Abstract
Filaggrin-2 (FLG2), a member of the S100-fused type protein family, shares numerous features with filaggrin (FLG), a key protein implicated in the epidermal barrier functions. Both display a related structural organization, an identical pattern of expression and localization in human epidermis, and proteolytic processing of a large precursor. Here, we tested whether FLG2 was a substrate of calpain 1, a calcium-dependent protease directly involved in FLG catabolism. In addition, deimination being critical for FLG degradation, we analyzed whether FLG2 deimination interfered with its proteolytic processing. With this aim, we first produced a recombinant form of FLG2 corresponding to subunits B7 to B10 fused to a COOH-terminal His tag. Incubation with calpain 1 in the presence of calcium induced a rapid degradation of the recombinant protein and the production of several peptides, as shown by Coomassie Blue-stained gels and Western blotting with anti-FLG2 or anti-His antibodies. MALDI-TOF mass spectrometry confirmed this result and further evidenced the production of non-immunoreactive smaller peptides. The degradation was not observed when a calpain 1-specific inhibitor was added. The calpain cleavage sites identified by Edman degradation were regularly present in the B-type repeats of FLG2. Moreover, immunohistochemical analysis of normal human skin revealed colocalization of FLG2 and calpain 1 in the upper epidermis. Finally, the FLG2 deiminated by human peptidylarginine deiminases was shown to be more susceptible to calpain 1 than the unmodified protein. Altogether, these data demonstrate that calpain 1 is essential for the proteolytic processing of FLG2 and that deimination accelerates this process.
Keywords: Calpain, Differentiation, Mass Spectrometry (MS), Post-translational Modification, Protease, Skin, Corneocytes, Epidermis, Keratinocytes, Peptidylarginine Deiminase
Introduction
In the epidermis, the program of keratinocyte terminal differentiation is an oriented process during which cells of the basal layer undergo a series of metabolic and structural changes throughout their migration to the surface of the tissue. The stratum corneum, the outermost layer of the epidermis, is formed by the stacking of so-called corneocytes, the end products of the process. The stratum corneum functions as an effective barrier between the body and its outside environment, limiting skin dehydration and preventing the penetration of outside pathogens, UV radiation, and exogenous chemicals. It also contributes to mechanical protection of the body. So that this function can be achieved, peculiar structures are formed, such as the cornified cell envelope, a resistant and insoluble protein shell that replaces the plasma membrane, and the intracorneocyte fibrous matrix made by the aggregation of keratin intermediate filaments.
The epidermal differentiation complex comprises a large number of genes that are of crucial importance for keratinocyte differentiation. So far, at least 45 genes encoding structural and regulatory proteins have been mapped within this 2-Mb region on chromosome 1q21 (1). Most of them were found to be organized in four clusters: (i) two clusters of genes encoding proteins of the cornified cell envelope, namely LCE (late cornified envelope) proteins (2) and SPR (small proline-rich) proteins (3); (ii) genes encoding EF-hand calcium-binding proteins of the S100A family (4); and (iii) genes of the seven known S100-fused type proteins: profilaggrin (5), hornerin (HRNR)2 (6–8), filaggrin-2 (FLG2) (9), repetin (10), cornulin (11), trichohyalin-like 1 (12), and trichohyalin (13).
Profilaggrin is the best characterized of the S100-fused type proteins (for a review, see Refs. 14 and 15). It consists of 10–12 tandem repeats of filaggrin (FLG) joined by a short hydrophobic linker peptide and flanked by NH2- and COOH-unique terminal domains. It is synthesized by granular layer keratinocytes, where it is stored in cytoplasmic granules, the so-called keratohyalin granules. During cornification, the transition from granular keratinocyte to corneocyte, profilaggrin is enzymatically proteolyzed into mature basic FLG monomers. Subsequently, FLG associates with intermediate filaments to promote the formation of macrofibrils and the fibrous matrix (16). Then, deimination of FLG arginyl residues by peptidylarginine deiminases types 1 and 3 (PAD1 and PAD3) results in the dissociation of FLG from the matrix (17–19) and promotes its degradation into free amino acids that are important for retaining water in the stratum corneum, in particular pyrollidone carboxylic acid derived from Gln, and for UV protection, in particular urocanic acid derived from His (20, 21). Several proteases, mainly calpain 1 and bleomycin hydrolase, are involved in this breakdown (22, 23). The importance of FLG in skin homeostasis was recently demonstrated when non-sense mutations of its gene were shown to be responsible for ichthyosis vulgaris and to be associated with atopic dermatitis, two frequent skin diseases (24–26).
FLG2, a more recently identified member of the family (9, 27) shares a number of common features with the other S100-fused type proteins. Its gene consists of three exons: a small 5′ non-coding exon, a small second exon containing the translation start site, and a very large third exon encoding most of the protein. FLG2 is mainly detected in skin, particularly in granular keratinocytes of the epidermis (9). Similarly to the FLG gene, FLG2 gene expression is probably regulated at the transcriptional level because the corresponding mRNA level is dramatically (800-fold) increased in granular compared with basal keratinocytes (27). FLG2 is a 2391-amino acid-long protein with a predicted NH2-terminal calcium binding domain homologous to S100 proteins followed by a large repetitive region containing two types of tandem repeats, each 75–77 amino acids long. The nine A-type repeats (A1–A9) are homologous (50–77% identity) to the repeats of HRNR (9), a component of cornified cell envelopes, as recently demonstrated in our laboratory (28). The fourteen B-type repeats (B1–B14) are closer to FLG units (28–39% identity) (9). The B-type repeats in particular are rich in Gln (8.4%) and in the basic amino acids Arg (8.4%) and His (14.9%), as FLG is. Furthermore, FLG2 has been shown, like FLG and HRNR, to accumulate in the keratohyalin granules and to be proteolytically processed later during cornification (9). To date, however, the biological function of FLG2 is unknown. Considering the essential role of FLG in keratinocyte terminal differentiation and the similarity of FLG2 to profilaggrin, it is important to determine the biochemical properties of FLG2.
In this study, we demonstrate that the FLG2 B-type repeat domain is maturated by proteolysis and that the serine protease, calpain 1, is involved in the processing. Furthermore, we also show that FLG2 is a substrate of PADs, the deimination promoting its proteolysis by calpain 1. Finally, we discuss the role of FLG2 in the stratum corneum.
EXPERIMENTAL PROCEDURES
Materials
Human calpain 1 was purchased from Calbiochem. N-Acetyl-Leu-Leu-norleucine-CHO (ALLN), a calpain inhibitor, was from Sigma-Aldrich. Recombinant human PADs were produced and purified as reported previously (19).
Antibodies
A polyclonal rabbit anti-FLG2 serum against a synthetic peptide (IP2; Fig. 1A) present on repeat B10, with an added NH2-terminal Cys residue for coupling purposes (CTVHKRHQTTHGQ) was produced by Génosphère (Paris, France). Anti-peptide anti-FLG2 antibodies were purified by affinity chromatography against the synthetic peptide covalently bound on a SulfoLink® Resin (SulfoLink® immobilization kit for peptides) as described by the manufacturer (Pierce). A goat polyclonal antibody directed against calpain 1 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and monoclonal antibodies to tetra-His (anti-His; Qiagen, Venlo, The Netherlands), actin (MAB1501; Chemicon International, Southampton, UK), and both profilaggrin and FLG (AHF3 (29)) were also used. Anti-mouse Flg2 antibodies were purified from a previously reported serum directed against the human FLG2 spacer (9) by affinity chromatography on a recombinant form of the mouse Flg2 NH2-terminal spacer domain (amino acids 96–231). The purified immunoglobulins were shown to specifically recognize both human FLG2 and mouse Flg2.3
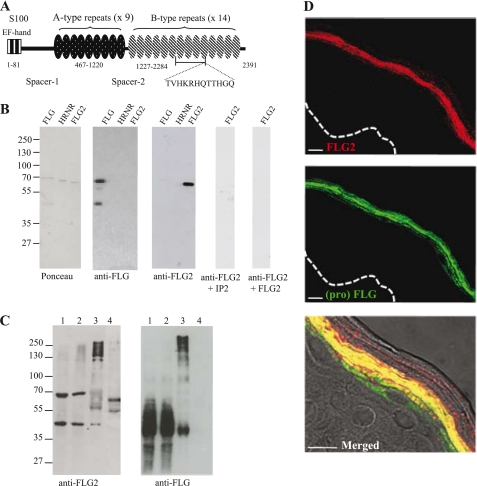
FIGURE 1.
Expression of FLG2 in normal human epidermis. A, schematic representation of human FLG2 shows the NH2 terminus EF-hand domain, the spacer 1 and 2 domains, and the repetitive region containing nine A-type repeats (A1–A9) and fourteen B-type repeats (B1–B14). The position and sequence of the peptide IP2 used for immunization is indicated. The underlined B7–B10 repeats correspond to the recombinant FLG2 produced. Numbers indicate amino acid residues. B, purified recombinant FLG, FLG2, and HRNR were separated by SDS-PAGE, Ponceau Red-stained, and analyzed by immunoblotting with the anti-FLG2 polyclonal and anti-FLG (namely AHF3) monoclonal antibodies. When indicated, the anti-FLG2 antibodies were preincubated with either IP2 or the recombinant GST-FLG2. C, epidermal proteins were sequentially extracted in different isotonic buffers containing EDTA (lane 1), Nonidet P-40 (lane 2), 8 m urea (lane 3), or urea and DTT (lane 4). Equal amounts of proteins were separated by SDS-PAGE, Ponceau Red stained and analyzed by immunoblot with the anti-FLG2 and AHF3 antibodies. Molecular masses of standards are indicated in kDa to the left. D, sections of paraffin-embedded normal human skin were doubly stained with the anti-FLG2 (red) and AHF3 (green) antibodies. The merged image shows the colocalization of FLG2 and either profilaggrin or FLG in the cytoplasm of the upper granular cells and in the matrix of the lower corneocytes, respectively. Scale bars, 10 μm.
Alexa Fluor® 488- and 555-conjugated donkey anti-rabbit, Alexa Fluor® 488-conjugated donkey anti-mouse, and Alexa Fluor® 555-conjugated donkey anti-goat secondary antibodies were purchased from Invitrogen. For immunoblotting analyses, peroxidase-conjugated goat anti-rabbit and anti-mouse secondary antibodies were purchased from Zymed Laboratories Inc. (San Francisco, CA).
Extraction of Epidermal Proteins
All experiments were performed according to the principles of the Declaration of Helsinki and with appropriate approval from the University of Toulouse ethics committee. Human abdominal epidermis from healthy donors undergoing plastic surgery was cleaved from the dermis, and proteins were sequentially extracted as described previously (30). Briefly, the epidermis was sequentially homogenized on ice in equal volumes of the following buffers: TE buffer (40 mm Tris-HCl, pH 7.4, 10 mm EDTA, 0.25 mm PMSF with 0.2% (v/v) protease inhibitor mixture (Sigma-Aldrich), TE-Nonidet P-40 buffer (TE buffer containing 0.5% Nonidet P-40), TEU buffer (TE buffer with 8 m urea), and TEUD buffer (TEU buffer with 50 mm dithiothreitol (DTT)). After each extraction, the homogenates were centrifuged for 15 min at 15,000 × g and the supernatants were kept at −20 °C until used.
Mouse skin samples were scraped off with a scalpel blade to get the epidermis. Epidermal proteins were then sequentially extracted as described above with TE-Nonidet P-40 and TEU buffers.
Immunoblotting Analyses
Samples were separated by 10% (or 7.5% when indicated) SDS-PAGE and transferred to nitrocellulose membrane. The blots were probed with primary antibodies and peroxidase-conjugated secondary antibodies. Detection was performed with ECL reagent (GE Healthcare, Little Chalfont, UK). The anti-FLG2 and AHF3 were diluted to 1:200 and to 1:5000, respectively. Anti-His and anti-mouse Flg2 were diluted to 1:1000. Anti-actin was used at 1:30,000. Peroxidase-coupled secondary antibodies were used according to the manufacturer's recommendations. Immunoreactive bands were scanned and quantified by densitometry using the National Institutes of Health ImageJ software available on the World Wide Web.
Expression and Purification of Recombinant COOH-terminal His6-tagged Proteins
Human FLG and FLG2 cDNAs were amplified by PCR with the following primers derived from the published sequences (GenBankTM accession numbers AF043380 and AY827490, respectively). 5′-CATATGCTATACCAGGTGAGCACTCATG-3′ (an added NdeI restriction site is underlined) and 5′-CTCGAGCCCTGAACGTCCAGACCGTCC-3′ (an added XhoI restriction site is underlined) were primers for FLG, and 5′-CATATGCAGTCAGAATCCATAGTTCC-3′ (an added NdeI restriction site is underlined) and 5′-CTCGAGTGGATAGTGAGATCCAGC-3′ (an added XhoI restriction site is underlined) were primers for FLG2. The resulting PCR products were cloned into TA vector (Invitrogen) and verified by sequencing. The PCR fragments were then subcloned into NdeI/XhoI-digested pET41b vector (Merck KGaA, Darmstadt, Germany), allowing inducible expression of the COOH-terminal His6-tagged proteins in the Escherichia coli strain BL21-Condon Plus (DE3+)-RiL (Stratagene, La Jolla, CA). Bacteria were transformed with the various recombinant expression plasmids and grown in LB broth medium (Invitrogen) supplemented with kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml). Cultures were incubated at 37 °C until cell density reached an A600 nm of 0.6–0.8. Then the bacteria were induced to express the recombinant proteins for 3 h by the addition of 0.5 and 0.1 mm IPTG to the culture medium for FLGHis and FLG2His, respectively. Cultures were centrifuged for 10 min at 6,000 × g, and recombinant proteins extracted from cell pellets with 8 m urea were affinity-purified from bacterial lysates using a Hi-Trap column (GE Healthcare), following the manufacturer's protocol. Protein expression was verified by SDS-PAGE and immunodetection with the anti-His antibody.
In Vitro Deimination Assays
Recombinant human FLGHis and FLG2His (100 ng/test) were incubated at 50 °C for 19 h in 100 mm Tris-HCl, pH 7.4, buffer containing 10 mm CaCl2, 5 mm DTT with or without 40 milliunits of human PAD1, PAD2, or PAD3 (19). The reactions were stopped by the addition of Laemmli's sample buffer and boiling for 3 min. Equal amounts of proteins were then separated by SDS-PAGE and immunodetected with the anti-His antibody.
In Vitro Calpain 1 Cleavage Assays
The cleavage of unmodified and deiminated proteins was performed at an enzyme/substrate molar ratio of 1:50. Purified recombinant FLGHis and FLG2His (15.8 μm) were incubated with calpain 1 (0.32 μm) in 100 mm Tris-HCl, pH 7.5, 2 mm CaCl2, and 10 mm DTT at 30 °C. A calpain 1 inhibitor, ALLN, was also present, when indicated, at a concentration of 50 or 100 μm. All reactions were stopped at the indicated time by the addition of Laemmli's sample buffer and boiling for 10 min. Proteins were then separated by 10% SDS-PAGE and stained with Coomassie Blue or assessed by immunoblotting.
Mass Spectrometry Analysis
Equal amounts of FLG2His and deiminated FLG2His were taken for matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) analyses. The samples were manipulated together with exactly the same procedure, and results were acquired in the same time. Calpain 1 digests were dried and reconstituted in 10 μl of a 0.3% trifluoroacetic acid, 30% acetonitrile solution. For each sample, a 0.75-μl drop of analytes solution was deposited on a MALDI plate, and a 0.75-μl drop of matrix (3,5-dimethoxy-4-hydroxy-cinnamic acid (sinapinic acid; Sigma-Aldrich)) dissolved at 30 mg/ml in a 0.3% trifluoroacetic acid, 30% acetonitrile aqueous solution was then added. The samples were then left to dry completely in air. MALDI-TOF analyses were performed on a Perseptive DE-STR Voyager, (Perseptive Biosystems, Framingham, MA) mass spectrometer in positive linear mode using an accelerating voltage of 20 kV and a 500 ns delay time. Mass spectra were acquired for m/z 500–50,000 (2,000 shots/spectrum). External calibration was performed with bovine serum albumin from the Sequazyme BSA test standard kit (Applied Biosystems) with a mass precision on the order of 50 Da after calibration. Spectra were analyzed using the Data Explorer software (Applied Biosystems).
NH2-terminal Amino Acid Sequencing
Recombinant FLG2His and calpain 1 were incubated as indicated above for 5 min, and the reaction was stopped by the addition of EDTA to obtain a final concentration of 10 mm. The reaction products were resolved by SDS-PAGE, transferred onto a polyvinylidene fluoride membrane (Applied Biosystems), and stained with Coomassie Blue. The membrane areas corresponding to some calpain-produced FLG2 peptides were excised. The NH2 terminus of the fragments was sequenced for up to 9 Edman degradation cycles on an Applied Biosystems 494A sequencer following the manufacturer's specifications in the Laboratoire de Microséquençage des Protéines of the Institut Pasteur (Paris, France).
Indirect Immunofluorescence Staining
The anti-FLG2 and AHF3 antibodies were diluted to 1:50 and 1:2,000, respectively. The anti-mouse Flg2 was diluted to 1:50. For the commercially available primary and secondary antibodies, we followed the manufacturer's recommendations. Staining intensities were quantified by densitometry using the NIH ImageJ software. Co-immunolocalization experiments were performed on sections of paraffin-embedded formaldehyde-fixed human skin. Deparaffinized and rehydrated 5-μm sections were processed for heat-induced epitope retrieval at 95 °C for 45 min using 50 mm glycine solution, pH 3.5. The sections were subsequently incubated overnight with either rabbit anti-FLG2 or mouse anti-FLG antibodies and then with goat anti-calpain 1 antibodies at 4 °C. Alexa Fluor-conjugated secondary antibodies were used for primary antibody detection. Fluorescence was analyzed using an inverted Zeiss LSM 710 confocal microscope (Carl Zeiss, Oberkochen, Germany).
Mice
Transgenic C57BL/6 mice constitutively overexpressing rabbit calpastatin (under the control of the cytomegalovirus immediate early enhancer/promoter region) have been described previously (31). Samples of back skin were obtained from 8-week-old transgenic and wild type mice.
Statistical Analysis
Unilateral Student's t test was performed for statistical evaluation of the data.
RESULTS
Expression and Localization of FLG2 in Normal Human Epidermis
To analyze the expression of FLG2 and a putative processing similar to that of FLG, sera were produced in rabbits and affinity-purified against a synthetic peptide (TVHKRHQTTHGQ; IP2) present within a B-type repeat of FLG2 (i.e. B10) (Fig. 1A). We previously checked in silico that this sequence did not show any similarities with other proteins, including the other S100-fused type proteins. The specificity of the affinity-purified anti-FLG2 antibodies was tested by immunoblotting on a FLG subunit, the FLG2 segment B7–B10 (residues 1683–1984), and HRNR, all produced in Escherichia coli as GST-fused recombinant proteins as described previously (28). The anti-FLG2 antibodies only recognized FLG2. Their reactivity was completely suppressed by preincubation with either the peptide used for immunization or the recombinant FLG2 (Fig. 1B) but not with two other synthetic peptides derived from the sequence of the B-type repeat domain of FLG2. Moreover, the reactivity of the monoclonal antibody AHF3, directed against FLG and profilaggrin, was not altered by preincubation with any of the three peptides. Further confirming the specificity of the anti-FLG2 antibodies, no reactivity was observed using the preimmune sera (data not shown).
To characterize the biochemical properties of FLG2, normal human epidermis was sequentially homogenized on ice in the presence of protease inhibitors, in equal volumes of a Tris-HCl buffer containing either EDTA (TE buffer), the non-ionic detergent Nonidet P-40 (TE-Nonidet P-40 buffer), urea (TEU buffer) or, finally, urea and the reducing agent DTT (TEUD buffer). Extracted proteins were then separated by SDS-PAGE and immunoblotted with the anti-FLG2 antibodies (Fig. 1C, left). In the first two extracts (TE and TE-Nonidet P-40 buffers), two proteins of ∼65 and ∼45 kDa were immunodetected (lanes 1 and 2). In addition, several immunoreactive proteins with a high molecular mass (from 250 to 130 kDa) were extracted only with TEU buffer (lane 3). The largest corresponded to the expected full size of FLG2 (∼248 kDa). Furthermore, several immunodetected proteins were extracted only in the presence of DTT (lane 4). The detection of multiple proteins smaller than the expected size of the full-length FLG2 indicates a possible proteolytic processing of a precursor as is already known for profilaggrin (32) and shown in Fig. 1C (right).
To locate FLG2 in the epidermis, 5-μm sections of formaldehyde-fixed paraffin-embedded healthy skins were analyzed by indirect immunofluorescence with both the anti-FLG2 antibodies and AHF3. FLG2 (red) was detected in the upper granular layer and in the lower stratum corneum (Fig. 1D). The labeling was completely suppressed by preincubation with either the peptide used for immunization or the purified recombinant FLG2 but not with the control peptides described above (data not shown). The merged image (Fig. 1D, bottom) clearly shows that FLG2 is expressed slightly later than profilaggrin (green) because it is not observed in the lowest granular keratinocyte in which profilaggrin is already evidenced. In the upper granular cells, FLG2 labeling was granular. In the lower stratum corneum, FLG2 and FLG were detected in the intracorneocyte fibrous matrix, whereas in the upper layer, they were not detected or only slightly detected. This parallel disappearance of the two proteins suggests that FLG2 may be degraded in the upper stratum corneum by one of the proteases involved in the degradation of FLG (e.g. calpain 1).
Proteolysis of FLG2 by Calpain 1
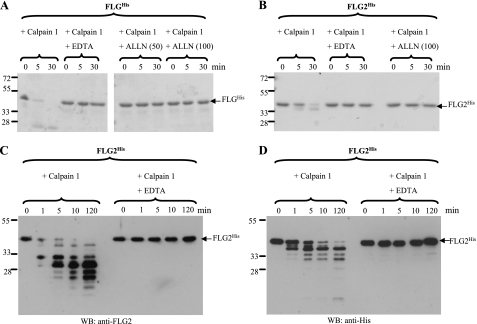
To test our hypothesis, a pure recombinant FLG2 fragment corresponding to repeat B7–B10 was produced with a COOH-terminal His tag (called FLG2His) and incubated with calpain 1. A recombinant FLG subunit with such a His tag (named FLGHis) was used as a positive control. The reaction products were then separated by SDS-PAGE followed by Coomassie Blue staining. As shown in Fig. 2, A and B, both FLGHis and FLG2His were rapidly degraded during the incubation with calpain 1 in the presence of 2 mm calcium ion. The calcium chelator EDTA and the calpain inhibitor ALLN prevented this degradation, indicating the specificity of the proteolysis. This result is consistent with the hypothesis that FLG2 is a direct substrate of calpain 1. The FLG2His proteolytic fragments were further examined by immunoblotting with the anti-FLG2 and anti-His antibodies (Fig. 2, C and D). This analysis confirmed the rapid and specific degradation of FLG2His by calpain 1 and highlighted the time-dependent appearance of many proteolytic fragments from 40 to 15 kDa. This indicated that multiple calpain 1 cleavage sites are present in the sequence of the FLG2 repeats B7–B10. Interestingly, the immunoblotting patterns produced by the anti-His and anti-FLG2 antibodies were different, suggesting that cleavage occurred at one site (at least) downstream of the epitope(s) recognized by the anti-FLG2 antibodies and very close to the COOH terminus of the protein, leading to the removal of the His6 epitope.
FIGURE 2.
Degradation of FLGHis and FLG2His by calpain 1 in vitro. A and B, recombinant FLGHis (A) and FLG2His (B) were incubated at 30 °C with calpain 1 in a calcium-containing buffer for the indicated time (min). When specified, 10 mm EDTA or the calpain 1 inhibitor ALLN at a final concentration of either 50 or 100 μm was added before incubations. The reactions were stopped by the addition of Laemmli's sample buffer. Samples were then subjected to SDS-PAGE, and proteins were stained with Coomassie Blue. C and D, recombinant FLG2His was incubated at 30 °C for the indicated time (min) with calpain 1 in the absence or presence of 10 mm EDTA. The reactions were stopped by the addition of Laemmli's sample buffer. Samples were then immunoblotted with anti-FLG2 (C) or anti-His (D) antibodies. Molecular masses of standards are indicated in kDa to the left. The arrows show the full-length FLGHis and FLG2His, as indicated. WB, Western blot.
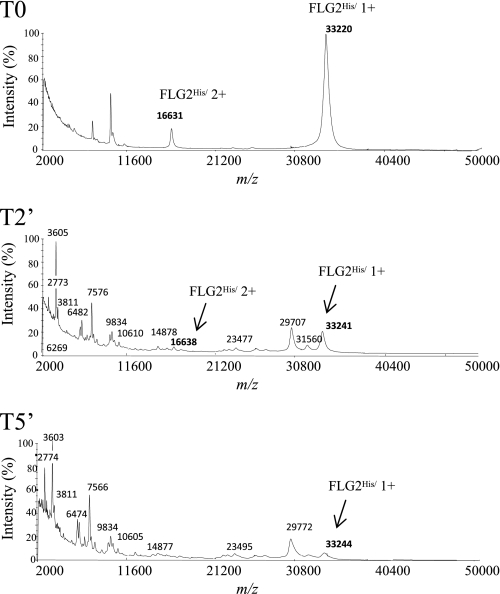
To further characterize the proteolysis of FLG2 by calpain 1, the intact recombinant FLG2His and a time course of degradation of the protein by this protease were analyzed by MALDI-TOF MS. The representative MS spectra obtained at m/z 2,000–50,000 are shown in Fig. 3. At the earliest time point (T0), two peaks at m/z 33,220 and 16,631, which can be assigned to singly and doubly charged molecular ions of recombinant FLG2His, were observed (the theoretical molecular mass of FLG2His is 33.13 kDa). These signals were dramatically reduced after a 2-min incubation of FLG2His with calpain 1, few peaks appearing at m/z between 33,000 and 20,000 (T2). The new peaks probably corresponded to some of the previously immunodetected FLG2His-derived fragments. Interestingly, several signals emerged at m/z 2,000–20,000, indicating that calpain 1 degraded FLG2His into several small peptides of different masses that were not immunodetected by the anti-His and anti-FLG2 antibodies in the immunoblotting conditions we used. In the spectra of the products of FLG2His proteolysis for 5 min (T5), the peaks at m/z 33,220 and 16,631 almost disappeared. Altogether, these data show that calpain 1 progressively processed the B-type repeats of FLG2 at multiple cleavage sites to generate numerous peptides.
FIGURE 3.
MALDI-TOF MS analysis of the peptides produced by calpain 1 digestion of FLG2His. Recombinant FLG2His was incubated with calpain 1 at 30 °C for 5 min. At the indicated times (0 min (T0), 2 min (T2′), and 5 min (T5′)), aliquots were removed and analyzed by MALDI-TOF MS as described under “Experimental Procedures.” Spectra at m/z 2,000–50,000 are shown. The m/z of the full-length FLG2His and calpain 1-produced fragments and peptides are indicated.
Identification of the Calpain 1 Cleavage Sites by Edman Degradation Sequencing
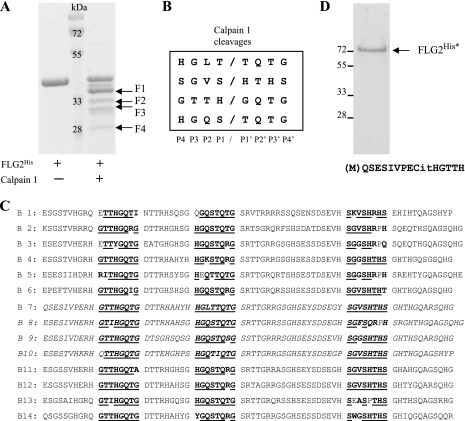
Because no consensus sequence has been clearly proposed for the calpain 1 cleavage site, we next defined the location of these sites within the B-type repeats of FLG2 by Edman degradation sequencing. In this analysis, we tried to identify the preferential rather than all cleavage sites. With this aim, a larger amount of FLG2His was digested with calpain 1 so as to obtain visible proteolytic fragments after SDS-PAGE and Coomassie Blue staining (Fig. 4A). The four most visible proteolytic fragments (F1, F2, F3, and F4), were sequenced. After nine cycles of Edman degradation, the sequences obtained were as follows: TQTGSRTTG for F1, HTHSGHTHG for F2, GQTGDTTRH for F3, and two sequences for F4: TGTGSRTTG (major) and MQSESIVPE (minor). Based on the peptide sequences and the molecular weights of the fragments, four cleavage sites were mapped on FLG2His as follows (from P4 to P4′): HGLT↓TQTG, SGVS↓HTHS, GTTH↓GQTG, and HGQS↓TQTG (Fig. 4B).
FIGURE 4.
Identification of calpain 1 cleavage sites by Edman degradation sequencing. A, recombinant FLG2His was separated by SDS-PAGE and Coomassie Blue-stained before and after digestion with calpain 1, as indicated. Four FLG2His fragments (F1–F4) produced by calpain 1 cleavage were selected for Edman degradation sequencing. Molecular masses of standards are indicated in kDa. B, from the NH2-terminal sequences of fragments F1–F4, four calpain 1 cleavage sites were identified. C, these and related calpain 1 cleavage motifs were shown to be present many times within the 14 B-type repeats of FLG2. Homologous amino acids are shown in boldface type, whereas identical amino acids are in boldface type and underlined. The amino acid sequence of B7–B10 repeats corresponding to the recombinant FLG2His is shown in italic type. D, the fully deiminated FLG2His (FLG2His*) was separated by SDS-PAGE and Coomassie Blue-stained, and its NH2 terminus was sequenced using Edman degradation. The obtained sequence is shown. The positions of molecular mass standards are indicated on the left.
On the basis of these four identified cleavage sites, we next searched for any other calpain 1 potential cleavage sites on the entire B-type repeat domain. Numerous identical or very similar amino acid motifs were shown to be regularly present within all B-type repeats (Fig. 4C). This result clearly explains that calpain 1 digestion of FLG2His produces many small proteolytic peptides, as shown by the MS analysis. It also suggests that calpain 1 could potentially cleave the entire B-type repeat domain of FLG2 into small peptides (from 17 to 30 amino acids), as already known for FLG (23). Curiously, such motifs were not observed in the FLG2 A-type repeats.
Localization of FLG2 and Calpain 1 in Human Epidermis
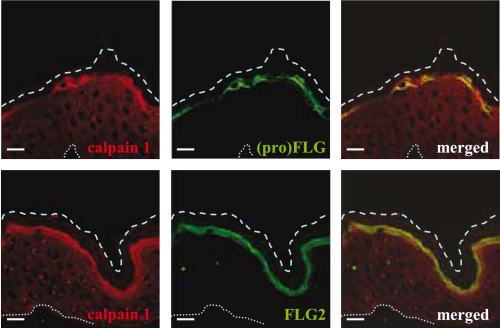
To better characterize the interaction between calpain 1 and FLG2, we next verified that they were colocalized in the upper part of the epidermis by indirect immunofluorescence analysis of human skin sections. We performed double-label staining and confocal microscopy analysis using a goat anti-calpain 1 antibody and either the anti-FLG2 or anti-FLG antibodies. On control sections incubated in the absence of primary antibody, no significant immunoreactivity was observed (data not shown). Calpain 1 was predominantly detected in the cytoplasm of granular keratinocytes and in the lower corneocytes (Fig. 5). It was shown to be colocalized with both (pro)filaggrin and FLG2, as pointed out by the merged images. This indicated a similar spatial distribution of the three proteins, supporting the idea that calpain 1 is involved in the degradation of both FLG and FLG2 in vivo.
FIGURE 5.
Indirect immunofluorescence localization of calpain 1 and FLG2 in human epidermis. Normal human skin sections were analyzed by confocal microscopy using anti-calpain 1, anti-FLG2, and AHF3 antibodies. The images show calpain 1 in red and either FLG2 or both FLG and profilaggrin in green. Comparative localizations of calpain 1 and its potential substrate are shown in the merged images. The upper dashed and the bottom dotted lines indicate the outer borders of the stratum corneum and the dermo-epidermal junction, respectively. Scale bars, 10 μm.
To further test for a possible cleavage of the epidermal FLG2 by calpain, an extract of human epidermis, prepared in the presence of a non-ionic detergent but in the absence of added protease inhibitors and known to contain partially processed FLG2, was incubated with or without calpain 1. The results (supplemental Fig. 1) showed that the epidermal FLG2 was proteolyzed in the presence of calpain 1 but not in the presence of both calpain 1 and ALLN. Moreover, the anti-FLG2-immunodetected peptides generated from the human epidermal and the recombinant forms of FLG2 presented similar molecular masses between 25 and 30 kDa.
FLG2 Is a Substrate of Calpain 1 in Vivo
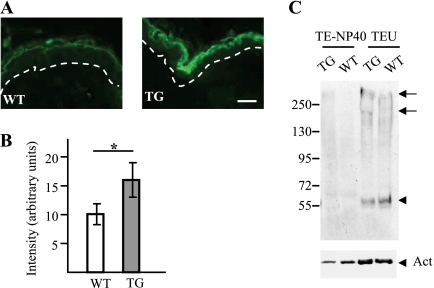
To test for the involvement of calpain 1 in vivo, the expression of Flg2 in the skin of wild type and transgenic mice overexpressing calpastatin, an inhibitor of calpain 1, was analyzed by indirect immunofluorescence and immunoblotting. The used affinity-purified antibodies recognized the NH2 terminus of Flg2 (spacer 1 region). The anti-Flg2 antibodies produced the same pattern of labeling when tested on wild type and transgenic mouse skin. Only the granular keratinocytes and the lower corneocytes were stained. However, the intensity of labeling was significantly higher on the epidermis of the transgenic mice (Fig. 6, A and B). The epidermal proteins were then sequentially extracted in equal volumes of TE-Nonidet P-40 and TEU buffers and immunoblotted with the anti-Flg2 antibodies (Fig. 6C). The full-size Flg2 (>250 kDa), a processed form of 200 kDa and a fragment of ∼55 kDa were essentially detected in the TEU buffer extracts obtained from both wild type and transgenic mice. However, in the extracts of transgenic mouse epidermis, the intensity of detection of the two larger bands was higher (by a mean factor of 2.12 after normalization against detection intensities of actin), whereas that of the 55 kDa band was lower (by a factor of 0.62). These data demonstrate an accumulation of the precursor form and a reduced processing of the protein in the transgenic mice. Unfortunately, no antibodies are yet available to analyze the processing of the repeat domain of mouse Flg2. Altogether, these data showed that calpain 1 is very likely involved in the processing of FLG2 in vivo.
FIGURE 6.
Expression of Flg2 in the epidermis of wild type and calpastatin transgenic mice. A, representative sections of paraffin-embedded skin of wild type (WT) and calpastatin-overexpressing transgenic (TG) mice were analyzed by indirect immunofluorescence with anti-Flg2 antibodies. Flg2 was located in the granular cells and the lower corneocytes. The dotted lines indicate the dermo-epidermal junction. Scale bars, 10 μm. B, the intensity of detection was quantified (n = 4) and shown to be higher in the TG mice. *, p < 0.05. C, epidermal proteins from WT and transgenic mice were sequentially extracted in isotonic buffers containing either Nonidet P-40 (TE-NP40) or 8 m urea (TEU). Equal volumes of each extracts were separated on 7.5% acrylamide gels, and proteins were immunoblotted with the anti-Flg2 antibodies (top) and a monoclonal antibody directed against actin (Act; bottom). A picture representative of at least three different experiments is shown. In the upper panel, the arrows indicate the full-length Flg2 and a processed form of 200 kDa, and the arrowhead indicates a fragment of 55 kDa. Molecular masses of standards are indicated in kDa to the left.
Deiminated FLG2 Is More Susceptible to Calpain 1 Cleavage
Deimination of FLG is a major step in its catabolism and allows its dissociation from the corneocyte matrix. In addition, FLG deiminated by mouse Pad3 has been shown to be more susceptible to calpain 1-mediated degradation than the unmodified form, possibly because of protein unfolding, one of the reported consequences of this post-translational modification. Because both FLG and the B-type repeats of FLG2 contain large amounts of Arg residues (10.8 and 8.4%, respectively), we predicted that FLG2 might be deiminated and that FLG2 deimination might modify its calpain susceptibility.
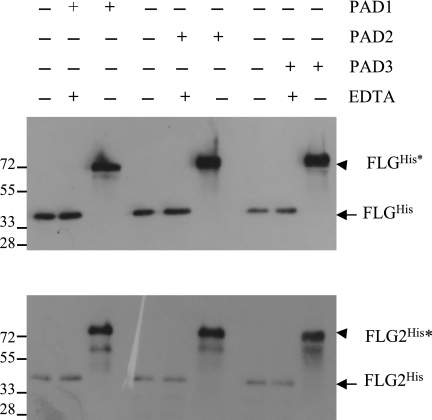
First, to test whether deimination is involved in the post-translational modifications of FLG2, purified recombinant human PAD1, -2, and -3 were used in deimination assays (Fig. 7). These are the three isotypes of PADs expressed in the epidermis, PAD1 and -3 being responsible for the deimination of FLG (18). The deimination of FLGHis, used as a positive control, induced a shift in its apparent molecular mass after SDS-PAGE. Such a shift has been described as a characteristic of deimination for several PAD substrates (19, 33, 34). Incubation of FLG2His with any of the three PADs also induced a shift in its migration in SDS gels from 40 to higher than 72 kDa. As expected, chelation of calcium, an ion essential for PAD activity, by the addition of an excess of EDTA during the incubation with the enzymes, completely inhibited the changes in mobility of both FLGHis and FLG2His (Fig. 7). Confirmation that FLG2His, like FLG, is deiminated by human PAD1, PAD2, and PAD3 in vitro was provided by its immunodetection with an antibody specific for citrulline, the AMC antibody (35), only after incubation with a PAD (supplemental Fig. 2).
FIGURE 7.
Deimination of FLG2His by human PAD1, -2, and -3. Recombinant FLGHis and FLG2His (100 ng/test) were incubated with or without human PAD1, PAD2, or PAD3 (40 milliunits/test) for 19 h at 50 °C in a 100 mm Tris-HCl (pH 7.6) buffer containing 10 mm CaCl2 and 5 mm DTT in the absence or presence of 15 mm EDTA, as indicated. The reactions were stopped by the addition of Laemmli's sample buffer. Samples were then subjected to SDS-PAGE and immunoblotting analysis with the anti-His antibody. Molecular masses of standards are indicated in kDa on the left. The arrows indicate the unmodified FLGHis and FLG2His, and the arrowheads show the fully deiminated proteins (FLGHis* and FLG2His*). Note that deimination induces a shift in the apparent molecular mass of both FLGHis and FLG2His.
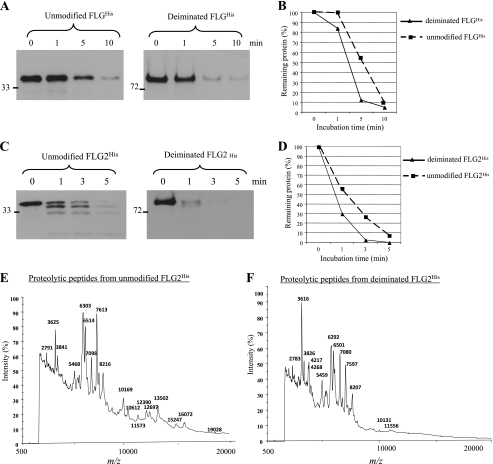
We next investigated the consequence of FLG and FLG2 deimination by human PADs on their proteolysis by calpain 1. Fig. 8A shows the time course of the decrease in the FLGHis and PAD1-deiminated FLGHis, as analyzed by SDS-PAGE and immunoblotting. Both proteins were almost completely degraded within 10 min. However, the deiminated FLGHis was degraded more rapidly than the unmodified protein. A densitometric analysis of scanned images confirmed these data and showed that 90% of degradation was achieved after 5 min of digestion for the former versus 10 min for the latter (Fig. 8B). Fig. 8C shows the time course of the degradation by calpain 1 of the unmodified and deiminated FLG2His. Both proteins were degraded very quickly, within 3–5 min. Whereas some full-length unmodified FLG2His and derived polypeptides were still detected after 5 min of incubation with the protease, the deiminated protein was completely degraded from 3 min of incubation. A densitometric analysis of scanned images convincingly showed that the deiminated FLG2His was degraded more quickly than the unmodified protein (Fig. 8D). It has been reported that the deiminated FLG is degraded by calpain 1 into shorter peptides than the non-deiminated form (23). Therefore, we further compared the proteolytic fragments generated from unmodified and deiminated FLG2His by calpain 1 digestion. Fig. 8, E and F, show the MS spectra at m/z 500–20,000 of the unmodified and deiminated FLG2His incubated with calpain 1 for 5 min. Compared with proteolysis of unmodified FLG2His, calpain 1-mediated cleavage of deiminated FLG2His produced shorter peptides with m/z <10,000. These spectral patterns supported the conclusion drawn from the immunoblotting analysis (i.e. that the deiminated FLG2His is more susceptible to calpain 1) and strongly suggested that the modified FLG2His was degraded into shorter fragments than the unmodified protein.
FIGURE 8.
Cleavage of deiminated FLG2His by calpain 1. A–D, the proteolysis of the unmodified (squares) and deiminated (triangles) FLGHis (A and B) and FLG2His (C and D) by calpain 1 was analyzed at an enzyme/substrate molar ratio of 1:50. Unmodified and deiminated FLGHis and FLG2His were incubated for the indicated times at 30 °C with calpain 1 in 100 mm Tris-HCl (pH 7.5) containing 2 mm CaCl2 and 10 mm DTT. The extent of cleavage was determined by immunoblotting with the anti-His antibody (A and C) followed by a densitometric analysis (B and D). E and F, the proteolysis of the unmodified (E) and deiminated (F) FLG2His by calpain 1 performed in the conditions described above for 5 min was analyzed by MALDI-TOF MS. Spectra at m/z 500–20,000 are shown. The m/z of the calpain 1-produced peptides are indicated.
We observed that most of the Arg residues within FLG2His are close to the calpain 1 cleavage sites. To test whether some of them could be deiminated, FLG2His was deiminated by PAD1, and its NH2 terminus was sequenced by Edman degradation (Fig. 4D). The obtained sequence (QSESIVPECitHGTTH) overlapping the cleavage site GTTH↓GQTG clearly demonstrates that it is the case. Taken together, these results indicate that calpain 1 acts preferentially on deiminated proteins, including FLG2, as compared with the unmodified forms.
DISCUSSION
To analyze the expression, localization, and processing of FLG2 in the human epidermis, we produced highly specific rabbit antibodies directed against an internal peptide corresponding to the B-type subunit B10. The antibodies were used in confocal microscopy analyses of breast skin samples and immunoblotting of epidermis sequential extracts. We detected the protein first in the cytoplasm of the upper granular keratinocytes, with a granular pattern of labeling, and then in the matrix of the lower corneocytes. Finally, the labeling disappeared from the upper corneocytes concomitantly with the degradation of FLG. The full-length 248-kDa protein was detected only in urea-containing extracts, suggesting that it is stored in keratohyalin granules. This hypothesis is consistent with the granular pattern of labeling and the observed colocation with profilaggrin. Multiple immunoreactive fragments from 250 to 40 kDa were also observed, indicating a proteolytic processing of this high molecular weight precursor during terminal differentiation. In particular, we observed a band with a molecular weight around 130,000, suggesting that a fragment corresponding to the whole B-type domain is produced. The domain is further proteolyzed, at least to 40-kDa fragments and possibly to other non-immunoreactive smaller peptides. These results are in accordance with previously published data obtained using antibodies specific for the NH2 terminus of FLG2, namely the spacer 1 region (9). In particular, the 130-kDa fragment described above has not been detected by these antibodies, showing that it does not comprise the NH2 terminus of FLG2. Such a proteolytic metabolism is well known for FLG. Numerous proteases, including calpain 1 (22), furin (36), profilaggrin endoproteinase 1 (37), matriptase (38), channel-activating serine protease 1 (39), and elastase-2 (40), have been identified as being involved either directly or indirectly in the profilaggrin processing to FLG monomers, whereas calpain 1, caspase-14, and bleomycin hydrolase (23) are required for further proteolysis of FLG monomers into smaller peptides and free amino acids. The latter are the major components of the so-called natural moisturizing factor (NMF), the main function of which is to contribute to water retention, flexibility of the stratum corneum, and UV protection (21, 41, 42). We speculated that the same enzymes could be involved in the processing of FLG2. We first focused on calpain 1.
Calpain 1 is a neutral calcium-activated protease present in a wide variety of tissues, including the epidermis. The protease has been detected in the upper part of the epidermis (43), and its expression has been shown to be up-regulated during keratinocyte differentiation in vitro (44). In our studies, in vitro cleavage assays revealed that calpain 1 was able to directly process a fragment of FLG2 consisting of B-type repeats B7–B10 into multiple peptides detected by both immunoblotting and mass spectrometry. Identification of the NH2 terminus of the major ones using Edman degradation sequencing provided evidence that at least four calpain 1 cleavage sites are present within the B-type repeats. Further detailed examination of FLG2 sequence highlighted many potential calpain 1 cleavage sites, regularly positioned along the whole B-type repeat domain. This explains the “ladder-like” cleavage pattern detected during the in vitro calpain 1 cleavage assays (Fig. 2, C and D). One of these potential cleavage sites is located in the epitope(s) recognized by the anti-FLG2 antibodies within repeat B10. This could also explain why we did not immunodetect fragments of FLG2 shorter than 40 kDa in epidermis protein extracts. Supporting an interaction between the two proteins in vivo, we showed that calpain 1 is colocated with FLG2 in the cytoplasm of granular keratinocytes and lower corneocytes and that the full-length Flg2 accumulates whereas a processed 55-kDa form is detected less in the epidermis of calpastatin-overexpressing mice as compared with wild type mice. Altogether, these data strongly suggest that calpain 1 is involved in vivo in the processing of the full-length FLG2 and in the proteolysis of the B-type repeat domain of FLG2 to small fragments, as has been shown for profilaggrin and the FLG subunits. To confirm this hypothesis, it will be of great interest in future experiments to identify the in vivo calpain 1 cleavage sites and to compare them with those determined in vitro. It is important to note that we did not find related potential calpain 1 cleavage sites within the A-type repeat domain, suggesting that this domain may undergo different processing if any.
During terminal differentiation, FLG monomers undergo deimination by PAD1 and PAD3 (18, 19). Deimination results in FLG being more susceptible to proteolysis and promotes its degradation into free amino acids (23). An increased susceptibility to degradation after deimination has already been described for another protein, the myelin basic protein (45, 46). We therefore examined whether FLG2 processing could also be regulated by deimination. First, our results showed that the Arg-rich B-type repeats were in vitro substrates of the three PAD isotypes expressed in the epidermis (i.e. PAD1, PAD2, and PAD3). Next, we observed that deimination of FLG2 facilitated its degradation by calpain 1. Importantly, most of the Arg residues within the B-type repeats are close to the calpain 1 cleavage sites. We demonstrated that at least one of them was modified by PAD1, a similar Arg residue being located in all B-type repeats in a conserved position just before a calpain 1 cleavage site. Based on previous studies (32, 47, 48), we believe that deimination of FLG2 alters its spatial conformation and consequently facilitates its proteolysis. The three-dimensional structure of neither FLG2 nor FLG is known; therefore, it is currently not possible to test this hypothesis in silico. New cleavage sites could also be recognized by calpain 1.
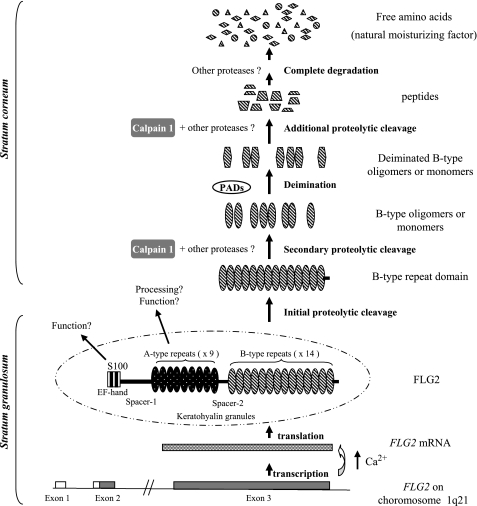
On the basis of these findings, we propose a model for FLG2 processing in the epidermis (Fig. 9). The FLG2 gene, located on 1q21 in the epidermal differentiation complex, encodes a large protein that comprises three domains: an NH2-terminal calcium binding domain homologous to the S100 proteins and two domains formed by repeated subunits, the A-type and B-type. FLG2 is transcribed in the granular keratinocytes, upon the Ca2+ concentration increase known to occur during the terminal differentiation process. FLG2 mRNAs are translated into a 248-kDa large precursor, which is stored in keratohyalin granules, in the cytoplasm of the granular cells. During the transition from granular to cornified layers, the precursor protein is subjected to an initial proteolysis by unknown protease(s). The S100-related NH2-terminal domain, the A-type repeat domain, and the B-type repeat domain are released. Further cleavage of the B-type repeat domain by calpain 1 leads to the production of B-type oligomers and/or monomers. Deimination of the B-type oligomers/monomers by PADs induces a decrease in their positive charges, changes their conformation, and promotes their subsequent degradation. Because the amino acid composition of FLG2 B-type repeats is similar to that of FLG, breakdown of FLG2 into free amino acids by other proteases, including caspase 14, may contribute to the NMF components. Until now, FLG has been considered as the only protein at the origin of the NMF and therefore responsible for stratum corneum hydration, flexibility, and UV protection (21, 40, 41). Based on our findings, FLG2 could contribute to stratum corneum hydration and barrier functions along with FLG. This is supported by the fact that homozygous carriers of FLG mutations have a low but non-null NMF content, suggesting that other proteins may contribute to NMF formation (49). The role and processing of the S100-related NH2-terminal domain and of the A-type repeat domain of FLG2 are unknown. The S100-related terminal domain of profilaggrin has been shown to contain a nuclear localization signal and to be translocated to transitional keratinocyte nucleus, where it could be implicated in the regulation of gene transcription through its capacity to bind calcium (50–52). We identified such a potential monopartite nuclear localization signal in the NH2 terminus of FLG2, KKHRR (residues 100–104), suggesting a similar function. This remains to be tested. Unlike the B-type repeats, the A-type repeats of FLG2 contain few Arg (2.6%) and no sequences related to calpain 1 cleavage sites. This suggests that the two repeat domains have different post-translational modifications and proteolytic processes. Interestingly, the A-type repeats display the highest homology to HRNR (8), an epidermal protein recently shown to be a component of cornified cell envelopes (28). A-type repeats of FLG2 and HRNR both contain high percentages of Gln (15 and 9.4%, respectively), which is a key amino acid for transglutaminase-mediated cross-linking to the cornified envelope (53, 54), suggesting that the A-type repeat domain is cross-linked to these corneocyte structures.
FIGURE 9.
Proposed model of the maturation and function of FLG2 in human epidermis. See “Discussion” for details.
FLG plays another important role in the stratum corneum as an intermediate filament-associated protein, promoting their aggregation and the formation of the corneocyte matrix (55, 56). Ionic interactions between the conserved positive charges of FLG and the negative charges distributed along the rod domains of intermediate filament proteins seem to be the mechanisms of FLG/filament binding (16). Regarding the high percentage of basic amino acids (His + Arg, 23.4%) within the B-type repeats, it is possible that human FLG2 may also have the ability to associate with intermediate filaments. In agreement, a recombinant part of mouse Flg2 has been shown to interact with and bundle keratin filaments extracted from bovine tongue (57).
Recently, many studies have provided evidence that proteins encoded by genes located in the epidermal differentiation complex are associated with several common skin disorders. In particular, loss-of-function mutations in the FLG gene have been shown to cause ichthyosis vulgaris and to predispose to atopic dermatitis. In addition, a deletion of the genes encoding the late cornified envelope LCE3B and LCE3C has been associated with psoriasis (24, 25, 58, 59). Therefore, a possible pathophysiological implication of FLG2 in skin diseases remains to be studied.
In summary, our present study provides the first deciphering of the complex processing of FLG2 during the terminal differentiation of the epidermis. It shows that calpain 1 is essential in its proteolytic maturation. Calpain 1 is involved in the proteolysis of the B-type repeat domain and probably in the release of free amino acids, components of NMF. Furthermore, deimination of FLG2 by PAD1, PAD2, and/or PAD3 was shown to promote its proteolysis by calpain 1. Further investigations of the proteolysis, post-translational modification(s), and biochemical property (or properties) of FLG2 will be helpful to understand the complete processing and biological function(s) of this particular protein during terminal differentiation.
Supplementary Material
Acknowledgments
We thank Prof. Jean-Pierre Chavoin (Plastic Surgery Department, Hospital of Toulouse, France) for the human skin samples, Prof. Laurent Baud (INSERM U702, University Paris 6, Paris, France) for the kind gift of mouse skin samples, Prof. Selim Aractingi (Department of Dermatology and Allergie, Hôpital Tenon, Paris) for help, Prof. Jens-Michael Schröder (University-Hospital Schleswig-Holstein, Kiel, Germany) and Dr. Rachida Nachat for helpful discussions and advice, Dr. Jacques D'Alayer (Institut Pasteur, Paris, France) for the microsequencing analyses, Sophie Allard from the confocal microscopy facility (INSERM, IFR150, Toulouse), and Catherine Bouchouata and Carole Pons for technical assistance.
This work was supported by CNRS, INSERM, Toulouse III University, Toulouse University Hospital, the “Région Midi-Pyrénées,” Pierre Fabre Dermo-Cosmétique and the French Society for Dermatological Research (SRD).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
B. Hansmann, unpublished data.
- HRNR
- hornerin
- ALLN
- N-acetyl-Leu-Leu-norleucine-CHO
- FLG
- filaggrin
- NMF
- natural moisturizing factor
- PAD
- peptidylarginine deiminase.
REFERENCES
- 1. Mischke D., Korge B. P., Marenholz I., Volz A., Ziegler A. (1996) J. Invest. Dermatol. 106, 989–992 [DOI] [PubMed] [Google Scholar]
- 2. Jackson B., Tilli C. M., Hardman M. J., Avilion A. A., MacLeod M. C., Ashcroft G. S., Byrne C. (2005) J. Invest. Dermatol. 124, 1062–1070 [DOI] [PubMed] [Google Scholar]
- 3. Cabral A., Voskamp P., Cleton-Jansen A. M., South A., Nizetic D., Backendorf C. (2001) J. Biol. Chem. 276, 19231–19237 [DOI] [PubMed] [Google Scholar]
- 4. Marenholz I., Heizmann C. W., Fritz G. (2004) Biochem. Biophys. Res. Commun. 322, 1111–1122 [DOI] [PubMed] [Google Scholar]
- 5. Presland R. B., Haydock P. V., Fleckman P., Nirunsuksiri W., Dale B. A. (1992) J. Biol. Chem. 267, 23772–23781 [PubMed] [Google Scholar]
- 6. Makino T., Takaishi M., Morohashi M., Huh N. H. (2001) J. Biol. Chem. 276, 47445–47452 [DOI] [PubMed] [Google Scholar]
- 7. Takaishi M., Makino T., Morohashi M., Huh N. H. (2005) J. Biol. Chem. 280, 4696–4703 [DOI] [PubMed] [Google Scholar]
- 8. Wu Z., Meyer-Hoffert U., Reithmayer K., Paus R., Hansmann B., He Y., Bartels J., Gläser R., Harder J., Schröder J. M. (2009) J. Invest. Dermatol. 129, 1446–1458 [DOI] [PubMed] [Google Scholar]
- 9. Wu Z., Hansmann B., Meyer-Hoffert U., Gläser R., Schröder J. M. (2009) PLoS One 4, e5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krieg P., Schuppler M., Koesters R., Mincheva A., Lichter P., Marks F. (1997) Genomics 43, 339–348 [DOI] [PubMed] [Google Scholar]
- 11. Contzler R., Favre B., Huber M., Hohl D. (2005) J. Invest. Dermatol. 124, 990–997 [DOI] [PubMed] [Google Scholar]
- 12. Marenholz I., Lovering R. C., Heizmann C. W. (2006) Biochim. Biophys. Acta 1763, 1282–1283 [DOI] [PubMed] [Google Scholar]
- 13. Lee S. C., Wang M., McBride O. W., O'Keefe E. J., Kim I. G., Steinert P. M. (1993) J. Invest. Dermatol. 100, 65–68 [DOI] [PubMed] [Google Scholar]
- 14. Dale B. A. (1985) Am. J. Dermatopathol. 7, 65–68 [DOI] [PubMed] [Google Scholar]
- 15. Sandilands A., Sutherland C., Irvine A. D., McLean W. H. (2009) J. Cell Sci. 122, 1285–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mack J. W., Steven A. C., Steinert P. M. (1993) J. Mol. Biol. 232, 50–66 [DOI] [PubMed] [Google Scholar]
- 17. Harding C. R., Scott I. R. (1983) J. Mol. Biol. 170, 651–673 [DOI] [PubMed] [Google Scholar]
- 18. Nachat R., Méchin M. C., Takahara H., Chavanas S., Charveron M., Serre G., Simon M. (2005) J. Invest. Dermatol. 124, 384–393 [DOI] [PubMed] [Google Scholar]
- 19. Méchin M. C., Enji M., Nachat R., Chavanas S., Charveron M., Ishida-Yamamoto A., Serre G., Takahara H., Simon M. (2005) Cell Mol. Life Sci. 62, 1984–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elias P. M. (2004) J. Invest. Dermatol. 122, xxxvi–xxxix [DOI] [PubMed] [Google Scholar]
- 21. Rawlings A. V., Harding C. R. (2004) Dermatol. Ther. 17, 43–48 [DOI] [PubMed] [Google Scholar]
- 22. Yamazaki M., Ishidoh K., Suga Y., Saido T. C., Kawashima S., Suzuki K., Kominami E., Ogawa H. (1997) Biochem. Biophys. Res. Commun. 235, 652–656 [DOI] [PubMed] [Google Scholar]
- 23. Kamata Y., Taniguchi A., Yamamoto M., Nomura J., Ishihara K., Takahara H., Hibino T., Takeda A. (2009) J. Biol. Chem. 284, 12829–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith F. J., Irvine A. D., Terron-Kwiatkowski A., Sandilands A., Campbell L. E., Zhao Y., Liao H., Evans A. T., Goudie D. R., Lewis-Jones S., Arseculeratne G., Munro C. S., Sergeant A., O'Regan G., Bale S. J., Compton J. G., DiGiovanna J. J., Presland R. B., Fleckman P., McLean W. H. (2006) Nat. Genet. 38, 337–342 [DOI] [PubMed] [Google Scholar]
- 25. Palmer C. N., Irvine A. D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S. P., Goudie D. R., Sandilands A., Campbell L. E., Smith F. J., O'Regan G. M., Watson R. M., Cecil J. E., Bale S. J., Compton J. G., DiGiovanna J. J., Fleckman P., Lewis-Jones S., Arseculeratne G., Sergeant A., Munro C. S., El Houate B., McElreavey K., Halkjaer L. B., Bisgaard H., Mukhopadhyay S., McLean W. H. (2006) Nat. Genet. 38, 441–446 [DOI] [PubMed] [Google Scholar]
- 26. Sandilands A., Terron-Kwiatkowski A., Hull P. R., O'Regan G. M., Clayton T. H., Watson R. M., Carrick T., Evans A. T., Liao H., Zhao Y., Campbell L. E., Schmuth M., Gruber R., Janecke A. R., Elias P. M., van Steensel M. A., Nagtzaam I., van Geel M., Steijlen P. M., Munro C. S., Bradley D. G., Palmer C. N., Smith F. J., McLean W. H., Irvine A. D. (2007) Nat. Genet. 39, 650–654 [DOI] [PubMed] [Google Scholar]
- 27. Toulza E., Mattiuzzo N. R., Galliano M. F., Jonca N., Dossat C., Jacob D., de Daruvar A., Wincker P., Serre G., Guerrin M. (2007) Genome Biol. 8, R107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henry J., Hsu C. Y., Haftek M., Nachat R., de Koning H. D., Gardinal-Galera I., Hitomi K., Balica S., Jean-Decoster C., Schmitt A. M., Paul C., Serre G., Simon M. (2011) FASEB J. 25, 1567–1576 [DOI] [PubMed] [Google Scholar]
- 29. Simon M., Sebbag M., Haftek M., Vincent C., Girbal-Neuhauser E., Rakotoarivony J., Sommé G., Schmitt D., Serre G. (1995) J. Invest. Dermatol. 105, 432–437 [DOI] [PubMed] [Google Scholar]
- 30. Simon M., Montézin M., Guerrin M., Durieux J. J., Serre G. (1997) J. Biol. Chem. 272, 31770–31776 [DOI] [PubMed] [Google Scholar]
- 31. Peltier J., Bellocq A., Perez J., Doublier S., Dubois Y. C., Haymann J. P., Camussi G., Baud L. (2006) J. Am. Soc. Nephrol. 17, 3415–3423 [DOI] [PubMed] [Google Scholar]
- 32. Presland R. B., Kimball J. R., Kautsky M. B., Lewis S. P., Lo C. Y., Dale B. A. (1997) J. Invest. Dermatol. 108, 170–178 [DOI] [PubMed] [Google Scholar]
- 33. Tarcsa E., Marekov L. N., Mei G., Melino G., Lee S. C., Steinert P. M. (1996) J. Biol. Chem. 271, 30709–30716 [DOI] [PubMed] [Google Scholar]
- 34. Kanno T., Kawada A., Yamanouchi J., Yosida-Noro C., Yoshiki A., Shiraiwa M., Kusakabe M., Manabe M., Tezuka T., Takahara H. (2000) J. Invest. Dermatol. 115, 813–823 [DOI] [PubMed] [Google Scholar]
- 35. Senshu T., Akiyama K., Kan S., Asaga H., Ishigami A., Manabe M. (1995) J. Invest. Dermatol. 105, 163–169 [DOI] [PubMed] [Google Scholar]
- 36. Pearton D. J., Nirunsuksiri W., Rehemtulla A., Lewis S. P., Presland R. B., Dale B. A. (2001) Exp. Dermatol. 10, 193–203 [DOI] [PubMed] [Google Scholar]
- 37. Resing K. A., Thulin C., Whiting K., al-Alawi N., Mostad S. (1995) J. Biol. Chem. 270, 28193–28198 [DOI] [PubMed] [Google Scholar]
- 38. List K., Szabo R., Wertz P. W., Segre J., Haudenschild C. C., Kim S. Y., Bugge T. H. (2003) J. Cell Biol. 163, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leyvraz C., Charles R. P., Rubera I., Guitard M., Rotman S., Breiden B., Sandhoff K., Hummler E. (2005) J. Cell Biol. 170, 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bonnart C., Deraison C., Lacroix M., Uchida Y., Besson C., Robin A., Briot A., Gonthier M., Lamant L., Dubus P., Monsarrat B., Hovnanian A. (2010) J. Clin. Invest. 120, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rawlings A. V., Scott I. R., Harding C. R., Bowser P. A. (1994) J. Invest. Dermatol. 103, 731–741 [DOI] [PubMed] [Google Scholar]
- 42. Rawlings A. V., Matts P. J. (2005) J. Invest. Dermatol. 124, 1099–1110 [DOI] [PubMed] [Google Scholar]
- 43. Miyachi Y., Yoshimura N., Suzuki S., Hamakubo T., Kannagi R., Imamura S., Murachi T. (1986) J. Invest Dermatol. 86, 346–349 [DOI] [PubMed] [Google Scholar]
- 44. Garach-Jehoshua O., Ravid A., Liberman U. A., Reichrath J., Glaser T., Koren R. (1998) Br. J. Dermatol. 139, 950–957 [DOI] [PubMed] [Google Scholar]
- 45. D'Souza C. A., Moscarello M. A. (2006) Neurochem. Res. 31, 1045–1054 [DOI] [PubMed] [Google Scholar]
- 46. Pritzker L. B., Joshi S., Gowan J. J., Harauz G., Moscarello M. A. (2000) Biochemistry 39, 5374–5381 [DOI] [PubMed] [Google Scholar]
- 47. Ishida-Yamamoto A., Senshu T., Eady R. A., Takahashi H., Shimizu H., Akiyama M., Iizuka H. (2002) J. Invest. Dermatol. 118, 282–287 [DOI] [PubMed] [Google Scholar]
- 48. Chavanas S., Méchin M. C., Nachat R., Adoue V., Coudane F., Serre G., Simon M. (2006) J. Dermatol. Sci. 44, 63–72 [DOI] [PubMed] [Google Scholar]
- 49. Kezic S., Kemperman P. M., Koster E. S., de Jongh C. M., Thio H. B., Campbell L. E., Irvine A. D., McLean W. H., McLean I. W., Puppels G. J., Caspers P. J. (2008) J. Invest. Dermatol. 128, 2117–2119 [DOI] [PubMed] [Google Scholar]
- 50. Ishida-Yamamoto A., Takahashi H., Presland R. B., Dale B. A., Iizuka H. (1998) Lab. Investig. 78, 1245–1253 [PubMed] [Google Scholar]
- 51. Pearton D. J., Dale B. A., Presland R. B. (2002) J. Invest. Dermatol. 119, 661–669 [DOI] [PubMed] [Google Scholar]
- 52. Zhang D., Karunaratne S., Kessler M., Mahony D., Rothnagel J. A. (2002) J. Invest. Dermatol. 119, 905–912 [DOI] [PubMed] [Google Scholar]
- 53. Kahlem P., Terré C., Green H., Djian P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14580–14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Candi E., Schmidt R., Melino G. (2005) Nat. Rev. Mol. Cell Biol. 6, 328–340 [DOI] [PubMed] [Google Scholar]
- 55. Kalinin A. E., Kajava A. V., Steinert P. M. (2002) BioEssays 24, 789–800 [DOI] [PubMed] [Google Scholar]
- 56. Manabe M., Sanchez M., Sun T. T., Dale B. A. (1991) Differentiation 48, 43–50 [DOI] [PubMed] [Google Scholar]
- 57. Listwan P., Rothnagel J. A. (2004) Methods Cell Biol. 78, 817–827 [DOI] [PubMed] [Google Scholar]
- 58. Stemmler S., Parwez Q., Petrasch-Parwez E., Epplen J. T., Hoffjan S. (2007) J. Invest. Dermatol. 127, 722–724 [DOI] [PubMed] [Google Scholar]
- 59. de Cid R., Riveira-Munoz E., Zeeuwen P. L., Robarge J., Liao W., Dannhauser E. N., Giardina E., Stuart P. E., Nair R., Helms C., Escaramís G., Ballana E., Martín-Ezquerra G., den Heijer M., Kamsteeg M., Joosten I., Eichler E. E., Lázaro C., Pujol R. M., Armengol L., Abecasis G., Elder J. T., Novelli G., Armour J. A., Kwok P. Y., Bowcock A., Schalkwijk J., Estivill X. (2009) Nat. Genet. 41, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.