Abstract
Hmgb1 (high mobility group box-1; amphoterin) is highly expressed in brain during early development of vertebrate and nonvertebrate species. However, its role in brain development remains elusive. Here we have cloned the zebrafish Hmgb1 and specifically manipulated Hmgb1 expression using injection of morpholino antisense oligonucleotides or Hmgb1 cRNA. The HMGB1 knockdown morphants produced by injection of three different morpholino oligonucleotides display a characteristic phenotype with smaller size, smaller brain width, and shorter distance between the eyes. Closer examination of the phenotype reveals severe defects in the development of the forebrain that largely lacks catecholaminergic neural networks. The HMGB1 morphant is deficient in survival and proliferation of neural progenitors and displays fewer cell groups expressing the transcription factor Pax6a in the forebrain and aberrant Wnt8 signaling. The mechanism of HMGB1-dependent progenitor survival involves the neuronal transmembrane protein AMIGO (amphoterin-induced gene and orf), the expression of which is regulated by HMGB1 in vivo. Our data demonstrate that HMGB1 is a critical factor for brain development, enabling survival and proliferation of neural progenitors that will form the forebrain structures.
Keywords: Brain, Cell Surface Protein, Cellular Regulation, Development, Neuroscience, Zebra Fish, Cell Survival and Proliferation, Dopaminergic Neurons, Neural Progenitors
Introduction
The high mobility group box-1 protein (HMGB13; also designated as HMG1 and amphoterin) is an abundantly occurring parental form of the HMG proteins (for review, see Ref. 1). HMGB1 is an exceptional member in the family of HMG-box proteins; depending on the cell type and its activation state, HMGB1 displays a nonnuclear localization and is secreted from cells, in contrast to most HMG-box proteins that are strictly bound to the cell nuclei (for recent reviews, see Refs. 2 and 3). During the last few years, the extensive literature dealing with HMGB1 functions has mainly focused on extracellular regulation of cells by HMGB1. HMGB1 can be passively released from injured cells, but it is also actively secreted due to several types of stimuli such as cell contact with extracellular matrix and cytokine stimulation of cells (2). Acetylation of lysine residues of HMGB1 has been shown to act as a signal leading to extracellular export via a non-classical secretory pathway (4). HMGB1 functions are currently mainly associated with binding to the cell surface receptor RAGE (receptor for advanced glycation end products), but Toll-like receptors have been increasingly suggested as membrane receptors of HMGB1 (for review, see Ref. 2).
Compared with the extensive recent literature dealing with the pathophysiological roles of HMGB1 in inflammation, much less attention has been paid to its physiological roles. The Hmgb1 knock-out mouse survives until early postnatal age, and problems in glucose homeostasis have been suggested to cause multiorgan failure in these mice (5).
HMGB1 was isolated from developing rat brain using neurite outgrowth in embryonic forebrain neurons as a readout in protein fractionation (6). These studies provided the initial evidence of HMGB1 as an extracellularly acting protein (6–8), which has become the major line of HMGB1 research during the last few years. HMGB1 was found to bind strongly to heparin/heparan sulfate and to be highly expressed in embryonic rat brain (6). RAGE acts as a receptor in HMGB1-induced extension of neurites (for review, see Ref. 2). Furthermore, exogenously added HMGB1 has been shown to enhance survival of neuronal cells in a RAGE-dependent manner (9). In cancer cells, HMGB1 has been shown to bind to RAGE and to enhance tumor growth and spread (10). Despite these in vitro and in vivo findings, which would be compatible with a developmental role for HMGB1, its possible role in brain development has not been explored.
The forebrain is the part of the nervous system that has undergone the most dramatic changes during vertebrate evolution. The early organization of the forebrain subdomains is conserved in all vertebrates. To understand the origins of the vertebrate forebrain, comparisons of gene expression patterns have been recently carried out in several nonvertebrate and early vertebrate organisms (see for example Refs. 11–13) representing species at the dawn of the vertebrate brain development. Hmgb1 is highly expressed in early embryonic brain and might, therefore, have a role in the development of ancestral forms of complex brain structures.
The findings explained above, neurite outgrowth-promoting and survival-enhancing effects on cultured neuronal cells and high expression in early brain structures, raise the question of whether Hmgb1 would be one of the genes required for brain development. In the current study we have addressed this question using zebrafish as a vertebrate model in which development of different nervous system structures can be easily followed. HMGB1 knockdown experiments in vivo using morpholino oligonucleotides clearly demonstrate defects in brain development, in particular in anterior neural structures. We suggest that in developing forebrain, HMGB1 is required for maintenance of proliferating cells/stem cells that give rise to neurons.
EXPERIMENTAL PROCEDURES
Experimental Animals
An outbred zebrafish (Danio rerio) strain from a local resource, Turku line, was used in this study for its steady yield of embryos (14, 15). Fish feeding, breeding, and maintenance were done according to Westerfield (16). The experiment permits were obtained from the University of Helsinki Committee for animal experiments and the Office of the Regional Government of Southern Finland, in agreement with the ethical guidelines of the European convention. We express the embryonic ages in hours post-fertilization (hpf) and days post-fertilization (dpf). To prevent pigment formation, 0.2 mm 1-phenyl-2-thiourea (Sigma) was added to the media of embryos to be studied before 3 dpf shortly after spawning.
RT-PCR and Quantitative RT-PCR
Total RNA from 2 dpf larvae was extracted with NucleoSpin RNA XS kit. RNA was reverse-transcribed in a reaction containing 1 μg of RNA, 0.25 mm dNTP-mix, 1 mm random nonamers, 20 units of recombinant RNasin® (Promega), 200 units of Moloney murine leukemia virus (MMLV)-RT (Promega) in 1× MMLV reaction buffer. 2 μl of the reverse transcription mixture was then used for polymerase chain reaction with gene-specific primers.
The primers 5′-ACA TCC ACA TAC AGC CAT TGC-3′ and 5′-GGC AAG GAT AGT GGT GTT GGA-3′ were used for cloning the 963-bp zebrafish HMGB1 full-length transcripts of coding sequence (supplemental Fig. S1). The primers 5′-AAG TCA CCG CCA TCA ACG AC-3′ and 5′-ACA ACG GAC ACA TCA ACG AC-3′ of zebrafish GAPDH (gene ID 317743) were used for relative PCR template control.
The subsequent PCR reaction was performed in a 50-μl PCR mix (2.5 μm dNTP, 150 mm KCl, 1.5 mm MgCl2, and 0.1% Triton X-100 in 10 mm Tris-HCl, pH 8.8) containing 0.2 μm 5′ primer and 3′ primer and 1 unit of Dy-NAzymeTM II DNA Polymerase or Phusion high fidelity polymerase (Finnzymes). The PCR cycling parameter was set at 95 °C for 5 min, denaturation at 95 °C for 30 s, annealing/detection at 60 °C for 30 s, elongation at 72 °C for 1 min, back to denaturation for 29 cycles and then at 72 °C for 5 min, and holding at 4 °C. The amplification products were separated on 1.0% agarose gel and stained with EtBr. The PCR products were purified with the MinElute Gel Extraction kit (Qiagen, Hilden, Germany), ligated to the pGEM-Teasy plasmid (Promega), and sequenced.
The zebrafish β-actin1 (AF057040.1, GI:3044209; primers are 5′-CGA GCA GGA GAT GGG AAC C-3′ and 5′- CAA CGG AAA CGC TCA TTG C-3′), elongation factor 1 α1 (zgc:109885, GI:90652818; primers are 5′-CCA ACT TCA ACG CTC AGG TCA-3′ and 5′-CAA ACT TGC AGG CGA TGT GA-3′), and ribosomal protein L13a (BC047855.1, GI:28838761; primers are 5′-AGA GAA AGC GCA TGG TTG TCC-3′ and 5′-GCC TGG TAC TTC CAG CCA ACT T-3′) were set as template quantity controls for quantitative real time PCR. These primers were obtained from the RT Primer Database. The following primers were obtained from the Zebrafish International Resource Center (ZIRC): Pas2a (ZIRC cb378; primers are 5′-ATC AGA GAC CGA CTC TTG GC-3′ and 5′-AAG GCT GCT GAA CTT TGG TT-3′), Pas6a (ZIRC cb280; primers are 5′-AAG AGG GAG TGT CCG TCA AT-3′ and 5′-CAG GTT GCG CAG TAC TCT GT-3′), and Krox20 (ZIRC cb427; primers are 5′-GAT GGC AAG TCA CAA AGA GC-3′ and 5′-CTT CTC CGT GCT CAT ATC CC-3′). Wnt8a1 (BC055535.1, GI:33416858; primers are 5′-TTG CGT CGT TGG TTA TGT CT-3′ and 5′-TAC ACT GCT GGT GTA TGC GA-3′), Wnt8a2 (U10869.2, GI:14574562; primers are 5′-TAA TGA AGC TGG ACG TTT GG-3′ and 5′-TAA TTG CCA ATC TCA CGG AA-3′), and Wnt8b (U10870.1, GI:968916; primers are 5′-TGC AGT GAT AAC GTG GGA TT-3′ and 5′-GTC CTC TGC ATG GTT CCT TT-3′) primers were designed with online tools (GenScript). The LuminoCtTM SYBR Green qPCR ReadyMixTM (Sigma) was used. The typical cycling parameters were as follows: initial denaturation at 95 °C for 30 s, denaturation at 95 °C for 5 s, annealing/detect at 62 °C for 30 s, back to denaturation for 39 cycles, hold at 4 °C. The PCR reactions were processed with the Bio-Rad CFX96 real time PCR machine via CFX96TM Real-Time PCR Detection System.
Production of HMGB1-GFP Fusion Proteins
The 615-bp zebrafish HMGB1 sequence encoding the full-length HMGB1 polypeptide from the start methionine was amplified by PCR with two pairs of primers with added restriction enzyme digestion sites required for ligation with double multi-clone sites pEGFP plasmid (derived from pEGFP-C1, Clontech). The first pair of primers was 5′-AGA ATG CGG CCG CCA CCA TGG GGA AGG ATC CGA CAA AAC C-3′ and 5′-AAG GCG CGC CCT ACT CGT CAT CCT CCT CCT CTT-C-3′ with NotI and AscI sites required for the cloning site at the N terminus of the GFP coding sequence. The second primers were 5′-TAC CCA AGC TTC GAT GGG GAA GGA TCC GAC AAA AC-3′ and 5′-GCG AAT TCC TAC TCG TCA TCC TCC TCC TCT TC-3′ with EcoRI and Hind sites required for the cloning site at the C terminus of GFP. The PCR reaction products were then digested as designed and ligated into Double MCS pEGFP plasmid for the construction of HMGB1-GFP recombinants. The digested products were purified by MinEluted kit (Qiagen) and then ligated by the T4 ligation system (Promega).The HMGB1-GFP fusion protein plasmids were transfected to 293T cells by using the FuGENETM 6 transfection reagent (Roche Applied Science). The cells were extracted with the SDS buffer (62.5 mm Tris, 1.8% SDS, 7.75% glycerol, and 4.4% 2-mercaptoethanol, pH 6.8), and the extracts were pressed several times through a needle. The extracts were boiled twice for 5 min and centrifuged at 10,000 × g for 10 min to remove insoluble material. Samples corresponding to the same wet weight were analyzed by Western blotting.
Preparation of Tissues
Zebrafish embryos were collected and transferred onto a Petri dish with E3 medium (miniQ water containing 5 mm NaCl, 0.17 mm KCl, 0.33 mm CaCl2, and 0.33 mm MgSO4). The fish were killed by keeping them on ice for more than 10 min followed by decapitation or fixation. Larval fish were fixed whole with 2% paraformaldehyde in 0.1 m phosphate buffer at room temperature for 2 h or at 4 °C overnight, and brains of 5-dpf larvae were dissected under a stereomicroscope after the fixation (15). The fixed samples could be saved in phosphate buffer for up to 2 weeks.
Whole-mount in Situ Hybridization
Whole-mount in situ hybridization was carried out as described previously (17) using the specific probes (18) of Pas2a (the ZIRC cb378), Pas6a (ZIRC cb280), and Krox20 (ZIRC cb427). Larvae at 30-hpf (Prim-5) and 48-hpf (Long-pec) stages were used in the experiments.
Antibodies, Western Blotting, and Immunocytochemistry
Affinity-purified anti-peptide IV and (against the peptide KFKDPNAPKRPPSA that corresponds to residues 87–100 of the rat HMGB1 sequence) and anti-peptide V (against the peptide KAEKSKKKKEE that corresponds to the residues 176–186 of the rat HMGB1 sequence) have been previously characterized (19) and were used at the concentration 250 ng/ml to detect the zebrafish and rodent HMGB1 in Western blotting. Binding of the anti-peptide IV antibodies to the zebrafish HMGB1 was verified using cells transfected with the recombinant zebrafish HMGB1-GFP fusion plasmid. Polyclonal anti-β-catenin antibodies (25 ng/ml; C2206 rabbit polyclonal, Sigma) were used to detect β-catenin in Western blotting. Mouse monoclonal anti-β-actin antibody (A2228 Sigma, 1:2000) was used as a control of sample loading.
For Western blotting, brains from more than 1-year-old adult zebrafish or lysates from 15–20 larvae were homogenized in 80 μl of SDS gel sample buffer (16); 15 μl of cell or tissue lysate was used for SDS-PAGE. Proteins were transferred to HybondTM nitrocellulose membrane (Amersham Biosciences) by a semidry blotting technique with transblotting cell (Bio-Rad) and detected with the antibodies as described (16). After the horseradish peroxidase-conjugated secondary antibody incubation, the samples were visualized with the ECL Western blotting detection system (Amersham Biosciences).
Mouse monoclonal anti-tyrosine hydroxylase (TH) antibody (Diasorin, Stillwater, MN) was used for the whole-mount staining of the catecholaminergic system in 5-dpf larval brains as descried earlier (15). The rabbit anti-HMGB1 peptide antibodies (see above, 250 ng/ml), chicken anti-AMIGO1 antibodies (1 μg/ml), and anti-active-β-catenin antibodies (mouse monoclonal, Millipore; the antibody epitope is 100% conserved in zebrafish; 1.1000) were used for whole-mount immunostaining of zebrafish larvae. The anti-AMIGO1 antibodies were generated against the AMIGO1 extracellular domain (20) using a GST-tagged zebrafish fusion protein as the antigen and affinity-purified with AMIGO1 extracellular domain-maltose affinity column. Specificity was shown by Western blotting and competition of binding to the 3 dpf larval samples by the AMIGO1 ectodomain.
For immunohistochemistry of the larvae, the samples were washed and preincubated in phosphate-buffered saline containing 0.1% Tween 20 or 0.25% Triton X-100 (PBS-T, pH 7.4) with 1% dimethyl sulfoxide (DMSO) and 4% natural goat serum at 4 °C overnight or longer. The specimens were incubated with the primary antibodies in the preincubation solution (PBS-T with 2% natural goat serum) for 1–3 days at 4 °C under slow stirring. The samples were then washed thoroughly with PBS-T and incubated with the Alexa®-conjugated donkey anti-rabbit (Alexa 546), goat anti-chicken (Alexa 488), or goat anti-mouse (Alexa 568) secondary antibodies (diluted 1:2000) in the preincubation solution for 1–2 days at 4 °C. The samples were washed with PBS-T twice for 30 min, once with PBS for 30 min, and once with 50% glycerol in PBS for 1 h, and the samples were then infiltrated overnight in 80% glycerol in PBS before mounting.
Morpholino Oligo and cRNA Injections
HMGB1 knockdown experiments were carried out with antisense morpholino oligonucleotides (MO) (Gene Tools) targeted to 5′ upstream sequences flanking the translation start site (MO1, 5′-CCT TCC CCA TCT TTG CCT AAA TAT C-3′) and to RNA splicing sites between the 3rd and 4th exons (MO2, 5′-AAC TGA TTT TCT CAC CTT CTC GTA C-3′; MO3, 5′-GCG ATG TCC TGA AAT AAG ATA TTC G-3′) of the Hmgb1 transcript. The Hmgb1 five mispaired MOs, corresponding to the sequence of MO1 but with five non-pairing nucleotides (5mis MO, CgT TCg CCA TgT TTc CCT AAt TAT C; the non-pairing nucleotides marked by lowercase letters), was used as a morpholino oligo injection control. Because a variety of MOs have the potential of inducing neural apoptosis caused by their off-targeting effects inducing the p53 apoptosis pathway, coinjection of p53 MO (21) with each HMGB1 MO was separately carried out to investigate whether the HMGB1 morphant phenotype can be rescued by p53 knockdown. Each HMGB1 MO was mixed with 4 ng of p53 MO to carry out microinjection. The transcription level of p53 in both single MO-injected and mixed MO-injected 2-dpf embryos was detected by quantitative RT-PCR with the primers reported (21).
The primers 5′-TAC CCA AGC TTC GAT GGG GAA GGA TCC GAC AAA AC-3′ and 5′-GCG AAT TCC TAC TCG TCA TCC TCC TCC TCT TC-3′ were used for adding EcoRI and HindI restriction enzyme digestion sites on both sides of the Hmgb1 first-strand cDNA. The primers 5′-ATA AGA TCT ATG CCC CCT TCC ATT AAT TG-3′ and 5′-AAG AAT TCC CGG TCA AAA GAT ACA CAT CCT C-3′ were used for adding BglΠ and EcoRI restriction enzyme digestion sites on both sides of the AMIGO1 first-strand cDNA. These restriction sites are required for insertion into the pMC expression vector, which caps the inserted fragment with the sequence elements of the β-globin 5′-UTR and 3′-UTR on both sides separately and SV40 poly(A) signal at the 3′ tail additionally. This elevates the transcribed cRNA stability and activity about 100-fold in cRNA injection experiments (22). GFP cRNA was used as a control in cRNA rescue experiments, and it was prepared using pEGFP-C1 vector. All cRNAs were prepared using mMessage mMachine kit (Ambion, Austin, TX) according to the manufacturer's instructions.
All MO aliquots were stored at 300 μm dilution in sterile distilled water at −70 °C. Capped Hmgb1 cRNA was stored at 1 μg/μl in sterile distilled water at −70 °C. For HMGB1 knockdown experiments, the injection solution containing 25% phenyl red with 50 μm HMGB1 MO1 or 100 μm concentrations of the other MOs (the concentration was varied in dosage experiments) was prepared and heated at 65 °C for 15min. The mixture was then cooled down at room temperature. An aliquot of 4 nl of the mixture (corresponding to 2 ng of MO1 and 4 ng of the other MOs when 50 or 100 μm solution was used correspondingly) was injected into the yolk of a 1–4-cell embryo that was allowed to develop at 28.5 °C. For cRNA rescue experiments, 0.1 μg/μl cRNA was mixed with MO injection solution after the heating. All injections were done with an injector (WPI PV830 Pneumatic Pico Pump) and a micromanipulator (Narishige, Tokyo, Japan).
Cell Death and Proliferation Detection
Whole-mount staining of 28-hpf larval apoptosis was performed using the DeadEndTM Fluorometric TUNEL system (Promega (23)), and whole-mount staining of 80-hpf larval cell proliferation was studied with Click-iT Edu Alexa® Fluor 555 imaging kit (Invitrogen) using the protocol recommended by the manufacturer. Unlike assays using bromodeoxyuridine, Click-iT® EdU assays are not antibody-based and, therefore, do not require DNA denaturation for detection of the incorporated nucleoside. Instead, Click-iT® EdU utilizes click chemistry for detection in a variety of Alexa Fluor® dye fluorescent readouts. (Invitrogen, #C10338).
Microscopy
Confocal imaging of 5-dpf brains stained with anti-TH antibodies was carried out as described (24). Zeiss LSM5 Pascal confocal microscopy system with air (10× dry) and water immersion objectives (high magnification, 25 or 63×) was used for imaging of 28-hpf larvae stained with anti-AMIGO1 and anti-HMGB1 antibodies. The fluorophores were excited with the 488- or 568-nm lines from an argon-krypton laser (Omnichrome; Melles Griot, Carlsbad, CA). Cross-talk between the channels and background noise was eliminated with sequential scanning and frame averaging as described earlier (15). Stacks of images taken at 1–1.2-μm intervals were compiled to make maximum intensity projection images. For each scan, uninjected larval samples were scanned first for imaging normalization. All morphants were then scanned using the same experimental details. Specimens for light microscopy were examined with inverted light microscopy using Olympus IX 70 connected through a CCD camera to the Analysis® software. High resolution whole-mount in situ images were obtained with a digital MicroFire S99808 camera (Optronics) attached to an Olympus BX51 epifluorescence microscope (Olympus, Tokyo, Japan).
The acquired images were further processed with Adobe Photoshop 8.0 software (Adobe Systems, San Jose, CA). Anatomical structures of larval brain were named and numbered using the neuroanatomical atlas of adult zebrafish brain (25, 26) and the atlas based on location of TH neurons in 5-dpf larval fish (14, 24).
Analytical Software
All statistical analyses were carried out using OriginPro 7.5. Intensities of the Western blot bands were examined with Quantityone 4.6.2 (Bio-Rad) software. HMGB1 protein sequence alignments of different species (Ensembl) and statistical analysis were carried out using Geneious pro trial 4.8.2 (Biomatters Ltd.).
RESULTS
Zebrafish Hmgb1
The zebrafish Hmgb1 gene has the same exon-intron organization (supplemental Fig. S2) as the mammalian Hmgb1. The full-length zebrafish Hmgb1 DNA was cloned by using mRNA from zebrafish larvae and primers designed according to the putative homologous sequence found in the Zv8 data base (Ensembl, search Danio rerio). Blast searches identified Hmgb1 as the closest homologue within the mammalian sequences. Comparisons of the deduced amino acid sequence (supplemental Fig. S1) to the human or mouse HMGB1 and HMGB2 sequences displayed 86 and 78% similarity, respectively. As in mammals, the zebrafish sequence corresponds to the protein having two homologous HMG boxes (HMG boxes A and B) followed by the acidic tail consisting of only glutamate and aspartate residues (Fig. 1A). The deduced amino acid sequence is slightly shorter (205 amino acids) than the mammalian one (214 amino acids).
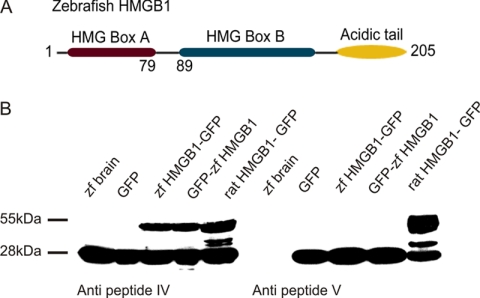
FIGURE 1.
Zebrafish HMGB1 and Western blotting using anti-HMGB1 antibodies. A, shown is the domain structure of the zebrafish HMGB1 consisting of 205 amino acids (for comparisons to HMGB1 sequences of other species, see supplemental Fig. S1). B, shown is Western blotting of zebrafish HMGB1 with rabbit anti-HMGB1 peptide antibodies. Anti-peptide IV antibodies recognize the endogenous HMGB1 of the zebrafish brain lysates (28-kDa band) and the fusion protein (55-kDa band) expressed in 293T cells that were transfected using the zebrafish HMGB1-GFP sequence. Anti-peptide V antibodies only recognize the rodent HMGB1. Zf brain, adult zebrafish brain tissue lysate; GFP, lysate of 293T cells transfected with the GPF construct; zf HMGB1-GFP, lysate of 293T cells transfected with the zebrafish HMGB1-GFP fusion plasmid (GFP at the C-terminal end); GFP zf-HMGB1, lysate of 293T cells transfected with the GFP-zebrafish HMGB1 fusion plasmid (GFP at the N-terminal end); rat HMGB1-GFP, lysate of 293T cells transfected with the rat HMGB1-GFP fusion plasmid. The fusion protein is detected in all experiments as the 55-kDa band; the endogenously occurring HMGB1 in zebrafish brain and in 293T cells used for transfection is detected as the 28-kDa band.
Detection of Zebrafish HMGB1 Using Specific Antibodies
We have previously produced a series of affinity-purified anti-protein and anti-synthetic peptide antibodies that detect the mammalian HMGB1 (19). Antibodies against a synthetic peptide (anti-peptide IV; see Ref. 19) corresponding to a highly conserved area immediately before and at the beginning of the B box (Fig. 1, A and B) were found to bind to the zebrafish HMGB1 in Western blotting. These antibodies detected the endogenously occurring HMGB1 of zebrafish brain lysate and of the cells used in transfection assays (the 28-kDa band in Fig. 1B). Furthermore, in transfection assays these anti-HMGB1 antibodies detected the zebrafish and rat fusion protein in a similar manner (the 55-kDa band in Fig. 1B). In contrast, antibodies against a sequence closer to the C-terminal end (anti-peptide V), where the zebrafish and the mammalian sequences display many differences, only detected the endogenously occurring HMGB1 of the cells used for transfection assays (the 28-kDa band in Fig. 1B) and the rat fusion protein (the 55-kDa band in Fig. 1B) but not the endogenously occurring zebrafish protein or the zebrafish fusion protein. Further evidence of the specificity of the antibodies (anti-peptide IV) is provided by the reduced or lacking detection of the zebrafish 28-kDa protein when translation or mRNA synthesis is disturbed in the morpholino antisense experiments specifically targeting HMGB1 (see e.g. the Western blots in Fig. 2).
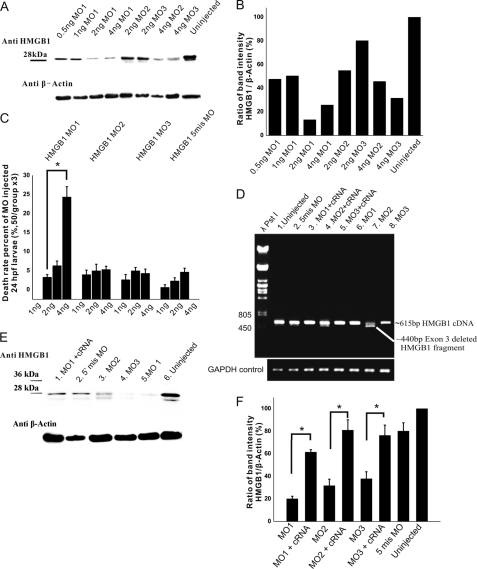
FIGURE 2.
Dosages of HMGB1 MOs required for specific knockdown of HMGB1 expression and rescue of the expression by coinjecting Hmgb1 cRNA. A, shown is Western blotting of HMGB1 from lysates of 3-dpf larvae that had been injected with different dosages of the MOs. MO1 causes a more prominent inhibition of HMGB1 expression than MO2 or MO3. B, shown is quantification of the Western blotting bands in A by plot density analyses. MO1 (2 ng) causes >80% knockdown of HMGB1 expression, and MO2 and MO3 (4 ng each) cause >50% inhibition of expression. C, statistics of the death rate in different MO-injected 24-hpf larvae are shown. By increasing the MO1 injection dosage from 1 to 4 ng, the death rate of the MO1-injected morphants displays a significant increase (compared by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.001), from less than 8% to around 20%. At the same doses, MO2 or MO3 do not cause an appreciable change. For each group, 300 larvae were used in 6 independent injections; 50 larvae were injected in each experiment. D, shown is RT-PCR of 2-dpf larval total RNA using primers from both ends of the coding sequence of the Hmgb1 cDNA (615 bp). MO1 does not cause an appreciable change in the analysis, whereas a band of 440 bp is found in the MO2-injected larval sample corresponding to exon 3 deletion. In the MO3-injected larval sample, the 615-bp Hmgb1 PCR product is clearly reduced compared with the controls. GADPH was used as the template control. E and F, inhibition of HMGB1 expression by the MOs and rescue of the expression by coinjection of the Hmgb1 cRNA are shown. The three groups of larvae injected with the morpholino oligonucleotides (2 ng of MO1; 4 ng of MO2 or MO3) display a clearly reduced expression of the HMGB1 protein compared with the uninjected group or the 5 mis MO-injected group. A Western blot is shown in E, and quantification of the Western blotting band intensities from four replicates is shown in F. In the groups of larvae injected with MO1, MO2, or MO3, the HMGB1 expression is significantly reduced compared with the MO/cRNA-coinjected larvae (compared by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.01). In the group with the best rescue effect (MO2/cRNA), the HMGB1 expression is ∼80% of uninjected controls. In the group with the lowest rescue effect (MO1/cRNA), the expression is >60% of uninjected controls. For Western blotting, 20 larvae of each injection group were selected randomly for sample preparation. The experiment was repeated three times by independent injections. The error bars in (C and F) indicate the S.E. values.
Inhibition of HMGB1 Expression in Zebrafish Larvae with Morpholino Oligonucleotides
Gene-specific antisense MOs have been widely used to inhibit gene expression in zebrafish larvae (27). We designed three MOs (see “Experimental Procedures” and supplemental Fig. S2) expected to interfere in Hmgb1 expression. The first one (MO1) targets a sequence that includes the ATG site (from −15 to +10 compared with the ATG site) and is expected to inhibit HMGB1 translation. The other two oligonucleotides (MO2 and MO3) target splice sites in the Hmgb1 pre-mRNA. MO2 is expected to cause an exon deletion, whereas MO3 is expected to cause an intron insertion. Noninjected larvae and larvae injected with the five mispaired MOs (5mis MO; having five non-pairing nucleotides compared with MO1) were used as controls.
In larval lysates, HMGB1 was specifically detected as the protein with a 28-kDa molecular mass (Fig. 2A). We used Western blotting of 3-dpf larval lysates to evaluate the effects of the MOs (at the doses 1–6 ng) on HMGB1 protein expression. Estimated from the Western blots (Fig. 2, A and B), 2 ng of MO1 were required to cause more than 50% inhibition of the protein expression, whereas somewhat higher doses of MO2 and MO3 (4 ng) were required for a similar inhibition. We selected these doses for further experiments aiming at studies on organ development as they allow early development and high proportions of viable larvae (Fig. 2C).
RT-PCR analysis (Fig. 2D) of the total RNA extracted from 2-dpf MO1-injected larvae revealed expression of Hmgb1 mRNA at the same level as in controls, as expected. In contrast, in MO2-injected larvae the normally occurring mRNA was strongly reduced, and a band corresponding to exon 3 deletion was found. In MO3-injected larvae, expression of the normally occurring mRNA was strongly reduced.
The MOs were designed so that they may not inhibit the capped Hmgb1 cRNA injected in rescue experiments together with the MOs. MO1 extends from −15 until +10 compared with the ATG site and MO2 and MO3 target splice sites of the pre-mRNAs (supplemental Fig. S2). Western blotting showed that in the selected doses (Fig. 2E, quantification from replicate filters in Fig. 2F) MO1 causes 80–90% inhibition in the protein expression, whereas MO2 and MO3 cause 70 and 50–60% inhibition correspondingly. Coinjection of the Hmgb1 mRNA together with MO1, MO2, or MO3 caused a significant rescue of the protein expression (Fig. 2, E and F).
Expression of HMGB1 during Early Development of Zebrafish
Expression of Hmgb1 mRNA has been mapped at different stages of zebrafish development (see The Zebrafish Model Organism Database). Hmgb1 is expressed in blastula, gastrula, and segmentation stages ubiquitously until 14 hpf. After this, HMGB1 is mainly expressed in brain and other parts of the nervous system until 5 dpf, when the expression is down-regulated. Ventral mesoderm also expresses Hmgb1 during organogenesis.
To gain further insight into HMGB1 expression in zebrafish embryos, we carried out whole-mount immunostaining experiments of 28-hpf larvae (Fig. 3) using antibodies against HMGB1 (anti-peptide IV; see above). The 28-hpf larvae have already completed primary neurogenesis and are entering into the stage of secondary neurogenesis (28). HMGB1 was found to be prominently expressed in forebrain, in particular in rostral telencephalon and telencephalon close to the ventricular wall, in pretectum, and at the anterior part of the diencephalic ventricular wall (Fig. 3 and supplemental Figs. S3–S5). Immunostaining on these regions is specific as it is strongly reduced or disappears in MO1 injected larvae. Furthermore, the coinjected Hmgb1 cRNA rescued immunostaining, although the immunostaining pattern is different from that caused by the endogenous HMGB1 expression (Fig. 3 and supplemental Figs. S3 and S4). The areas where HMGB1 is expressed are populated by brain stem cells/neuronal progenitor cells (29) during the primary and secondary neurogenesis (28). In addition to the forebrain, HMGB1 was found to be expressed symmetrically along the brain midline in the spinal cord and in the notochord (supplemental Fig. S6).
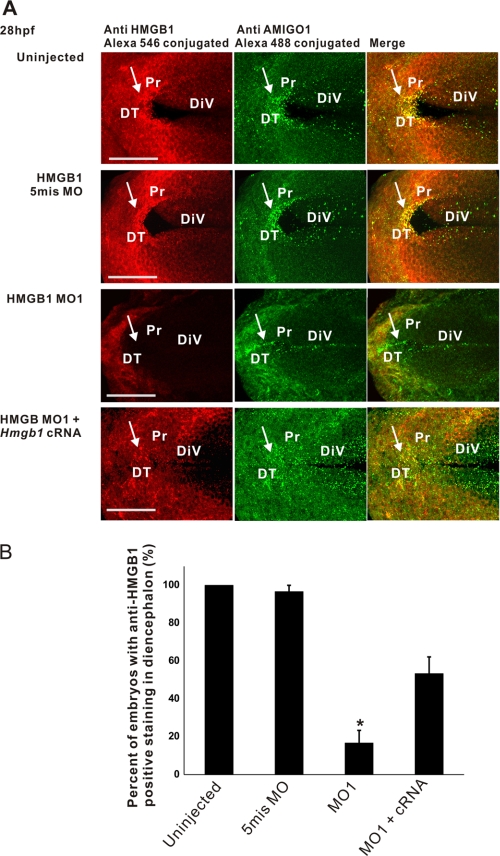
FIGURE 3.
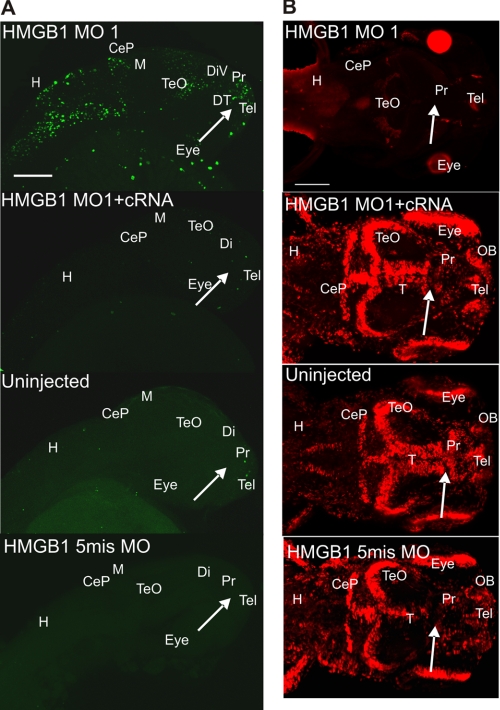
Whole-mount immunostaining of HMGB1 (Alexa 546-conjugated) and AMIGO1 (Alexa 488-conjugated) in 28-hpf (Prim-15) larvae (dorsal view). A, the red color shows the HMGB1 immunostaining, and the green color shows the AMIGO1 immunostaining. The uninjected and the 5mis MO-injected larvae show intense immunostaining of HMGB1 in the anterior diencephalon and in diencephalon close to the ventricular wall (arrows), which is essentially absent in the MO1-injected larvae. In the MO1/cRNA-injected larvae, HMGB1 is clearly immunostained in diencephalon and other parts of the forebrain but in a more diffuse pattern compared with the endogenously expressed HMGB1. AMIGO1 displays a similar distribution compared with HMGB1, and the two proteins colocalize on the area where HMGB1 is intensely immunostained (the merged figures on the right). In MO1 morphants, AMIGO1 expression is down-regulated and restricted along the base of the diencephalic ventricle. In the MO1/Hmgb1 cRNA coinjected larvae, AMIGO1 expression is rescued and displays a diffuse pattern in the forebrain. Maximum intensity projection images using the optical sections throughout the diencephalon are displayed in the panels. The stack size is 40–50 μm covering the sample thickness, and the step interval is 1 μm. DiV, diencephalic ventricle; DT, dorsal thalamus; Pr, pretectum. Scale bars, 50 μm. B, shown are statistics of larvae in which HMGB1 expression is detected in diencephalon. The HMGB1 MO1 morphants display strongly reduced numbers of larvae expressing HMGB1 in diencephalon compared with MO1/cRNA-coinjected or the 5mis MO-injected groups (one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction have been done; *, p < 0.001). The error bars indicate the S.E. values. For every injection group, 10 larvae were randomly selected for whole-mount immunostaining. Three independent injections were carried out.
AMIGO1 (amphoterin-induced gene and orf) is a nervous system specific transmembrane protein whose expression is induced in neuronal cells by extracellular matrix-associated HMGB1 (20). We used AMIGO1 as a neuronal plasma membrane marker in double-immunostaining with HMGB1. Immunostaining using the anti-HMGB1 and anti-AMIGO1 ectodomain antibodies revealed partial colocalization in developing forebrain (Fig. 3 and supplemental Figs. S3 and S4). A high resolution view of HMGB1/AMIGO1 double-immunostaining in diencephalon revealed a patchy detection of both proteins that partially colocalize at the plasma membrane (Fig. 3 and supplemental Fig. S5). Visualization of membrane structures with the lipid-soluble dye DiI revealed a similar localization at the membrane level (not shown).
Furthermore, immunostaining experiments showed that MO1 injection down-regulates both the HMGB1 and AMIGO1 expression, whereas expressions of both proteins are up-regulated by coinjection of the Hmgb1 cRNA (Fig. 3 and supplemental Figs. S3 and S4). The expression and function of AMIGO1 may, therefore, be linked to those of HMGB1 during forebrain development.
Morphological Characteristics of the HMGB1 Knockdown Morphants
Each MO caused phenotypic changes at the dose range of 1–6 ng per embryo. To analyze the phenotypic effects of the MOs, we selected the lowest doses causing a 50% or higher inhibition of HMGB1 protein expression as evidenced by Western blotting analysis but not compromising the viability of the embryos (see above and Fig. 2).
Examination of gross morphology of MO-injected zebrafish revealed prominent changes starting at the early stages of development (1–2 dpf). The morphant phenotype can be easily recognized based on smaller size, curling tails, smaller brain width, and shorter distance between the eyes (Fig. 4A and supplemental Fig. S7). They are immobile and stay alive until about 1 week. All three MOs (MO1, MO2, and MO3) produced a very similar change, but there was a difference in the frequency of the characteristic phenotype observed among the three MOs. Analysis of 3-dpf larvae revealed that injection of MO1 (2 ng) caused the characteristic phenotype in 80–90% of the injected zebrafish with somewhat lower frequencies for MO2 and MO3 injections (4 ng of each MO; Fig. 4B). This finding agrees with the Western blot analysis showing that MO1 causes a more prominent and effective down-regulation of HMGB1 expression compared with MO2 or MO3 (Fig. 2F). In more than 90% of the experiments, the 5-misMO (4 ng) injected larvae did not display any morphological changes. Coinjection of the Hmgb1 cRNA was able to rescue the MO-induced phenotypic change (Fig. 4B). Because MO1 proved to be the most reliable one in causing the characteristic phenotype and the phenotype was shown to be rescued by mRNA injection and MO1 caused the strongest down-regulation of HMGB1 expression in Western blotting, we mainly used this antisense oligonucleotide in further characterization of the developmental role of HMGB1.
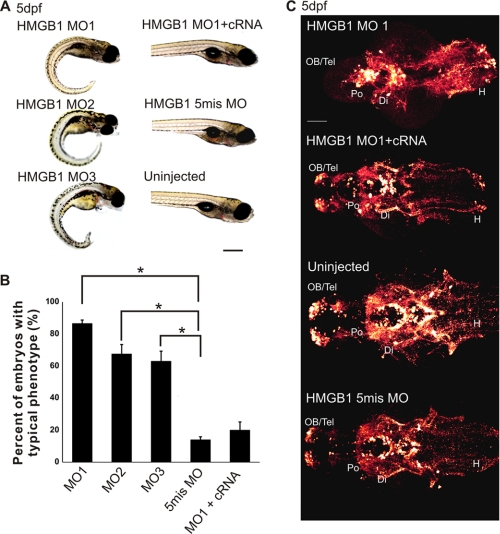
FIGURE 4.
Typical phenotypes of HMGB1 knockdown morphants. A, shown is light microscopy of 5 dpf larvae. All three HMGB1 MOs cause a similar phenotype displaying a shorter trunk with curled tail and a smaller head part. Compared with the HMGB1 knockdown morphants, the HMGB1 MO1/Hmgb1 cRNA-coinjected and the 5mis MO-injected larvae do not show any severe morphological defects and have the same outlook as the uninjected larvae. The scale bar indicates 1 mm. B, shown is are statistics of the morphants with typical HMGB1 knockdown phenotype (shown in panel A). All three HMGB1 MO morphants show a significant difference compared with the 5mis MO morphants (compared by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.001). The phenotype is rescued in the HMGB1 MO1/Hmgb1 cRNA-coinjected larvae. For each group, 200 larvae were used in 4 independent injections; 50 larvae were injected in each experiment. The error bars indicate the S.E. values. C, catecholaminergic networks of 5-dpf larval brains detected by TH1 immunostaining (Alexa 568 conjugated; ventral view) are shown. In the HMGB1 MO1-injected larvae, catecholaminergic networks are not detected in the telencephalon, and the staining is reduced in the diencephalon. Coinjection of Hmgb1 cRNA with MO1 partially rescues the phenotype. The HMGB1 5mis MO morphants show a similar staining pattern as the uninjected controls. The stack size is about 100 μm throughout the brain thickness, and the step interval is 1 μm. Di, diencephalon; H, hindbrain; OB, olfactory bulb; Po, preoptic region; Tel, telencephalon. The scale bar indicates 50 μm.
Closer examination of the MO1-injected larvae during an early developmental stage (30 hpf) displayed a disordered pattern of prosomeres and a reduced size of the forebrain due to perturbed formation of both diencephalon and telencephalon (supplemental Fig. S7). To further characterize structural changes in the nervous system development in the HMGB1 knockdown, we used tyrosine hydroxylase1 (TH1) immunostaining as a marker to detect catecholaminergic, mainly dopaminergic neuronal networks that form an important and well characterized structure in the early zebrafish brain (24, 30, 31). Immunostaining of TH1-positive networks revealed prominent changes in the morphants (shown for 5-dpf morphants in Fig. 4C, at the stage when the networks have been essentially fully formed). The most striking change in HMGB1 knockdown zebrafish was the nearly complete absence of TH1-positive neuron clusters in the telencephalon and the anterior basal diencephalon (Fig. 4C). Furthermore, the HMGB1 morphants had fewer TH1 neurons in the hypothalamus and disordered distribution of TH1 reactive axon fibers in the hindbrain along the midline. The phenotype was essentially rescued in MO1/Hmgb1 cRNA-coinjected larvae, with only slight changes compared with wild-type controls (Fig. 4C).
Cell Survival and Proliferation in the HMGB1 Knockdown Zebrafish
Previous studies have shown that HMGB1 enhances survival of cultured embryonic cells (9). Because HMGB1 is robustly expressed close to ventricular walls populated by stem cells/proliferating cells, we reasoned that HMGB1 might be required for survival and proliferation in early forebrain. TUNEL staining revealed abundantly apoptotic cells in forebrain and hindbrain in 28-hpf MO1-injected zebrafish larvae (Fig. 5A). At this stage, apoptosis has been shown to be very rare in the normal developing brain (32), in agreement with our analysis of the wild-type larvae used as controls (Fig. 5A). Coinjection of Hmgb1 cRNA was essentially able to reverse the effect (Fig. 5A), suggesting that apoptosis was specifically caused by inhibition of HMGB1 expression.
FIGURE 5.
Detection of embryonic apoptotic and proliferating activity. A, shown is TUNEL staining (Alexa 488 conjugated) of 28-hpf larvae. The HMGB1 MO1 morphants display widespread apoptosis in the forebrain (arrow) and the hindbrain. In the HMGB1 MO1/Hmgb1 cRNA-coinjected, mis MO-injected, or the uninjected groups, apoptosis is quite limited. The stack size is about 150 μm throughout the sample thickness, and the step interval is 1.2 μm. The scale bar indicates 100 μm. B, detection of proliferation activity by EdU-Alexa 555 staining of 3-dpf larvae (dorsal view; the arrow shows the forebrain/midbrain boundary) is shown. In the HMGB1 MO1 morphants, very few proliferating cells are detected. In the HMGB1 MO1/Hmgb1 cRNA-coinjected morphants, the pattern of proliferating cells in the brain is similar to that found in the uninjected and the 5mis MO-injected controls. On some areas of the brain, the cRNA induces a more intense proliferation compared with the controls. The stack size is about 150 μm throughout the sample thickness, and step interval is 1.2 μm. The scale bar indicates 150 μm. CeP, cerebellar plate; Di, diencephalon; DiV, diencephalic ventricle; DT, dorsal thalamus; H, Hindbrain; M, midbrain; OB, olfactory bulb; Pr, pretectum; Tel, telencephalon; TeO, tectum opticum.
Based on TUNEL labeling, the HMGB1 knockdown larvae display apoptosis both in forebrain and hindbrain, which is somewhat unexpected as the morphological changes were largely restricted to the forebrain compared with other parts of the nervous system (see above). Yet the cRNA rescue experiments clearly argued for a specific effect (Fig. 5A). As an additional set of control experiments, we therefore studied the possible p53 activation that is suggested as a control of off-target effects in siRNA and MO knockdown technologies (21). We did not find any evidence of p53 activation that would explain apoptosis or other effects in the knockdown experiments using any of the three HMGB1 MOs (supplemental Fig. S8, A and B). Furthermore, inclusion of p53 MO is recommended to reduce the risk of off-target effects in knockdown experiments, but this approach did not change the characteristic larval phenotype of the HMGB1 knockdown (supplemental Fig. S8C) or the numbers of TUNEL-positive larvae (not shown).
If HMGB1 is required to maintain proliferating cells/stem cells in developing brain, one would expect to see a difference in DNA synthesis in brain regions populated by such cells in experiments where the HMGB1 expression level is manipulated. EdU labeling revealed prominent staining that was strongly reduced in the brain of MO1-injected larvae (shown for 3-dpf larvae in Fig. 5B). The staining suggesting cell proliferation was rescued by injecting Hmgb1 cRNA together with MO1. Interestingly, EdU labeling was somewhat more widespread along the midline close to the brain ventricles in forebrain and midbrain compared with the controls without cRNA injection (Fig. 5B). Correspondingly, Hmgb1 cRNA-injected larvae also showed diffuse staining with the HMGB1 antibody (Fig. 3). This is to be expected as the injected coding sequence lacks regulatory elements and may, therefore, cause ubiquitous HMGB1 expression. HMGB1 thus appears to affect cell proliferation on regions where it is not even endogenously expressed in the nervous system.
Regionally Expressed Markers in the HMGB1 Knockdown Zebrafish
Examination of gross morphology and immunohistochemistry using TH1 staining as a marker suggested prominent changes in telencephalon and diencephalon of the HMGB1 knockdowns. To get further insight into the regional role of HMGB1 in brain development, we studied the expression of the transcription factors Pax6a, Pax2a, and Krox20 that have been implicated in regional development of brain. Pax6 is essential for early development of many regions of the central nervous system, especially for the early forebrain patterning (33, 34). Furthermore, Pax6 activity has been found to be important for catecholaminergic organization during the early forebrain development (36). Pax2a was used as the embryonic regional marker of diencephalon-derived optic stalk and midbrain-hindbrain boundary and Krox20 as the marker of early developing hindbrain rhombomeres (18, 35).
The most prominent change found in these studies was the virtual absence of the Pax6a-expressing cell group in telencephalon of HMGB1 knockdown morphants, which can be rescued by Hmgb1 cRNA coinjection in most of the embryos studied (see Fig. 6 for morphology and the percentages of embryos lacking Pas6-positive cells in telencephalon). In contrast to the telencephalic area, Pax6a expression appeared normal in more posterior areas of the central nervous system. We did not detect any major differences in the expression patterns of Pax2a or Krox20 (Fig. 6). These results are in agreement with the morphological findings (see above), suggesting that forebrain development is especially vulnerable to down-regulation of HMGB1 expression.
FIGURE 6.
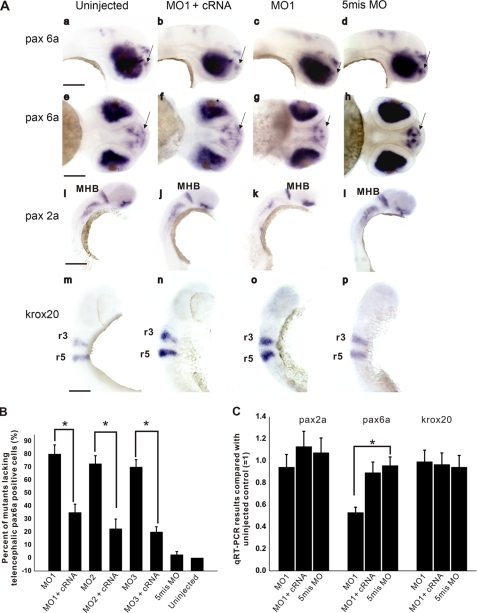
Whole-mount in situ hybridization to detect Pas6a, Pas2a, and Krox20 expression in the uninjected and the MO-injected brains. Panel, a–h, shown is Pas6a in 48-hpf larvae (the pharyngula period from High-pec to Long-pec) as a marker of forebrain development; a–d show the lateral view in which the scale bar indicates 100 μm; e–h show the ventral view in which the scale bar indicates 60 μm. The arrow shows the telencephalic Pas6a-positive cell group, which disappears in the HMGB1 MO1 morphants. i–l, shown is Pas2a in 48-hpf larvae (the pharyngula period from High-pec to Long-pec in) as a marker of the midbrain-hindbrain barrier (MHB) development; the scale bar indicates 250 μm. m–p, Krox20 in 24–30-h larvae (the pharyngula period from Prim 5 to Prim 15) detect development of the hindbrain rhombomere3 (r3) and rhombomere5 (r5); the scale bar indicates 100 μm. Panel B, percentages of the mutants lacking the Pas6a-positive cell group in telencephalon (the arrow in a–h). The chart shows that the HMGB1 MO1, MO2, and MO3 morphants have a high percentage of mutants lacking the telencephalic Pas6a-positive cell group compared with the HMGB1 5mis MO morphants. Coinjection of the Hmgb1 cRNA with the MOs causes a significant rescue effect (compared by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.001; the error bars indicate the S.E.). Each group contains 40 larvae from 4 independent injections; 10 larvae were selected randomly from every injection for whole-mount in situ hybridization. Panel C, quantification of Pas2a, Pas6a, and Krox20 in 2-dpf larvae by qRT-PCR. The transcription level of Pas6a, but not of Pas2a or Krox20, is significantly down-regulated by the MO1 injection (compared by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.01; the error bars indicate the S.E.). The result is based on five independent injections. Every group has 50 larvae in each injection, and 10 larvae were randomly collected and prepared for the qRT-PCR analysis.
Wnt Signaling in the HMGB1 Knockdown Zebrafish
Wnt signaling plays a key role in the development of vertebrates and nonvertebrates (for review, see Ref 37). In particular, Wnt8 signaling has been connected to forebrain development and is known to become dysregulated in Pax6−/− embryos (34, 38). We, therefore, decided to study whether HMGB1 knockdown affects Wnt8 signaling. Surprisingly, Wnt8 a1 mRNA was up-regulated 4–5-fold in MO1-injected larvae, and the effect was rescued by injection of the Hmgb1 cRNA. In addition, Wnt8a2 and Wnt8b mRNAs were up-regulated about 2-fold and 2–3-fold, respectively (Fig. 7A).
FIGURE 7.
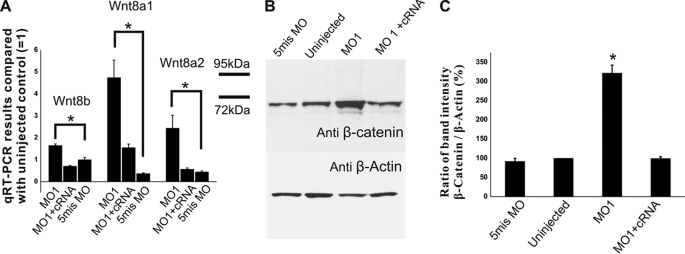
Up-regulation of Wnt-β-catenin signaling by HMGB1 knockdown. A, shown is quantification of Wnt8a1, Wnt8a2, and Wnt8b in 2-dpf larvae by qRT-PCR. HMGB1 MO1 morphants show a significantly increased expression of Wnt8a1, Wnt8a2, and Wnt8b compared with the other groups (compared by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.001; the error bars indicate the S.E.). In the HMGB1 MO1 morphants, Wnt8a1 expression is about 4–5-fold higher than in 5mis MO-injected controls (=1), and Wnt8a2 and Wnt8b are about 2-fold higher than in 5mis MO-injected controls (=1). The morphants injected with MO1/Hmgb1 cRNA do not show an increase in Wnt8 signals. In Wnt8b qRT-PCR, 60 larvae were used in each group, and analysis of each group is based on 6 independent injections. In Wnt8a1 and Wnt8a2 qRT-PCR, 30 larvae were used in each group, and analysis of each group is based on 3 independent injections. From each injection, 10 larvae were randomly collected and separately prepared for qRT-PCR template. B, shown is Western blotting of 3-dpf larval lysates with anti-β-catenin antibody shows an increased β-catenin expression in the MO1 morphants, which confirms the qRT-PCR results shown in A. C, shown is quantification of the Western blotting bands from four anti-β-catenin antibody Western-blotting experiments. The HMGB1 MO1 morphants have about 3-fold β-catenin expression compared with uninjected or 5mis MO-injected controls (the significance confirmed by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.001). The error bars indicate the S.E. In every experiment, 20 larvae of each injection group were randomly collected for Western blotting.
A key question is, therefore, whether the changes in mRNA levels reflect on protein levels and cell signaling. Because the Wnt proteins are hard to detect, we chose to take an indirect approach by following expression of the β-catenin protein that is known to become stabilized and, therefore, up-regulated during canonical Wnt signaling. Western blotting showed that the β-catenin level is indeed specifically up-regulated about 3-fold in the HMGB1 morphants (shown for 3-dpf morphants in Fig. 7, B and C).
Role of AMIGO1 in HMGB1-dependent Cell Survival and Development
Immunostaining studies (Fig. 3 and supplemental Figs. S3 and S4) suggested coexpression of HMGB1 and AMIGO1 in brain. AMIGO1 was previously cloned as a gene that is robustly induced by cell matrix-bound HMGB1 in neuronal cells in vitro (20). We, therefore, examined more closely whether HMGB1 regulates AMIGO1 expression in zebrafish in vivo. Western blotting experiments revealed that the expression of AMIGO1 is indeed closely linked to the HMGB1 expression; in MO1-injected larvae both proteins are clearly down-regulated, and they both reappear when Hmgb1 cRNA is coinjected with MO1 (Fig. 8, A and B).
FIGURE 8.
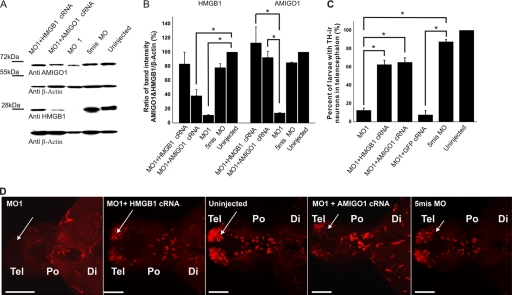
Inhibition of AMIGO1 expression in the HMGB1 knockdown morphants and the rescue effect of the AMIGO1 and Hmgb1 cRNA on the development of catecholaminergic neural networks in the HMGB1 knockdown morphants. A, Western blotting of AMIGO1 and of HMGB1 using β-actin as the control for each protein is shown. B, quantification of the Western blotting result shown in A is based on four independent injections; 20 larvae of each injection group were randomly selected for sample preparation. The AMIGO1 and HMGB1 expression level of uninjected larvae has been set as 100%. Expression of both HMGB1 and AMIGO1 is strongly inhibited by MO1, whereas the 5mis MO does not cause a significant inhibition. The HMGB1 cRNA rescues the expression of both HMGB1 and of AMIGO1, whereas the AMIGO1 cRNA rescues expression of AMIGO1 but not of HMGB1. All groups were compared by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.001. The error bars indicate the S.E. values. C and D, both Hmgb1 and AMIGO1 cRNA have a rescue effect on the development of catecholaminergic neural networks of telencephalon in the HMGB1 morphant. C shows statistics of larvae with positive TH1 staining in the 5-dpf larval telecephalon. Four independent injections were carried out, and 10 larvae from each group (50 larvae per injection group) were randomly selected for detection. >90% of the HMGB1 morphants have no TH1-positive cells in telencephalon. The rescue effects of Hmgb1 or AMIGO1 cRNA coinjection with HMGB1 MO1 are from >50% to even more than 70%, whereas the GFP cRNA does not display a significant rescue effect. The groups were analyzed by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.001. The error bars indicate the S.E. values. D shows a ventral forebrain view of 5-dpf larvae immunostained with anti-TH antibody, which was used as the basis of counting the larvae with positive anti-TH immunostaining in the telencephalon. The HMGB1 MO1 morphant has no catecholaminergic networks in telencephalon (arrow). Partial rescue in the development of the catecholaminergic pathways is seen in the Hmgb1 and AMIGO1 cRNA-injected groups compared with the uninjected or the 5mis MO-injected larvae. The stack size is about 100 μm throughout the brain thickness, and the step interval is 1 μm. Di, diencephalon; Po, preoptic region; Tel, telencephalon. Scale bar, 80 μm.
If AMIGO1 is involved in the mechanism through which HMGB1 regulates brain development, AMIGO1 cRNA might have a rescue effect in the HMGB1 knockdown. We used immunostaining of TH1-expressing neuronal networks to evaluate the possible rescue effect. In more than 60% of larvae, TH1-positive networks became detectable in telencephalon when AMIGO1 cRNA was coinjected with MO1 (Fig. 8, C and D). The corresponding value for the MO1/Hmgb1 cRNA-coinjected groups was about the same, whereas GFP cRNA coinjected control did not cause a significant rescue effect (Fig. 8D).
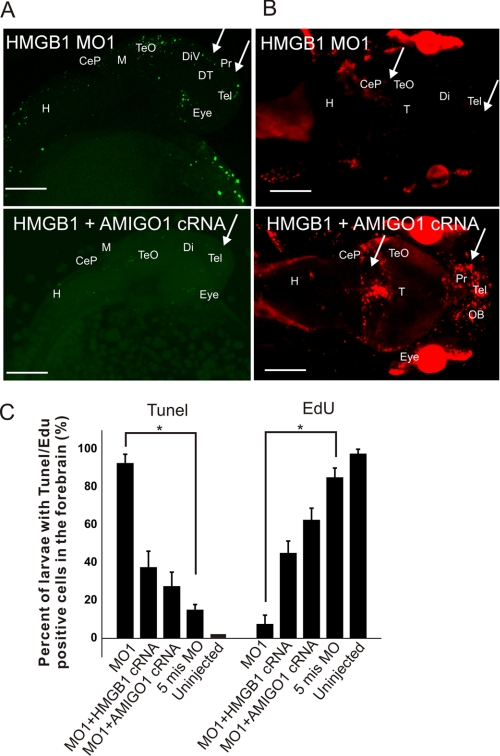
Furthermore, rescue experiments suggested that AMIGO1 is involved in the survival and proliferation-enhancing mechanism of HMGB1. Abundant TUNEL staining was found in a high proportion of MO1-injected larvae compared with mis MO-injected or uninjected larvae; injection of Hmgb1 cRNA or AMIGO1 cRNA both caused a clear rescue effect (see the qualitative comparison in Fig. 9A and a comparison from replicate experiments in Fig. 9C). Coinjection of p53 MO or GFP cRNA did not change the numbers of TUNEL-positive larvae (not shown). As expected, a corresponding rescue effect was observed in EdU staining of proliferating cells (Fig. 9, B and C).
FIGURE 9.
Rescue of cell survival and proliferation in the HMGB1 knockdown morphants by AMIGO1 and Hmgb1 cRNA coinjection. A, AMIGO1 cRNA rescues 28-hpf larvae from apoptosis due to HMGB1 knockdown. The forebrain apoptotic area in MO1 morphants and the corresponding area in MO1/AMIGO1 cRNA morphants have been indicated by arrows. Stack size is about 150 μm throughout the sample thickness, and the step interval is 1.2 μm. The scale bar indicates 100 μm. B, AMIGO1 cRNA rescues 3-dpf larval proliferating activity in the telencephalon, diencephalon, and tectum opticum (shown by arrows). Stack size is about 150 μm throughout the sample thickness, and the step interval is 1.2 μm. The scale bar indicates 150 μm. CeP, cerebellar plate; Di, diencephalon; DiV, diencephalic ventricle; DT, dorsal thalamus; H, hindbrain; M, midbrain; OB, olfactory bulb; Pr, Pretectum; Tel, telencephalon; T, tectum; TeO, tectum opticum. C, statistics of TUNEL and EdU staining are shown in A and B. For both TUNEL and EdU staining, the number of larvae with staining in the forebrain area was counted in each group. The HMGB1 5mis MO-injected and uninjected larvae were used as the controls. The HMGB1 MO1 morphants showed a significantly higher TUNEL staining and a significantly lower EdU staining in 3-dpf larval forebrain (compared by one-way ANOVA followed by Fisher's LSD post hoc test and Bonferroni correction; *, p < 0.001; the error bars indicate the S.E. values). Both Hmgb1 and AMIGO1 cRNA display a clear rescue effect on cell survival and proliferation. The experiment was repeated 4 times by independent injections. In every injection 10 larvae from each group (50 larvae per injection group) were randomly selected for the TUNEL or EdU staining.
DISCUSSION
Previous in situ hybridization studies have shown that during organogenesis,HMGB1 mRNA accumulates in early brain structures in many nonvertebrate species, such as amphioxus (12) and Xenopus (13) and the basal vertebrate lamprey (11). In vertebrates, a high expression level of HMGB1 (amphoterin) has been demonstrated in embryonic rat brain compared with the adult brain (6), and mapping studies using in situ hybridization have shown that during organogenesis in zebrafish HMGB1 is essentially a nervous system protein that is abundantly expressed in brain (see The Zebrafish Model Organism Database). However, excluding in vitro studies using primary forebrain cells and neuroblastoma cells (6, 8), the possible role of HMGB1 in brain development remains to be explored.
HMGB1 Regulates Forebrain Development
The current study shows that HMGB1 is essential for forebrain development in zebrafish. Several lines of experiments argue for specificity in our knockdown experiments. Western blotting shows that the HMGB1 protein is down-regulated by three different HMGB1 antisense MOs, and the phenotypic changes parallel the HMGB1 down-regulation. Furthermore, similar morphants were observed for all three antisense MOs but not for the control MO (5mis MO). Finally, clear rescue effects can be observed when the Hmgb1 cRNA is injected in knockdown experiments.
Because HMGB1 is highly expressed in the central nervous system during early development in all species studied so far, we suggest that the role in brain development is a conserved phenomenon in evolution. In particular, the diencephalic and telencephalic forebrain structures appear vulnerable to the down-regulation of HMGB1 expression, and we suggest that this highly conserved gene has an important function enabling survival and proliferation of stem cells/precursor cells that will form the forebrain structures. It is still unclear to us why the forebrain appears especially vulnerable to the down-regulation of HMGB1 expression. A possible reason to this is that, compared with other parts of the brain, high numbers of cells have to be produced and added to the complex network structures in the developing forebrain. Therefore, the forebrain would be more critically dependent on survival/proliferation enhancing factors than lower parts of the nervous system, leading to the situation that down-regulation of only one factor causes massive perturbation of forebrain structures.
HMGB1 as an Extracellular and Intracellular Factor Regulating Forebrain Development
Proliferation and differentiation of neural progenitors is heavily dependent on the specialized microenvironment, the niche in which the cells reside. The cells are regulated within these niches by soluble and membrane-bound molecules and by extracellular matrix. One should, therefore, consider whether HMGB1 participates in cell-to-cell signaling within the niches that regulate neural progenitors. It is currently generally accepted within the HMG field that HMGB1 can be actively secreted upon cell stimulation by cytokines/growth factors, and its secretion can be even induced by cell contact with extracellular matrix (for review, see Ref 2). Because the stem cells and progenitor cells reside within niches that are areas where many types of cytokines/growth factors and matrix factors regulate cells, one would expect HMGB1 to become secreted on such areas. In fact, in whole-mount immunostaining, HMGB1 can be detected in wild-type but not in knockdown zebrafish larvae at the plasma membrane level, resembling a matrix-type structure that surrounds the cells. We have previously shown that HMGB1 can regulate neural cells and other cell types in vitro as a cell matrix-associated molecule (for review, see Ref 2), which would be consistent with a role as a factor surrounding neural progenitors and regulating them through transmembrane signaling. However, one should keep in mind that HMGB1 may have important intracellular functions in neural progenitor regulation, a possibility that clearly warrants further studies. In the immune cell regulation, HMGB1 was recently shown to participate in both extracellular and intracellular mechanisms (39).
HMGB1 has several cell surface receptors, and further work is warranted to elucidate the relative importance of different membrane receptors of HMGB1 in neural progenitors. Regulation of many cell types by HMGB1 is generally suggested to depend, at least partially, on binding to RAGE. A search of the zebrafish genome data base does not identify any obvious RAGE homologue, although hypothetical protein structures displaying some similarity in the domain structure (for example, an Ig domain structure with ∼30% similarity compared with the HMGB1 binding Ig domain of mammalian RAGE) can be found in the data base. Interestingly, Toll-like receptors that were initially identified as proteins guiding neuronal development in Drosophila have been recently implicated in neurogenesis in mouse brain (40). Toll-like receptors have been identified as HMGB1 receptors in the immune system in several studies (for review, see Ref 2), and their possible role as HMGB1 receptors in brain development needs to be elucidated.
A search of genes regulated by HMGB1 in embryonic neural cells using ordered differential display analysis was the basis of cloning of a novel adhesion protein designated as AMIGO1 (20). In these in vitro studies regulation of AMIGO1 expression was shown to depend on extracellular matrix-bound HMGB1 and transmembrane signaling in embryonic neural cells. The finding that HMGB1 regulates AMIGO1 expression in a similar manner in vivo in zebrafish larvae provides further evidence for a role of HMGB1 as an extracellular factor that would mediate cell-to-cell communication within the niches that are required for survival/proliferation of neural progenitors.
HMGB1, Pax6, and Wnt Signaling
Of the regional markers of brain development analyzed in the current study, Pax6 expression is of particular interest in the HMGB1 knockdown zebrafish. Lack of HMGB1 clearly results in reduction of Pax6a-expressing cells in the forebrain. Interestingly, Wnt signaling has been shown to become up-regulated in Pax6−/− mutants and may be involved in the mechanism of Pax6 in forebrain development (33, 38). Our finding of enhanced Wnt signaling in the HMGB1 knockdown may, therefore, be due to down-regulation of Pax6 activity.
Wnt signaling has a complex role from very early to late stages of the nervous system development (for review, see Ref. 41). In particular, Wnt8 has been reported to play a key role in brain development (42, 43) and to affect development of catecholaminergic neural networks (44) that is compromised in the HMGB1 morphants. Wnt8 and β-catenin were even dramatically up-regulated in the HMGB1 knockdown, in contrast to AMIGO1 that essentially disappears in the knockdown. Wnt8 has been shown to act as a posteriorizing factor that is expressed in posterior parts of the central nervous system and is suggested to diffuse to anterior areas inhibiting its development (42, 43). Furthermore, Wnt8 has been shown to restrict the number of catecholaminergic progenitors during neurogenesis in diencephalon (44). Therefore, up-regulation of Wnt8 expression likely contributes to perturbed forebrain development in the HMGB1 morphants. The finding that HMGB1 signaling connects to Wnt signaling opens up novel views of the roles of HMGB1 in development and diseases such as inflammatory conditions and cancer. Further in vitro and in vivo studies on HMGB1/Pax6/Wnt connections are, therefore, warranted.
AMIGO1 in the Mechanism through Which HMGB1 Regulates Forebrain Development
The key question as regarding the regulation of AMIGO1 expression by HMGB1 is whether AMIGO1 should be included in the HMGB1-dependent survival/proliferation mechanism. In our previous studies we cloned two other proteins that are about 50% homologous as compared with AMIGO1 and designated these proteins as AMIGO2 and AMIGO3 (20). However, AMIGO2 was in addition independently cloned using neuron survival as a readout and is designated as Alivin-1 (45). We, therefore, decided to study whether regulation AMIGO1 expression by HMGB1 contributes to the survival/proliferation mechanism of HMGB1. Experiments in zebrafish in vivo using AMIGO1 cRNA injection in the HMGB1 knockdown clearly demonstrate that AMIGO1 should be included in the signaling pathway through which HMGB1 regulates neural progenitors. Furthermore, based on our in vitro studies, AMIGO1 has a similar cell survival enhancing effect on embryonic rat brain neurons as AMIGO2.4
Concluding Remarks
The current study shows that HMGB1 is critically important to construct forebrain structures in zebrafish, a phenomenon that is likely to be conserved across many vertebrate and nonvertebrate species. It appears clear that at the cellular level, regulation of survival and proliferation of neural progenitors is intimately involved in the cellular mechanism through which HMGB1 regulates brain development. As regarding the molecular mechanism through which HMGB1 regulates neural progenitors, further work is clearly warranted. Our findings suggesting roles for Pax6, Wnt8, and AMIGO1 in the molecular network affected by HMGB1 provide clues for further mechanistic studies on the role of HMGB1 in brain development. Furthermore, it seems clear to us that HMGB1 should be considered in regenerative medicine as a factor that regulates neural progenitors during regeneration.
Supplementary Material
Acknowledgments
The technical assistance of Henri Koivula is gratefully acknowledged. We thank Dr. Henri Huttunen for critical reading of the manuscript.
This work was supported by the Academy of Finland and the Sigrid Jusélius Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
J. Kuja-Panula, X. Zhao, and H. Rauvala, unpublished observations.
- HMGB1
- high mobility group (HMG) box-1 protein
- hpf
- hours post-fertilization
- dpf
- days post-fertilization
- ZIRC
- Zebrafish International Resource Center
- TH
- tyrosine hydroxylase
- MO
- morpholino oligonucleotides
- 5mis MO
- five mispaired MOs
- EdU
- 5-ethynyl-2′-deoxyuridine.
REFERENCES
- 1. Hock R., Furusawa T., Ueda T., Bustin M. (2007) Trends Cell Biol. 17, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rauvala H., Rouhiainen A. (2010) Biochim. Biophys. Acta 1799, 164–170 [DOI] [PubMed] [Google Scholar]
- 3. Stros M. (2010) Biochim. Biophys. Acta 1799, 101–113 [DOI] [PubMed] [Google Scholar]
- 4. Bonaldi T., Talamo F., Scaffidi P., Ferrera D., Porto A., Bachi A., Rubartelli A., Agresti A., Bianchi M. E. (2003) EMBO J. 22, 5551–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calogero S., Grassi F., Aguzzi A., Voigtländer T., Ferrier P., Ferrari S., Bianchi M. E. (1999) Nat. Genet. 22, 276–280 [DOI] [PubMed] [Google Scholar]
- 6. Rauvala H., Pihlaskari R. (1987) J. Biol. Chem. 262, 16625–16635 [PubMed] [Google Scholar]
- 7. Merenmies J., Pihlaskari R., Laitinen J., Wartiovaara J., Rauvala H. (1991) J. Biol. Chem. 266, 16722–16729 [PubMed] [Google Scholar]
- 8. Rauvala H., Merenmies J., Pihlaskari R., Korkolainen M., Huhtala M. L., Panula P. (1988) J. Cell Biol. 107, 2293–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huttunen H. J., Kuja-Panula J., Sorci G., Agneletti A. L., Donato R., Rauvala H. (2000) J. Biol. Chem. 275, 40096–40105 [DOI] [PubMed] [Google Scholar]
- 10. Taguchi A., Blood D. C., del Toro G., Canet A., Lee D. C., Qu W., Tanji N., Lu Y., Lalla E., Fu C., Hofmann M. A., Kislinger T., Ingram M., Lu A., Tanaka H., Hori O., Ogawa S., Stern D. M., Schmidt A. M. (2000) Nature 405, 354–360 [DOI] [PubMed] [Google Scholar]
- 11. Guérin A., d'Aubenton-Carafa Y., Marrakchi E., Da Silva C., Wincker P., Mazan S., Rétaux S. (2009) PLoS ONE 4, e5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang X., Wang L., Zhang H. (2005) Int. J. Dev. Biol. 49, 49–52 [DOI] [PubMed] [Google Scholar]
- 13. Kinoshita M., Hatada S., Asashima M., Noda M. (1994) FEBS Lett. 352, 191–196 [DOI] [PubMed] [Google Scholar]
- 14. Chen Y. C., Priyadarshini M., Panula P. (2009) Histochem. Cell Biol. 132, 375–381 [DOI] [PubMed] [Google Scholar]
- 15. Kaslin J., Panula P. (2001) J. Comp. Neurol. 440, 342–377 [DOI] [PubMed] [Google Scholar]
- 16. Westerfield M. (2000) The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th Ed., University of Oregon Press, Eugene, OR [Google Scholar]
- 17. Thisse C., Thisse B. (2008) Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- 18. Thisse C., Thisse B. (2005) High Throughput Expression Analysis of ZF-Models Consortium Clones, ZFIN Direct Data Submission [Google Scholar]
- 19. Parkkinen J., Raulo E., Merenmies J., Nolo R., Kajander E. O., Baumann M., Rauvala H. (1993) J. Biol. Chem. 268, 19726–19738 [PubMed] [Google Scholar]
- 20. Kuja-Panula J., Kiiltomäki M., Yamashiro T., Rouhiainen A., Rauvala H. (2003) J. Cell Biol. 160, 963–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. (2007) PLoS Genet. 3, 787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fink M., Flekna G., Ludwig A., Heimbucher T., Czerny T. (2006) Dev. Dyn. 235, 3370–3378 [DOI] [PubMed] [Google Scholar]
- 23. Blin M., Norton W., Bally-Cuif L., Vernier P. (2008) Mol. Cell. Neurosci. 39, 592–604 [DOI] [PubMed] [Google Scholar]
- 24. Sallinen V., Torkko V., Sundvik M., Reenilä I., Khrustalyov D., Kaslin J., Panula P. (2009) J. Neurochem. 108, 719–731 [DOI] [PubMed] [Google Scholar]
- 25. Mueller T., Wullimann M. F. (2005) Atlas of Early Zebrafish Brain Development: A Tool for Molecular Neurogenetics, pp. 29–120, Elsevier, Amsterdam [Google Scholar]
- 26. Rink E., Wullimann M. F. (2002) Brain Res. Dev. Brain Res. 137, 89–100 [DOI] [PubMed] [Google Scholar]
- 27. Eisen J. S., Smith J. C. (2008) Development 135, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 28. Wullimann M. F., Mueller T. (2004) J. Comp. Neurol. 475, 143–162 [DOI] [PubMed] [Google Scholar]
- 29. Appel B. (2000) Dev. Dyn. 219, 155–168 [DOI] [PubMed] [Google Scholar]
- 30. Guo S., Wilson S. W., Cooke S., Chitnis A. B., Driever W., Rosenthal A. (1999) Dev. Biol. 208, 473–487 [DOI] [PubMed] [Google Scholar]
- 31. Holzschuh J., Ryu S., Aberger F., Driever W. (2001) Mech. Dev. 101, 237–243 [DOI] [PubMed] [Google Scholar]
- 32. Cole L. K., Ross L. S. (2001) Dev. Biol. 240, 123–142 [DOI] [PubMed] [Google Scholar]
- 33. Manuel M., Price D. J. (2005) Brain Res. Bull. 66, 387–393 [DOI] [PubMed] [Google Scholar]
- 34. Hébert J. M., Fishell G. (2008) Nat. Rev. Neurosci. 9, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oxtoby E., Jowett T. (1993) Nucleic Acids Res. 21, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wullimann M. F., Rink E. (2002) Brain Res. Bull. 57, 363–370 [DOI] [PubMed] [Google Scholar]
- 37. Martin B. L., Kimelman D. (2009) Curr. Biol. 19, R215–R219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muzio L., DiBenedetto B., Stoykova A., Boncinelli E., Gruss P., Mallamaci A. (2002) Cereb. Cortex 12, 129–139 [DOI] [PubMed] [Google Scholar]
- 39. Yanai H., Ban T., Wang Z., Choi M. K., Kawamura T., Negishi H., Nakasato M., Lu Y., Hangai S., Koshiba R., Savitsky D., Ronfani L., Akira S., Bianchi M. E., Honda K., Tamura T., Kodama T., Taniguchi T. (2009) Nature 462, 99–103 [DOI] [PubMed] [Google Scholar]
- 40. Rolls A., Shechter R., London A., Ziv Y., Ronen A., Levy R., Schwartz M. (2007) Nat. Cell Biol. 9, 1081–1088 [DOI] [PubMed] [Google Scholar]
- 41. Ciani L., Salinas P. C. (2005) Nat. Rev. Neurosci. 6, 351–362 [DOI] [PubMed] [Google Scholar]
- 42. Houart C., Caneparo L., Heisenberg C., Barth K., Take-Uchi M., Wilson S. (2002) Neuron 35, 255–265 [DOI] [PubMed] [Google Scholar]
- 43. Kim J. D., Chun H. S., Kim S. H., Kim H. S., Kim Y. S., Kim M. J., Shin J., Rhee M., Yeo S. Y., Huh T. L. (2009) Biochem. Biophys. Res. Commun. 381, 717–721 [DOI] [PubMed] [Google Scholar]
- 44. Russek-Blum N., Gutnick A., Nabel-Rosen H., Blechman J., Staudt N., Dorsky R. I., Houart C., Levkowitz G. (2008) Development 135, 3401–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ono T., Sekino-Suzuki N., Kikkawa Y., Yonekawa H., Kawashima S. (2003) J. Neurosci. 23, 5887–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.