H]+ at m/z 343 and fragment ions at 213 and 231). Quantitation of succinyl residues indicated the molar ratio of succinyl residues to arabinosyl residues to be 1:175. Thus, the succinylation in M. leprae AG was 4.4-fold less than that of M. tuberculosis where the ratio of succinyl to arabinosyl residues was 1:41 (16) in agreement with the mass spectral data presented in Fig. 2.
The MALDI-TOF MS analyses of the endogenous arabinase released arabinans revealed that the smaller arabino-oligosaccharide fragments such as Ara7-8 did not show any succinylation (Fig. 2). This observation implies that the succinates are probably located on the internal α-3,5-arabinan chain, as in M. tuberculosis, although definitive data such as NMR done for M. tuberculosis (16) are lacking.
The MALDI-TOF spectrum of the arabinans containing a GalNH2 residue is presented in Fig. 3. In this case, oligoarabinosides up to Ara31(GalNH2)1 are seen, exactly as was found for M. tuberculosis arabinan (16). Their presence, in contrast to the lack of nongalactosaminylated arabinans containing greater than 21 residues (Fig. 2), may be due to MS detection issues as the GalNH2 residue allows for stronger ionization or it may reflect a genuine difference in arabinan elongation. No succinyl groups are present on the GalNH2-substituted arabinans in M. leprae as in M. tuberculosis (16). The reason for this is likely that both the first succinyl and the GalNH2 are attached to the 2-position of the interior branched 3,5-Araf.
Data from C. glutamicum suggest that up to three arabinans can be attached per galactan chain (17). We had assumed the same number for mycobacteria (16), but the presence of three arabinan chains does not fit the data of Table 1 or 2 for either M. tuberculosis or M. leprae. For M. tuberculosis, the glycosyl composition data of Table 1 suggest about 53 arabinosyl residues per AG molecule. The MS analysis contained M + Na+ ions for galactosaminylated arabinosides only up to Ara31 suggesting the lack of arabinans longer than Ara31. Thus, two arabinans with an average of 26 arabinosyl residues are most likely present. For M. leprae, the glycosyl linkage data (Table 2) require about 45 arabinosyl residues, which is consistent with two arabinans averaging 22 arabinosyl residues each. In both cases, the data suggest heterogeneity in the length of the α-1,5-Araf linear interior region of the arabinan. One possible caveat is the presence of more branched 5,6-Galf in both cases than expected, as indicated in Table 2. It is possible that additional α-1,5-Araf linear arabinan chains with no Ara17 branched reducing end, perhaps only one or two Araf units long, are also present.
Attachment of Mycolic Acids to the Arabinan Chains
Previous data from M. tuberculosis, using selective alkylation of arabinan (14), revealed that approximately two-thirds of the sites for mycolate attachment (C-5 of the T-Araf and C-5 of the 2-Araf as depicted in Figs. 2 and 3) were occupied with mycolate residues. This is consistent with the data presented in Table 1 where a fully substituted AG molecule with two arabinans (in the case of M. tuberculosis) has 16 potential sites for mycolylation, and if two-thirds are occupied, the ratio of AG to mycolate would be 1:10.7, which is reasonably consistent with the found ratio of 1:12.6 or 79% occupied (Table 1). For M. leprae, Table 1 reveals a ratio of AG to mycolate of one AG molecule for every 6.9 mycolic acids or 42% of the possible mycolylation sites being occupied. However, as shown in Table 1, in the wall of M. leprae there are considerably more AG molecules per 10 PG repeating units (3.0 versus 1.3 as shown in Table 1), and thus the ratio of mycolate to PG is higher in M. leprae (20.8 mycolates/10 PG repeat units) than in M. tuberculosis (16.4 mycolates/10 PG repeat units) even though the arabinans themselves are not as fully substituted.
The AG, in turn, is attached to the peptidoglycan. In the case of M. tuberculosis AG, our data calculate to one AG molecule per eight disaccharide repeat units of PG (Table 1), which agrees reasonably well with earlier work (13, 30). In the case of the more abundant AG molecules in M. leprae, our data calculate to one AG molecule for every 3.3 repeat units of PG (Table 1).
DISCUSSION
Primary Structural Considerations
The PG of M. leprae differs from the PG of M. tuberculosis only by the lack of N-glycolylated muramic acid in its peptidoglycan and the substitution of glycine for l-alanine (11). Our results show that the AG structures of M. tuberculosis and M. leprae are similar with each AG molecule containing ∼79 and 59 glycosyl residues, respectively. The galactan is consistently shorter in M. leprae than in M. tuberculosis (13 versus 23 residues). The basic architecture of the arabinan is the same with a doubly branched structure as shown by the data in Figs. 2 and 3. The composition and methylation data fit best with approximately two arabinan chains per AG molecule in both M. tuberculosis and M. leprae AG (Tables 1 and 2), but the chains are shorter in M. leprae. One of the largest differences between the two species is that more AG molecules are present per PG molecule in the M. leprae cell wall (3.0 AG molecules/10 repeating units of PG) than in M. tuberculosis (1.3 AG molecules/10 repeating units of PG) with a p value of less than 0.001 in the Tukey-Kramer t test comparing the two ratios. Thus, the cell wall of M. leprae is poised for a higher degree of mycolylation and indeed does have a higher degree of mycolylation per 10 repeating units of PG (20.8 for M. leprae versus 16.4 for M. tuberculosis) with a p value of less than 0.05 in the Tukey-Kramer t test comparing the two ratios. The fact that the AG molecules are somewhat shorter in M. leprae may result in a smaller periplasmic space.
Modeling the Mycobacterial Cell Wall
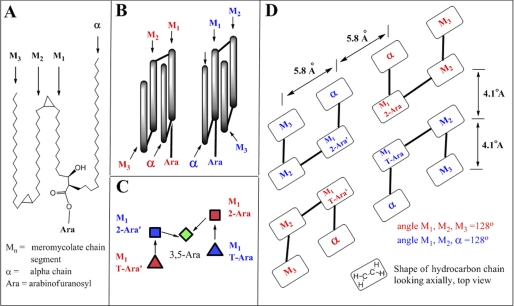
Recent innovations of the conformation of mycolic acids and the packing of hydrocarbon chains allow modeling of the conformation of the mycolates as they are attached to AG. Thus, two studies using cryo-electron microscopy have shown that the inner half of the outer membrane of mycobacteria is consistent with the mycolic acids occurring in a folded configuration (18, 19). Earlier and independently, Villeneuve et al. (31, 32) also proposed a folded mycolic acid structure based on Langmuir monolayer packing. They proposed that essentially four columns are present that are not in a single plane, but rather they extend outward from the apexes of a parallelogram as shown in Fig. 4, B and D (32). A lipid bilayer of C-20 fatty acids revealed unit cell dimensions of 5.8 × 4.1 Å when analyzed by scanning tunneling microscopy (34), suggesting similar dimensions for each mycolate column (Fig. 4D).
FIGURE 4.
A, two-dimensional representation of an α-mycolic acid folded to form four hydrocarbon domains. Three of these domains, M1, M2, and M3, are in the meromycolate portion of the molecule, and one, labeled “α,” is the α chain region of the molecule. The esterification to the 5-position of either the β-T-Araf or the α-2-Araf at the carboxyl group between the α and M1 domain is also shown. B, three-dimensional representation of two α-mycolic acids folded to form four hydrocarbon domains each. The conformation designated with red and blue labeling are, at the level of the four chains, mirror images, and both conformations are needed to fit the mycolates onto the arabinan (as shown in C and D). C, approximate conformation the nonreducing terminal pentaarabinosyl unit must adopt to accommodate complete mycolic acid substitution. The pentaarabinosyl unit is viewed from the top; the two T-Ara and the two 2-Ara are residues are roughly in the same vertical plane, and the 3,5-Ara unit is below them. The colored symbols representing the arabinosyl residues correlate to the two conformations shown in B. D, schematic top view of a tetramycolylated nonreducing terminal pentaarabinosyl unit.
The structure of the arabinan and the confirmation of the mycolic acid present considerable steric restraints. Thus, given the ∼5-Å size of a mycolate column and the molecular dimensions of the terminal pentaarabinoside, the attachment of four mycolates can only take place in a unique conformation of the mycolates and the terminal pentaarabinoside as illustrated in Fig. 4, B–D. This is because the primary hydroxyl groups on the T-Araf and the 2-Araf groups can only be spaced 5–8 Å apart as shown clearly with molecular models. Because each mycolate molecule carries four columns with it, the pentaarabinoside cannot be in a linear arrangement as the mycolate columns will interfere with each other, but it must be as shown in Fig. 4C leading to the mycolates adopting a conformation similar to that shown in Fig. 4B. The overall arrangement is shown in Fig. 4D.
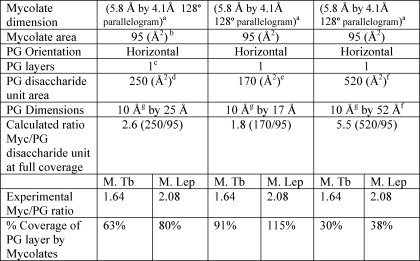
In addition, the data of Tables 1 and 2 can be used to estimate what percentages of the fatty acyl groups in the inner leaflet of the outer membrane are mycolic acids covalently linked to AG and thus what percentage of the PG surface area is covered with mycolic acids covalently linked to it (via AG). Thus (see Table 3), it is reasonable to assume that the four columns of each mycolate when viewed from above cover about 95 Å2 given their four-column structure (32) and the dimensions of a single fatty acid chain of 19.4 Å2 (34). To calculate the area of the PG, its orientation to the cell must be known as well as the dimensions of the glycan and connecting peptides. A consensus is emerging that PG glycan strains are mostly circumferential (35), that the peptide cross-links are not fully extended (36), and that in Gram-negative bacteria (and by implication in mycobacterial PG) there is only a single layer of PG present, parallel to the plasma membrane (35, 36). Quantitative analysis of PG and the surface area in E. coli suggests that the unit cell of PG (disaccharide repeat with its half of a connecting peptide cross-link) has an area of 250 Å2 in one study (36) and 170 Å2 in an earlier study (37). In contrast, direct visualization of the PG glycan chains by electron cryotomography suggested separation of the glycan chains by 50–80 Å (35). (Separation by greater than 52 Å is beyond the theoretical limit if the glycans are perfectly parallel, which they may not be (38).) Using these different areas for the disaccharide unit surfaces areas, the coverage of the PG layer varies from 29 to 88% for M. tuberculosis and from 39 to 119% for M. leprae (Table 3). Clearly, more precise data on the area of the PG monomers are required, but in all estimates, even with the larger PG monomeric unit, a substantial portion of the inner leaflet is covalently attached to mycolic acids in contrast to typical Gram-negative bacteria where only a few percent of the fatty acyl groups of the inner leaflet of the outer membrane are covalently attached to PG via the cell wall lipoprotein.
TABLE 3.
Calculated percent coverage of the PG layer by the mycolate layer for M. leprae and M. tuberculosis cell wall at different PG conformations
a Data are based on measurement for C-20 fatty acids by tunneling microscopy (34).
b Data are calculated from mycolate dimensions and are reasonably consistent with studies on Langmuir monolayers (31), which yields 78 Å2 for four chains.
c Data are based on electron cryotomography in E. coli (35).
d Data indicate the measured area in E. coli (37).
e Data indicate the measured area in a different strain of E. coli (38).
f 52 Å is the distance of a fully extended peptide chain and the associated glycan (39); glycan chains (which may not be perfectly parallel) were found to be 50–80 Å apart by electron cryotomography (35).
g Data are calculated from the known 10 Å disaccharide length.
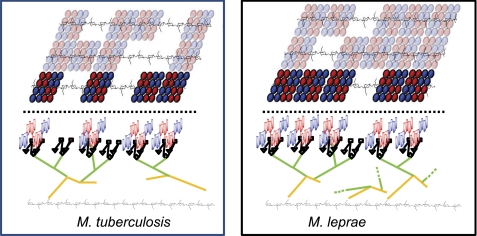
A model of the covalently attached mycolate “patches” assuming a PG unit area of 250 Å2 (∼63% PG coverage for M. tuberculosis and 80% PG coverage for M. leprae) is shown in Fig. 5. The empty regions are presumably filled with noncovalently attached lipids such as trehalose mono- and di-mycolates. The structure of a single arabinan is such that the two “outer arms” can readily accommodate mycolate patches that are next to each other or as far apart as 45 Å. Some attempt was made for proper scaling. Thus, one AG molecule for eight PG repeat units is shown for M. tuberculosis, whereas three AG molecules per 10 AG repeat units is shown for the M. leprae cell wall (see Table 1). Also, approximately 79% of the AG from M. tuberculosis is substituted with mycolic acids as compared with 42% for M. leprae (Table 1). It is readily apparent that the AG from M. leprae is more abundant (i.e. more molecules per PG repeating unit) and also that the AG is capable of accepting additional mycolic acid groups.
FIGURE 5.
Left, model of M. tuberculosis cell wall; right, model of M. leprae cell wall. Top portion of figures (above dashed line) shows an arrangement of the covalently bound mycolates present in the inner leaflet of the outer membrane as viewed looking down at the cell wall. The peptidoglycan beneath the mycolates is shown, but the connecting AG is not. The orientation of PG versus the mycolates is arbitrary; the flexibility of the AG would allow any orientation. The positions of the “blank” areas devoid of covalently linked mycolates are also arbitrary, but the percent coverage of the PG layer is ∼63% for M. tuberculosis and ∼80% for M. leprae (see Table 3). The blank areas are presumably filled with noncovalently attached lipids such as trehalose mono- and di-mycolates. Bottom portion of figures (below dashed line) show a side view of the mycolates attached to PG for the mycolate clusters in bright colors shown in the top view. The PG is shown in black with only the GlcNAc-MurNAc repeat units designated. The AG is shown in gold (galactan) and green (arabinan). The nonreducing pentaarabinoside is shown with diamonds, squares, and triangles as in Fig. 4C. The mycolates are as shown in Fig. 4, B and D, but are not in scale with the rest of the drawing; rather they are shown much smaller for clarity.
The conversion of M. tuberculosis from an actively growing acid-fast form to a nonactively growing non-acid-fast form during the course of infection is well documented (2). Although the mechanism of acid-fastness is subtle and not understood, it is clear that a change in cell wall must occur in the nongrowing M. tuberculosis. The difference between the cell walls of M. leprae grown in vivo versus M. tuberculosis grown in vitro suggests that a possible change might be the presence of additional AG molecules and covalently linked mycolic acids rendering these bacteria less permeable in general. Clearly what is required at this stage is a careful analysis of acid fast M. tuberculosis cell walls isolated from animals during logarithmic growth and comparison to the non-acid-fast bacilli evident during the dormancy phase of tuberculosis; such studies are in progress.
Acknowledgments
We gratefully acknowledge Dr. John Spencer for providing the M. leprae cell wall core via National Institutes of Health NIAID Contract AI 25469 and Dr. Alan Schenkel for help with statistical analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants AI-33706 (to M. R. M.), AI049151 (to D. C. C.), and AI018357 (to P. J. B.) from USPHS NIAID. This work was also supported by Grant 42775 from the Bill and Melinda Gates Foundation (to J. T. B. and D. C. C.).
M. S. Scherman and M. R. McNeil, unpublished observation.
- DAP
- diaminopimelic acid
- PG
- peptidoglycan
- AG
- arabinogalactan
- mAGP
- mycolyl arabinogalactan peptidoglycan complex
- GalNH2
- galactosaminosyl.
REFERENCES
- 1. Crick D. C., Brennan P. J., McNeil M. R. (2004) in Tuberculosis (Rom W. N., Garay S. M. eds) pp. 115–134, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 2. Seiler P., Ulrichs T., Bandermann S., Pradl L., Jörg S., Krenn V., Morawietz L., Kaufmann S. H., Aichele P. (2003) J. Infect. Dis. 188, 1326–1331 [DOI] [PubMed] [Google Scholar]
- 3. Vijayaraghavan R. (2009) Scand. J. Lab. Anim. Sci. 36, 167–176 [Google Scholar]
- 4. Levy L. (1976) Lepr. Rev. 47, 103–106 [DOI] [PubMed] [Google Scholar]
- 5. Ordway D., Henao-Tamayo M., Harton M., Palanisamy G., Troudt J., Shanley C., Basaraba R. J., Orme I. M. (2007) J. Immunol. 179, 522–531 [DOI] [PubMed] [Google Scholar]
- 6. Muñoz-Elías E. J., Timm J., Botha T., Chan W. T., Gomez J. E., McKinney J. D. (2005) Infect. Immun. 73, 546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cummins C. S., Atfield G., Rees R. J., Valentin R. C. (1967) J. Gen. Microbiol. 49, 377 [Google Scholar]
- 8. David H. L., Rastogi N. (1983) Curr. Microbiol. 9, 269–274 [Google Scholar]
- 9. Draper P., Kandler O., Darbre A. (1987) J. Gen. Microbiol. 133, 1187–1194 [DOI] [PubMed] [Google Scholar]
- 10. Torrelles J. B., Khoo K. H., Sieling P. A., Modlin R. L., Zhang N., Marques A. M., Treumann A., Rithner C. D., Brennan P. J., Chatterjee D. (2004) J. Biol. Chem. 279, 41227–41239 [DOI] [PubMed] [Google Scholar]
- 11. Mahapatra S., Crick D. C., McNeil M. R., Brennan P. J. (2008) J. Bacteriol. 190, 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Daffe M., Brennan P. J., McNeil M. (1990) J. Biol. Chem. 265, 6734–6743 [PubMed] [Google Scholar]
- 13. McNeil M., Daffe M., Brennan P. J. (1990) J. Biol. Chem. 265, 18200–18206 [PubMed] [Google Scholar]
- 14. McNeil M., Daffe M., Brennan P. J. (1991) J. Biol. Chem. 266, 13217–13223 [PubMed] [Google Scholar]
- 15. Dong X., Bhamidi S., Scherman M., Xin Y., McNeil M. R. (2006) Appl. Environ. Microbiol. 72, 2601–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bhamidi S., Scherman M. S., Rithner C. D., Prenni J. E., Chatterjee D., Khoo K. H., McNeil M. R. (2008) J. Biol. Chem. 283, 12992–13000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alderwick L. J., Radmacher E., Seidel M., Gande R., Hitchen P. G., Morris H. R., Dell A., Sahm H., Eggeling L., Besra G. S. (2005) J. Biol. Chem. 280, 32362–32371 [DOI] [PubMed] [Google Scholar]
- 18. Hoffmann C., Leis A., Niederweis M., Plitzko J. M., Engelhardt H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zuber B., Chami M., Houssin C., Dubochet J., Griffiths G., Daffé M. (2008) J. Bacteriol. 190, 5672–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunter S. W., Rivoire B., Mehra V., Bloom B. R., Brennan P. J. (1990) J. Biol. Chem. 265, 14065–14068 [PubMed] [Google Scholar]
- 21. Ciucanu I., Kerek F. (1984) Carbohydr. Res. 131, 209–217 [Google Scholar]
- 22. Sweet D. P., Shapiro R. H., Albersheim P. (1975) Carbohydr. Res. 40, 217–225 [Google Scholar]
- 23. Amar-Nacasch C., Vilkas E. (1970) Bull. Soc. Chim. Biol. 52, 145–151 [PubMed] [Google Scholar]
- 24. Shui G., Bendt A. K., Pethe K., Dick T., Wenk M. R. (2007) J. Lipid Res. 48, 1976–1984 [DOI] [PubMed] [Google Scholar]
- 25. Song S. H., Park K. U., Lee J. H., Kim E. C., Kim J. Q., Song J. (2009) J. Microbiol. Methods 77, 165–177 [DOI] [PubMed] [Google Scholar]
- 26. Minnikin D. E., Dobson G., Goodfellow M., Draper P., Magnusson M. (1985) J. Gen. Microbiol. 131, 2013–2021 [DOI] [PubMed] [Google Scholar]
- 27. Kusaka T., Mori T. (1986) J. Gen. Microbiol. 132, 3403–3406 [DOI] [PubMed] [Google Scholar]
- 28. Watanabe M., Aoyagi Y., Mitome H., Fujita T., Naoki H., Ridell M., Minnikin D. E. (2002) Microbiology 148, 1881–1902 [DOI] [PubMed] [Google Scholar]
- 29. Minnikin D. E., Polgar N. (1967) Chem. Commun. 18, 916–918 [Google Scholar]
- 30. Cunto G., Kanetsuna F., Imaeda T. (1969) Biochim. Biophys. Acta 192, 358–360 [DOI] [PubMed] [Google Scholar]
- 31. Villeneuve M., Kawai M., Kanashima H., Watanabe M., Minnikin D. E., Nakahara H. (2005) Biochim. Biophys. Acta 1715, 71–80 [DOI] [PubMed] [Google Scholar]
- 32. Villeneuve M., Kawai M., Watanabe M., Aoyagi Y., Hitotsuyanagi Y., Takeya K., Gouda H., Hirono S., Minnikin D. E., Nakahara H. (2007) Biochim. Biophys. Acta 1768, 1717–1726 [DOI] [PubMed] [Google Scholar]
- 33. Deleted in proof.
- 34. Smith D. P., Bryant A., Quate C. F., Rabe J. P., Gerber C., Swalen J. D. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 969–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gan L., Chen S., Jensen G. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18953–18957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wientjes F. B., Woldringh C. L., Nanninga N. (1991) J. Bacteriol. 173, 7684–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zaritsky A., Woldringh C. L., Mirelman D. (1979) FEBS Lett. 98, 29–32 [DOI] [PubMed] [Google Scholar]
- 38. Vollmer W., Höltje J. V. (2004) J. Bacteriol. 186, 5978–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dmitriev B. A., Toukach F. V., Schaper K. J., Holst O., Rietschel E. T., Ehlers S. (2003) J. Bacteriol. 185, 3458–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sartain M. J., Dick D. L., Rithner C. D., Crick D. C., Belisle J. T. (2011) J. Lipid Res. 52, 861–872 [DOI] [PMC free article] [PubMed] [Google Scholar]