Abstract
Many regulatory processes in biology involve reversible association of proteins with membranes. Clotting proteins bind to phosphatidylserine (PS) on cell surfaces, but a clear picture of this interaction has yet to emerge. We present a novel explanation for membrane binding by GLA domains of clotting proteins, supported by biochemical studies, solid-state NMR analyses, and molecular dynamics simulations. The model invokes a single “phospho-l-serine-specific” interaction and multiple “phosphate-specific” interactions. In the latter, the phosphates in phospholipids interact with tightly bound Ca2+ in GLA domains. We show that phospholipids with any headgroup other than choline strongly synergize with PS to enhance factor X activation. We propose that phosphatidylcholine and sphingomyelin (the major external phospholipids of healthy cells) are anticoagulant primarily because their bulky choline headgroups sterically hinder access to their phosphates. Following cell damage or activation, exposed PS and phosphatidylethanolamine collaborate to bind GLA domains by providing phospho-l-serine-specific and phosphate-specific interactions, respectively.
Keywords: Blood Coagulation Factors, Membrane Bilayer, Molecular Dynamics, NMR, Phosphatidylserine, Phospholipid, Protein-Membrane Interactions
Introduction
Proteins that bind to phosphatidylserine (PS)2 are implicated in diverse processes such as intracellular signal transduction cascades, vesicle fusion and neurotransmitter release, plasma membrane-cytoskeleton interactions, phagocytosis of apoptotic cells, and blood clotting (1). Indeed, most of the steps in the coagulation cascade require the reversible Ca2+-dependent association of clotting proteins with membrane bilayers containing exposed PS (2). Releasing these proteins from the membrane surface renders them thousands of times less active. An exemplar is the initiation of clotting, triggered when the plasma protease factor VIIa (fVIIa) binds to the integral membrane protein tissue factor (TF). The resulting TF-fVIIa complex activates two membrane-bound zymogens, factors IX (fIX) and X (fX), by limited proteolysis (3). fVIIa, fIX, and fX are homologous proteins that interact with PS-containing membranes via their GLA domains, so named because they are rich in γ-carboxyglutamate. GLA domains are the most common membrane-binding motif in blood clotting (2), but despite their importance, we lack a detailed understanding of their interaction with the membrane at atomic resolution. This study provides a new view of how lipids collaborate to promote the binding of GLA domains to membrane surfaces and trigger blood clotting.
A puzzling feature of TF-fVIIa is its requirement for unphysiologically high PS levels (∼30% PS) for optimal procoagulant activity when TF is incorporated into liposomes with mixtures of PS and phosphatidylcholine (PC). The answer may come from phosphatidylethanolamine (PE). By itself, PE (in PE/PC liposomes) supports little to no clotting activity, but when PE is incorporated into PS/PC liposomes, it dramatically decreases the PS requirement for optimal activity of membrane-bound blood clotting reactions, including proteolytic activation of fX by the TF-fVIIa complex (4), inactivation of factor Va by activated protein C (5), activation of prothrombin by the factor Va-factor Xa (prothrombinase) complex (6), and activation of fX by the factor VIIIa-factor IXa (intrinsic tenase) complex (7). Of the four major plasma membrane phospholipids, PC and sphingomyelin are abundant on the outer surface, whereas PS and PE are actively sequestered to the inner leaflet (see Fig. 1A). Cell lysis, damage, or activation causes externalization of PS and PE, which promotes clotting reactions (8). PE is considerably more abundant than PS, so the ability of small amounts of PS to support blood clotting reactions in the presence of larger amounts of PE represents a more physiologic environment for assembling clotting reactions.
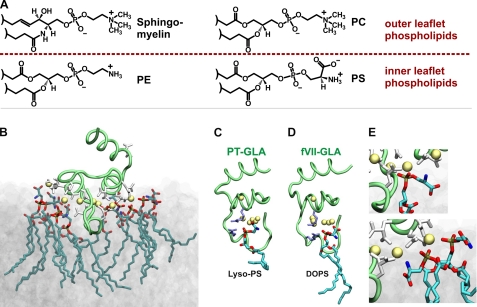
FIGURE 1.
GLA domain-PS interactions. A, the four most abundant plasma membrane phospholipids. Sphingomyelin and PC are abundant in the outer leaflet, whereas PS and PE are largely restricted to the inner leaflet; this membrane asymmetry is lost following platelet activation or cellular trauma (32). B, membrane-bound model of the human factor VII GLA domain obtained from MD simulations on the surface of a PS bilayer (12). The GLA domain backbone is a green tube, the γ-carboxyglutamate residues are white, tightly bound Ca2+ are yellow spheres, and the interacting PS is shown in stick representation. C and D, the proposed phospho-l-serine-specific binding site independently observed in the crystal structure of bovine prothrombin fragment 1 (PT; Protein Data Bank code 1NL2) (14) (C) and in our MD simulations of the fVIIa GLA domain-membrane complex (12) (D). In C and D, two Arg residues contributing to the site are shown as purple sticks, and the uniquely bound lyso-PS and dioleoyl-PS molecules are drawn using stick representations. E, examples of two phosphate-specific interactions from B.
How PE “synergizes” with PS is not known, but Zwaal et al. (2) and Gilbert and Arena (7) have summarized the following hypotheses from the literature. 1) Being a non-lamellar phospholipid, PE induces the formation of PS-rich microdomains, which are more efficient in supporting clotting reactions. 2) Clotting proteins contain binding site(s) for PE (or a combination of PE and PS). 3) PE has hydrogen bond donors, but PC does not. When PE forms hydrogen bonds with adjacent PS molecules, it induces PS headgroup conformations that are more favorable for binding to clotting proteins. 4) The PC headgroup is so bulky and highly hydrated (9, 10) that it sterically hinders access of GLA domains to adjacent PS molecules.
Our previous studies demonstrated approximately one fX membrane-binding site for every six to eight PS molecules, which is about equal to a GLA domain's membrane footprint (11). Here, we present a novel general explanation for GLA domain interactions with membranes that we term the “Anything But Choline” (ABC) hypothesis, triggered by detailed molecular dynamics (MD) simulations of GLA domains associating with PS-containing bilayers (Fig. 1B) (12, 13). The ABC hypothesis proposes two key types of GLA domain-phospholipid interactions: a single “phospho-l-serine-specific” binding site per GLA domain (Fig. 1, C and D) and multiple “phosphate-specific” interactions, in which phospholipid headgroups bend to allow their phosphates to form coordination complexes with tightly bound Ca2+ in the GLA domain (Fig. 1E). The unique phospho-l-serine-specific binding site was independently identified in the crystal structure of lyso-PS bound to the prothrombin GLA domain (Fig. 1C) (14) and in MD simulations of fVIIa GLA domain binding to PS bilayers (Fig. 1D) (12). The proposed phosphate-specific interactions have been repeatedly observed in our MD simulations of GLA domains associating with PS-containing bilayers (12, 13). We propose that PS can provide both interactions, whereas PC can provide neither (because its bulky choline headgroup sterically hinders access of proteins to its own phosphate moiety). PE can provide phosphate-specific but not phospho-l-serine-specific interactions, explaining why PE/PC bilayers poorly support clotting, whereas PE synergizes with small amounts of PS.
A strong prediction of the ABC hypothesis is that there is nothing unique about the ethanolamine headgroup in PE other than that it is not as bulky as PC. Thus, any phospholipid whose headgroup can bend to allow its phosphate to contribute phosphate-specific interactions with GLA domains should synergize with PS, a prediction inconsistent with the PE-specific hypotheses outlined above. In this study, we used a combination of biochemical techniques, solid-state NMR (SSNMR) analyses, and MD simulations to compare the predictions of the ABC hypothesis versus PE-specific hypotheses to explain how phospholipids synergize to support fX activation by TF-fVIIa.
EXPERIMENTAL PROCEDURES
Materials
Diacylglycerol (DAG) and phospholipids, except for PS with d-serine (d-PS) and isotopically labeled PS (PS*), were from Avanti Polar Lipids (Alabaster, AL). Bio-Beads® SM-2 adsorbent was from Bio-Rad. d-Serine was from Sigma. l-[U-13C,15N]Serine was from Cambridge Isotope Laboratories (Andover, MA). Human fVIIa was from American Diagnostica Inc. (Stamford, CT). Bovine prothrombin was from Hematologic Technologies (Essex Junction, VT). Human fX and α-thrombin were from Enzyme Research Laboratories (South Bend, IN). Nitrilotriacetic acid Biacore sensor chips were from GE Healthcare. Recombinant membrane-anchored TF (11) and the membrane scaffold protein for Nanodisc production (15) were expressed in Escherichia coli and purified as described. d-PS and PS* were synthesized from natural isotopic abundance palmitoyloleoylphosphatidylcholine (POPC) via phospholipase D-catalyzed headgroup exchange using either d-serine (for d-PS) or l-[U-13C,15N]serine (for PS*) as described previously (16). Prothrombin fragment 1 was prepared by digestion of bovine prothrombin with human α-thrombin and purified by ion-exchange and size-exclusion chromatography essentially as described (17).
TF-Liposome Preparation and Measurement of fX Activation
TF was incorporated into liposomes of varying phospholipid composition as described previously (18) using Bio-Beads® SM-2 and 20 mm sodium deoxycholate. The initial rates of fX activation by TF-fVIIa assembled on TF-liposomes were quantified as described previously (11, 19), typically using 100 nm fX, 5 pm fVIIa, and 500 pm TF.
Surface Plasmon Resonance (SPR) Analyses of fX Binding to Nanoscale Bilayers
TF-free Nanodiscs of varying phospholipid composition were prepared as described (11). fX binding affinities for nanoscale bilayers were quantified by SPR analyses (on a Biacore 3000 instrument) as described with Nanodiscs immobilized on Ni2+-nitrilotriacetic acid sensor chips via the oligohistidine tag on the membrane scaffold protein (11). As reported previously (11), binding isotherms were plotted from maximal steady-state response units versus the fX concentration flowed over the chip surface, from which Kd values were derived by fitting the single-site ligand binding equation to the data.
Magic Angle Spinning (MAS)-SSNMR Spectroscopy
SSNMR studies were performed on Varian InfinityPlus spectrometers. Palmitoyloleoylphosphatidylserine (POPS)*/POPC Nanodisc spectra were acquired at 600 MHz with a Varian T3 HXY 3.2-mm probe at an MAS rate of 10,000 ± 3 or 13,333 ± 2 Hz. The variable temperature gas was maintained at a flow of 90 ± 10 standard cubic feet/h, and the reported sample temperatures take into account thermocouple calibration and frictional heating due to MAS, calibrated with ethylene glycol (20). All experiments utilized tangent-ramped cross-polarization (21) with two-pulse phase-modulated proton decoupling (22) at ∼80-kHz nutation frequency.
MD Simulations
Four bilayers each composed of 1188 lipids and solvated by water at an ∼43:1 water/lipid ratio were simulated. The bilayers were all composed of palmitoyloleoyl-phospholipids but with different headgroups, viz. PE, CH3-PE, (CH3)2-PE, and PC, respectively. All simulations were performed using NAMD2 (23) utilizing the CHARMM22 force field with ϕ/ψ cross-term map (CMAP) corrections (24) for proteins and CHARMM36 parameters for lipids (25). The TIP3P model (26) was used for water. All simulations were performed using an NPT ensemble at 1.0 atm and 298 K with a time step of 2 fs. Constant pressure was maintained using the Nosé-Hoover Langevin piston method (27, 28), whereas constant temperature was maintained by Langevin dynamics using a damping coefficient (γ) of 0.5 ps−1 applied to all heavy atoms. Non-bonded interactions were cut off after 12 Å with a smoothing function applied after 10 Å. The particle mesh Ewald method (29) was used for long-range electrostatic calculations with a grid density >1 Å−3. As a measure of accessibility, the solvent-accessible surface area of the lipid phosphates was calculated in VMD (30), where a probe of a specified radius representing molecular species of various sizes is rolled over the phosphate groups, and the surface available for direct contact with the probe is integrated. Molecular images were generated using VMD (30).
RESULTS
Many Phospholipids Synergize with PS to Enhance fX Activation by TF-fVIIa
The ABC hypothesis predicts that many phospholipids can synergize with PS to enhance fX activation by TF-fVIIa, whereas PE-centric hypotheses rely on specific properties of the ethanolamine headgroup. (Here, we refer to PS with the usual l-serine in its headgroup as l-PS and to PS with d-serine as d-PS.) To test these hypotheses, we measured the rates of fX activation by TF-fVIIa on TF-liposomes containing binary lipid mixtures (0–30% l-PS, balance = PC) or ternary lipid mixtures in which the sum of l-PS plus the other test lipid equaled 30% (balance = 70% PC). The lipids tested included PE, phosphatidylglycerol (PG), phosphatidylinositol (PI), phosphatidic acid (PA), and d-PS.
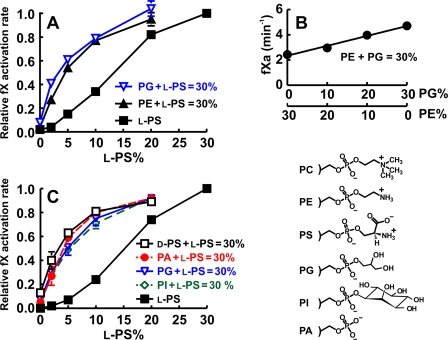
Consistent with our previous report (4), PE strongly synergized with l-PS to support fX activation, left-shifting the l-PS response relative to l-PS/PC liposomes (Fig. 2A). PG also strongly synergized with l-PS while supporting very low rates of fX activation in the absence of l-PS (Fig. 2A). On the other hand, PE did not synergize with PG, as the effects of these lipids were strictly additive (Fig. 2B). This is consistent with the ABC hypothesis, which predicts the same role for PE and PG, i.e. providing phosphate-specific interactions. In Fig. 2 (A and B), we employed dioleoyl-phospholipids to ensure that they were always above the phase transition because palmitoyloleoyl-PE has a relatively high transition temperature. For other phospholipid mixtures, we used palmitoyloleoyl-phospholipids; the results (cf. Fig. 2, A and C) confirm that PG synergized with l-PS equally well whether palmitoyloleoyl- or dioleoyl-phospholipids were employed. Phospholipid synergy is thus independent of any subtle bilayer packing differences due to dioleoyl- versus palmitoyloleoyl-phospholipids. PA and PI also strongly synergized with l-PS while supporting very low rates of fX activation in the absence of l-PS (Fig. 2C). Taken together, these results demonstrate that the PE ethanolamine moiety is not required for phospholipid synergy, as lipids with very different headgroups synergized equally well with l-PS.
FIGURE 2.
All tested glycerophospholipids synergize with l-PS to enhance fX activation by TF-fVIIa on liposomes, but PE does not synergize with PG. A, normalized rates of fX activation by fVIIa on TF-liposomes prepared with binary lipid mixtures (0–30% l-dioleoyl-PS, balance = dioleoyl-PC) or ternary lipid mixtures in which the sum of l-dioleoyl-PS plus dioleoylphosphatidylethanolamine (DOPE) or dioleoyl-PG equaled 30% (balance = 70% dioleoyl-PC). B, rates of fX activation by fVIIa on TF-liposomes prepared with ternary lipid mixtures in which the sum of dioleoyl-PG + DOPE equaled 30% (balance = 70% dioleoyl-PC). C, normalized rates of fX activation by fVIIa on TF-liposomes prepared with binary lipid mixtures (0–30% l-POPS, balance = POPC) or ternary lipid mixtures in which the sum of l-POPS plus the other test lipid equaled 30% (balance = 70% POPC). Lipids tested included PG, PI, PA, and d-POPS. fX activation rates for each TF-liposome preparation in A and C were normalized to the rate with 30% l-PS and 70% PC. Data are means ± S.E. (n = 3–10). Headgroup structures for lipids in this experiment are included on the right.
Components of the prothrombinase and factor VIIIa-factor IXa complexes bind much more weakly to membranes if l-PS is replaced by d-PS (reviewed by Zwaal et al. (2)). To our knowledge, PS headgroup stereospecificity has not been examined for fX activation by TF-fVIIa. We prepared TF-liposomes with binary lipid mixtures (0–30% l-PS, balance = PC) or ternary lipid mixtures of a total of 30% l-PS + d-PS (balance = 70% PC). d-PS poorly supported fX activation on its own, but it strongly synergized with l-PS (Fig. 2C). This is fully consistent with the ABC hypothesis because d-PS should not support phospho-l-serine-specific interactions but should provide phosphate-specific interactions with GLA domains.
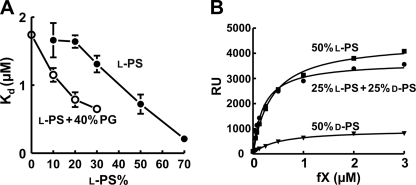
PG and d-PS Synergize with l-PS to Enhance fX Binding to Membranes
Multiple protein-phospholipid interactions are present when TF-fVIIa activates fX, but the dominant membrane contribution to catalysis appears to be fX binding to membranes adjacent to TF-fVIIa (11). We therefore quantified fX binding to nanobilayers using SPR as described previuosly (11). Consistent with our previous findings, the Kd for fX binding to Nanodiscs decreased with increasing l-PS content. The l-PS dose response was left-shifted in the presence of 40% PG (Fig. 3A), demonstrating that PG synergizes with l-PS to support fX binding to bilayers. We also examined fX binding to nanobilayers containing 50% l-PS, 50% d-PS, or 25% l-PS + 25% d-PS (balance in all cases = 50% PC) (Fig. 3B). fX bound less to bilayers with 50% d-PS compared with 50% l-PS, whereas bilayers with 25% l-PS + 25% d-PS performed as well as those with 50% l-PS. Thus, d-PS synergizes with l-PS to enhance membrane binding of fX.
FIGURE 3.
PG and d-PS synergize with l-PS to enhance fX membrane binding. A, binding affinities of fX for Nanodiscs with varying l-POPS content with or without 40% palmitoyloleoyl-PG (balance = POPC). Nanodiscs were immobilized on Biacore sensor chips, after which steady-state levels of fX binding were quantified. Kd values were derived from the resulting binding isotherms. Data are means ± S.E. (n = 3). B, steady-state binding of fX to nanoscale bilayers quantified by SPR using Nanodiscs with 50% POPC and 50% l-POPS, 50% d-POPS, or 25% l-POPS + 25% d-POPS.
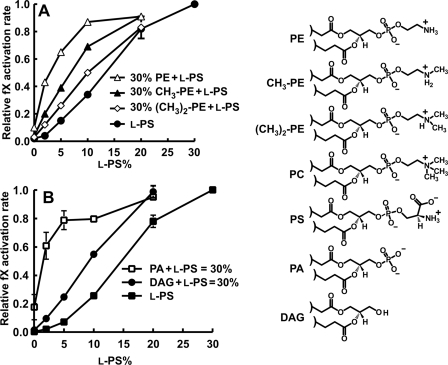
Methylation of PE Headgroups Diminishes Synergy
PC differs from PE by three N-methyl groups (cf. Fig. 4). We therefore asked how the presence of one or two N-methyl groups on PE would affect synergy with l-PS using TF-liposomes containing binary lipid mixtures (0–30% l-PS, balance = PC) or ternary lipid mixtures of 0–30% l-PS plus 30% PE, 30% N-monomethyl-PE, or 30% N-dimethyl-PE (balance = PC). The results (Fig. 4A) show a progressive diminution in synergy when one or two N-methyl groups were added to PE.
FIGURE 4.
Influence of PC methyl groups and PA phosphate on fX activation by TF-fVIIa. A, rates of fX activation by fVIIa on TF-liposomes prepared with varying l-dioleoyl-PS and no DOPE, 30% DOPE, 30% CH3-DOPE, or 30% (CH3)2-DOPE (balance = dioleoyl-PC). B, rates of fX activation by fVIIa on TF-liposomes prepared with binary lipid mixtures (0–30% l-POPS, balance = POPC) or ternary lipid mixtures in which the sum of l-POPS plus either palmitoyloleoyl-PA or DAG equaled 30% (balance = 70% PC). (DAG had palmitoyloleoyl acyl chains.) In both panels, fX activation rates for each TF-liposome preparation were normalized to the rate with 30% l-PS and 70% PC). Data are means ± S.E. (n = 3–7). Headgroup structures for lipids in this experiment are included on the right.
Importance of the Phosphate Groups of Phospholipids for Synergy
The experiment in Fig. 2C showed that PA synergized well with l-PS to support fX activation. We next investigated whether the PA phosphate is required by comparing the ability of PA and DAG to synergize with l-PS to enhance fX activation by TF-fVIIa. We prepared TF-liposomes containing binary lipid mixtures (0–30% l-PS, balance = PC) or ternary lipid mixtures of a total of either 30% PA + l-PS or 30% DAG + l-PS (balance = PC). DAG, which lacks a phosphate group, weakly synergized with l-PS compared with robust synergy with PA (Fig. 4B).
MD Simulations of Phosphate Accessibility in Bilayers
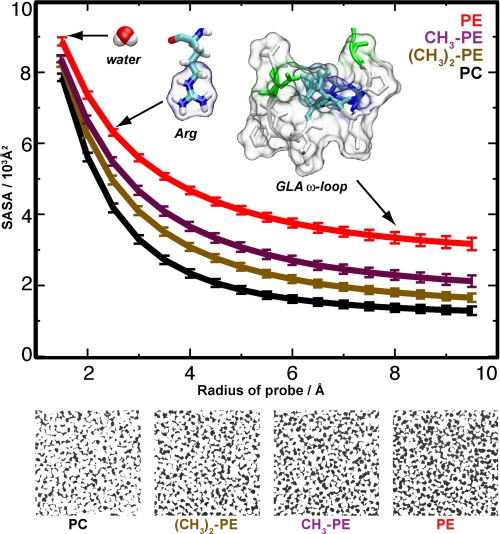
To investigate the molecular basis of phospholipid synergy, especially in terms of the accessibility of lipid phosphate groups, four independent 40-ns MD simulations of bilayers were performed in which the lipid headgroups were progressively transformed from PE to N-monomethyl-PE, N-dimethyl-PE, and PC, paralleling the experimental setting in Fig. 4. Each simulation included 1188 lipids distributed equally in the two leaflets to provide statistically meaningful data. Analyses of solvent-accessible surface areas (Fig. 5) clearly showed that each N-methyl group diminished the accessibility of the phosphate group for all probes with radii >1.5 Å (approximately the size of water), with the difference in phosphate exposure also being evident from top view snapshots shown for representative frames taken from the simulations (Fig. 5, lower). Although protein binding to membranes may induce lipid rearrangement, these measurements clearly show that the choline headgroup of PC sterically hinders access to the PC phosphate group.
FIGURE 5.
MD simulations show that PC has restricted phosphate accessibility. Upper, MD simulations showing that adding N-methyl groups to PE reduces the solvent-accessible surface areas (SASA) of the phosphate group: PE (red), CH3-PE (magenta), (CH3)2-PE (brown), and PC (black). The solvent-accessible surface areas were calculated using varying probe radii for the last 20 ns of 40-ns MD simulations of 1188 lipids. Water (radius of 1.4 Å), Arg (guanidinium group radius of ∼2.5 Å), and the fVIIa GLA domain ω-loop (radius of ∼8 Å) are shown to compare the results with the approximate sizes of functional groups that could interact with the phosphate groups. Lower, visual comparison of phosphate exposure in the bilayer simulations. Phosphate is black; the remainder of the lipid is white.
SSNMR Evidence for a New PS Chemical Environment Induced by GLA Domains
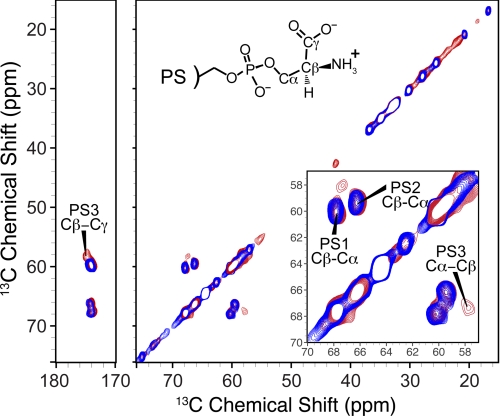
Using MAS-SSNMR, we recently showed that Ca2+ induces two distinct, equally populated conformations of l-PS headgroups in close spatial proximity, consistent with Ca2+ promoting l-PS clusters (16). In the present study, we employed this approach to investigate PS-GLA domain binding interactions using PS with uniformly 13C,15N-labeled l-serine (termed PS*) incorporated into Nanodiscs. When l-PS engages the GLA domain's proposed phospho-l-serine-specific binding site, its headgroup should take up a unique conformation observable by SSNMR. Two-dimensional 13C-13C spectra were acquired of 30% PS* and 70% PC Nanodiscs prepared with Ca2+ in the absence (Fig. 6, blue trace) or presence (red trace) of saturating levels of prothrombin fragment 1 (which contains the GLA domain). In the presence of fragment 1, we observed a new correlation pattern at 58, 67, and 175 ppm, corresponding to a new chemical environment for l-PS headgroups, termed PS3 (Fig. 6). This is in addition to the two nearly equally populated Ca2+-induced headgroup configurations (termed PS1 and PS2) that we also observed in our previous study (16). The individual cross-peaks made up the following fractions of the total l-PS intensities: PS1, 42 ± 3%; PS2, 41 ± 2%; and PS3, 17 ± 2%. SPR studies and MD simulations suggest that six to eight l-PS molecules constitute a GLA domain-binding site (11, 12). The observation that 17 ± 2% of the l-PS headgroups are found in the PS3 chemical environment upon GLA domain binding is consistent with the idea that one l-PS molecule per GLA domain is involved in the phospho-l-serine-specific binding interaction, with the rest of the PS molecules participating in phosphate-specific interactions (as depicted in Fig. 1).
FIGURE 6.
SSNMR spectroscopy demonstrates a novel PS headgroup environment induced upon GLA domain binding to bilayers. A 13C-13C two-dimensional SSNMR spectrum of 30% POPS* and 70% POPC Nanodiscs (25-ms dipolar assisted rotational resonance mixing, 2.6 h at 13 °C, 13.333-kHz MAS rate) is shown in blue. Overlaid in red is a spectrum of 30% POPS* and 70% POPC Nanodiscs in the presence of prothrombin fragment 1 (50-ms dipolar assisted rotational resonance mixing, 50 h at 13 °C, 10.000-kHz MAS rate). Both spectra were acquired on a 600-MHz (1H frequency) spectrometer. Inset, expansion of serine Cα-Cβ regions. PS1 and PS2 represent equally abundant chemical environments for PS headgroups in the presence of Ca2+, whereas PS3 represents a novel PS headgroup environment induced by Ca2+ plus prothrombin fragment 1.
DISCUSSION
The impetus for this study was to explain why PE, a relatively abundant plasma membrane phospholipid, poorly supports clotting reactions in bilayers composed of binary mixtures of PE and PC but strongly promotes clotting reactions when small amounts of PS are also included. We reasoned that solving this conundrum would allow us to develop a general mechanism for how GLA domains interact with PS-containing membranes. To investigate this question, we studied fX activation by TF-fVIIa assembled on liposomes with a wide variety of lipid compositions. Existing hypotheses to explain PE/PS synergy (hypotheses 1–4 in the Introduction) focus on specific properties of the PE ethanolamine headgroup. In contrast, the ABC hypothesis proposes that GLA domains bind to bilayers via a single phospho-l-serine-specific binding interaction together with multiple phosphate-specific interactions. In binary PS/PC mixtures, PS has to provide both types of interaction because the bulky choline moiety of PC sterically hinders GLA domains from interacting with its own phosphate group. (The same situation would also apply to sphingomyelin, whose phosphocholine headgroup is identical to that of PC. Because PC and sphingomyelin are the predominant phospholipids on the outer leaflet of the plasma membrane and because PS is sequestered to the inner leaflet, this explains the generally anticoagulant surface of healthy intact cells.) PE collaborates with PS to create membrane-binding sites for GLA domains because PE provides the more numerous phosphate-specific interactions, freeing up limited amounts of PS to engage in the single phospho-l-serine-specific interaction per GLA domain. We tested these hypotheses by examining how various phospholipids synergize with PS to enhance fX binding to bilayers and to enhance fX activation by TF-fVIIa in liposomes. Glycerophospholipids with a wide variety of different headgroups were fully as effective as PE in synergizing with PS, including those with myo-inositol (PI), glycerol (PG), d-serine (d-PS), or even just a bare phosphate (PA). We also found that PE did not synergize with PG in the absence of l-PS; instead, the effects of PE and PG were strictly additive. This is consistent with the ABC hypothesis, which predicts that although PE and PG can both provide phosphate-specific interactions, they cannot provide the essential phospho-l-serine-specific interaction. The fact that TF-liposomes containing binary mixtures of d-PS and PC poorly supported fX activation by TF-fVIIa recapitulates similar findings with prothrombinase and factor VIIIa-factor IXa (2). On the other hand, d-PS synergized strongly with l-PS to support TF-fVIIa activity, which is fully consistent with the ABC hypothesis because d-PS should retain the ability to enter into phosphate-specific interactions with GLA domains.
Hypotheses 1–4 are inconsistent with our findings. 1) Hypothesis 1 (i.e. non-lamellar PE promotes PS-rich microdomains) cannot explain our results because both lamellar and non-lamellar phospholipids synergized with PS. Furthermore, the nanoscale bilayers in Nanodiscs preclude long-range clustering of PS into microdomains, yet they strongly supported PG/PS and d-PS/l-PS synergy for fX binding. 2) Hypothesis 2 (i.e. PE-specific binding sites in GLA domains) likewise cannot explain our finding that phospholipids with headgroups structurally unrelated to ethanolamine all strongly synergized with PS. In addition, progressive diminution of PE/PS synergy as one or two N-methyl groups were added to PE (by progressively inhibiting access to its own phosphate) is consistent with the ABC hypothesis but inconsistent with hypothesis 2, which predicts an abrupt loss of synergy when the PE ethanolamine is chemically altered. Using MD simulations, we also showed that accessibility of the phosphate groups to objects the size of amino acids or the ω-loop of GLA domains is progressively diminished as the ethanolamine moiety of PE contains one, two, or three N-methyl groups. 3) Hypothesis 3 (i.e. hydrogen bonding between PE and adjacent phospholipids alters PS headgroup conformations) is also inconsistent with our findings because the various PS-synergizing headgroups tested cannot engage in the same kind of hydrogen bonding interactions that ethanolamine can. 4) Hypothesis 4 is the most similar to the ABC hypothesis, the chief difference being that hypothesis 4 proposes that bulky PC sterically hinders access of GLA domains to adjacent PS molecules, whereas the ABC hypothesis proposes that the PC headgroup sterically hinders access of GLA domains to PC's own phosphate. Hypothesis 4 is not completely ruled out by many of the experiments presented here, but DAG (which contains no phosphate group) synergized weakly with PS compared with full synergy observed with PA. Weak PS synergy with DAG is consistent with the notion that simply removing the bulky choline reduces some steric hindrance to the bilayer in general. However, this experiment argues that lipids need to retain their phosphate moieties to synergize fully with PS, in agreement with the ABC hypothesis.
Electrostatics per se cannot explain PE/PS synergy because we observed identical PS synergy with phospholipids having very different net charges, including PE (neutral); d-PS, PI, and PG (−1); and PA (−1 to −2). It is not necessary for the synergizing phospholipid to be zwitterionic or even to have an amino group, as PA, PI, and PG lack these features. The common feature required for a phospholipid to synergize with PS appears to be a phosphate unshielded by a bulky headgroup.
It is possible that when l-PS engages the phospho-l-serine-specific binding site in a GLA domain, it induces a conformational change in the GLA domain that promotes the phosphate-specific interactions. (In fact, PS has been proposed to be an allosteric regulator of clotting protein function (31).) This may help explain why GLA domains bind so weakly to membranes containing relatively high amounts of anionic phospholipids other than l-PS.
Our SSNMR results provide direct evidence for a unique PS headgroup environment induced when a GLA domain binds to PS-containing bilayers. The finding that approximately one-sixth of the total PS* signal intensity is in this chemical environment is fully consistent with the ABC hypothesis, which predicts that about this fraction of the PS molecules in a PS/PC bilayer will be engaged in one single phospho-l-serine-specific binding interaction per GLA domain provided that a GLA domain-binding site consists of about six or so PS molecules, a notion that is supported by both our MD simulations (Fig. 1B) (12, 13) and SPR-based binding studies (11). On the basis of these stoichiometries from Nanodisc-based studies, one might expect maximal rates of fX activation on liposomes with ∼5% l-PS + 25% synergistic phospholipid. When we examined fX activation by TF-fVIIa on TF-liposomes, maximal fX activation generally required somewhat more than 5% l-PS in combination with non-PC phospholipids. One explanation could be that l-PS is more readily clustered into nanodomains in the presence of Ca2+ than are other non-PC phospholipids.
This study provides a new explanation for how plasma membrane phospholipids synergize to create binding sites for GLA domain-containing blood clotting proteins using fX activation by TF-fVIIa as the exemplar. In future studies, it will be interesting to test the predictions of the ABC hypothesis versus PE-specific hypotheses on other membrane-dependent clotting reactions, e.g. activities of the prothrombinase, factor VIIIa-factor IXa, and protein S-activated protein C complexes. The nature of the proposed phospho-l-serine-specific and phosphate-specific binding sites deserves considerably greater scrutiny and is the subject of our ongoing studies aimed at achieving an atomic-scale understanding of the binding of GLA domains to membrane bilayers.
Acknowledgments
We thank Julie Collins and Jessica Luzwick for excellent technical contributions, Dr. John Boettcher for help with NMR experiments, and Dr. Stephen G. Sligar for kindly providing membrane scaffold protein.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL47014, R01 HL103999, R01 GM086749, R01 GM067887, R01 GM075937, and P41 RR05969. This work was also supported by American Heart Association Postdoctoral Fellowship 0920045G to R. L. D.-H. All simulations were performed using computer time on TeraGrid resources (Grant MCA06N060).
- PS
- phosphatidylserine
- fVIIa
- factor VIIa
- TF
- tissue factor
- fIX
- factor IX
- fX
- factor X
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- ABC
- Anything But Choline
- MD
- molecular dynamics
- SSNMR
- solid-state NMR
- DAG
- diacylglycerol
- POPC
- palmitoyloleoylphosphatidylcholine
- SPR
- surface plasmon resonance
- MAS
- magic angle spinning
- POPS
- palmitoyloleoylphosphatidylserine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PA
- phosphatidic acid
- DOPE
- dioleoylphosphatidylethanolamine.
REFERENCES
- 1. Stace C. L., Ktistakis N. T. (2006) Biochim. Biophys. Acta 1761, 913–926 [DOI] [PubMed] [Google Scholar]
- 2. Zwaal R. F., Comfurius P., Bevers E. M. (1998) Biochim. Biophys. Acta 1376, 433–453 [DOI] [PubMed] [Google Scholar]
- 3. Morrissey J. H. (2004) Int. J. Hematol. 79, 103–108 [DOI] [PubMed] [Google Scholar]
- 4. Neuenschwander P. F., Bianco-Fisher E., Rezaie A. R., Morrissey J. H. (1995) Biochemistry 34, 13988–13993 [DOI] [PubMed] [Google Scholar]
- 5. Smirnov M. D., Esmon C. T. (1994) J. Biol. Chem. 269, 816–819 [PubMed] [Google Scholar]
- 6. Smeets E. F., Comfurius P., Bevers E. M., Zwaal R. F. A. (1996) Thromb. Res. 81, 419–426 [DOI] [PubMed] [Google Scholar]
- 7. Gilbert G. E., Arena A. A. (1995) J. Biol. Chem. 270, 18500–18505 [DOI] [PubMed] [Google Scholar]
- 8. Zwaal R. F., Comfurius P., Bevers E. M. (2005) Cell. Mol. Life Sci. 62, 971–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgess S. W., McIntosh T. J., Lentz B. R. (1992) Biochemistry 31, 2653–2661 [DOI] [PubMed] [Google Scholar]
- 10. Murzyn K., Zhao W., Karttunen M., Kurdziel M., Róg T. (2006) Biointerphases 1, 98–105 [DOI] [PubMed] [Google Scholar]
- 11. Shaw A. W., Pureza V. S., Sligar S. G., Morrissey J. H. (2007) J. Biol. Chem. 282, 6556–6563 [DOI] [PubMed] [Google Scholar]
- 12. Ohkubo Y. Z., Tajkhorshid E. (2008) Structure 16, 72–81 [DOI] [PubMed] [Google Scholar]
- 13. Ohkubo Y. Z., Morrissey J. H., Tajkhorshid E. (2010) J. Thromb. Haemost. 8, 1044–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang M., Rigby A. C., Morelli X., Grant M. A., Huang G., Furie B., Seaton B., Furie B. C. (2003) Nat. Struct. Biol. 10, 751–756 [DOI] [PubMed] [Google Scholar]
- 15. Bayburt T. H., Grinkova Y. V., Sligar S. G. (2002) Nano Lett. 2, 853–856 [Google Scholar]
- 16. Boettcher J. M., Davis-Harrison R. L., Clay M. C., Nieuwkoop A. J., Ohkubo Y. Z., Tajkhorshid E., Morrissey J. H., Rienstra C. M. (2011) Biochemistry 50, 2264–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mann K. G. (1976) Methods Enzymol. 45, 123–156 [DOI] [PubMed] [Google Scholar]
- 18. Smith S. A., Morrissey J. H. (2004) J. Thromb. Haemost. 2, 1155–1162 [DOI] [PubMed] [Google Scholar]
- 19. Waters E. K., Morrissey J. H. (2006) Biochemistry 45, 3769–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Geet A. L. (1968) Anal. Chem. 40, 2227–2229 [Google Scholar]
- 21. Hediger S., Meier B. H., Kurur N. D., Bodenhausen G., Ernst R. R. (1994) Chem. Phys. Lett. 223, 283–288 [Google Scholar]
- 22. Bennett A. E., Rienstra C. M., Auger M., Lakshmi K. V., Griffin R. G. (1995) J. Chem. Phys. 103, 6951–6958 [Google Scholar]
- 23. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mackerell A. D., Jr., Feig M., Brooks C. L., 3rd (2004) J. Comput. Chem. 25, 1400–1415 [DOI] [PubMed] [Google Scholar]
- 25. Klauda J. B., Venable R. M., Freites J. A., O'Connor J. W., Tobias D. J., Mondragon-Ramirez C., Vorobyov I., MacKerell A. D., Jr., Pastor R. W. (2010) J. Phys. Chem. B 114, 7830–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Phys. 79, 926–935 [Google Scholar]
- 27. Feller S. E., Zhang Y., Pastor R. W. (1995) J. Chem. Phys. 103, 4613–4621 [Google Scholar]
- 28. Martyna G. J., Tobias D. J., Klein M. L. (1994) J. Chem. Phys. 101, 4177–4189 [Google Scholar]
- 29. Darden T., York D., Pedersen L. (1993) J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 30. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 31. Lentz B. R. (2003) Prog. Lipid Res. 42, 423–438 [DOI] [PubMed] [Google Scholar]
- 32. Schroit A. J., Zwaal R. F. (1991) Biochim. Biophys. Acta 1071, 313–329 [DOI] [PubMed] [Google Scholar]