Abstract
Cytoplasmic lipid droplets (CLD) in mammary epithelial cells undergo secretion by a unique membrane envelopment process to produce milk lipids. Adipophilin (ADPH/Plin2), a member of the perilipin/PAT family of lipid droplet-associated proteins, is hypothesized to mediate CLD secretion through interactions with apical plasma membrane elements. We found that the secretion of CLD coated by truncated ADPH lacking the C-terminal region encoding a putative four-helix bundle structure was impaired relative to that of CLD coated by full-length ADPH. We used homology modeling and analyses of the solution and membrane binding properties of purified recombinant ADPH C terminus to understand how this region possibly mediates CLD secretion. Homology modeling supports the concept that the ADPH C terminus forms a four-helix bundle motif and suggests that this structure can form stable membrane bilayer interactions. Circular dichroism and protease mapping studies confirmed that the ADPH C terminus is an independently folding α-helical structure that is relatively resistant to urea denaturation. Liposome binding studies showed that the purified C terminus binds to phospholipid membranes through electrostatic dependent interactions, and cell culture studies documented that it localizes to the plasma membrane. Collectively, these data provide direct evidence that the ADPH C terminus forms a stable membrane binding helical structure that is important for CLD secretion. We speculate that interactions between the four-helix bundle of ADPH and membrane phospholipids may be an initial step in milk lipid secretion.
Keywords: Lipid Droplet, Lipid Transport, Lipoprotein, Mammary Gland, Secretion, Adipophilin, Domain, Structure
Introduction
Milk lipids are an essential source of neonatal calories and provide nutrients in the form of fatty acids and bioactive lipids that are required for growth and development (1). Milk lipids are delivered to the newborn as milk fat globules (MFG)4 derived from triglyceride-rich cytoplasmic lipid droplets (CLD) (2). Adipophilin (ADPH/ADRP/Adfp/Plin2), a member of the perilipin/PAT (perilipin/adipophilin/TIP47) family of proteins (3), is an abundant MFG protein (4–6) and a prominent CLD-associated protein of mammary epithelial cells (7, 8). PAT proteins have been shown to promote the formation and accumulation of CLD in a variety of tissues (9, 10) and are also implicated in the trafficking and secretion of CLD (2, 9). The lactating mammary gland is an ideal system to study the recurrent cycles of CLD formation, accumulation, and secretion, and previous work from our laboratory has implicated ADPH in each of these processes (5, 8, 11).
CLD are secreted during lactation by an apocrine process where the apical plasma membrane engulfs associated CLD, which are then released by a budding process to form MFG (2). Two key regulators of CLD secretion are the transmembrane protein butyrophilin (BTN) and the cytoplasmic homodimer, xanthine oxidoreductase (XOR). Mice deficient in either of these proteins display impaired CLD envelopment and secretion (12, 13). In addition to being an abundant protein on mammary epithelial CLD and secreted MFG (7), ADPH co-localizes with BTN and XOR at sites of CLD secretion on the apical membrane in lactating mammary glands (5), and it is isolated as a detergent-stable complex with BTN and XOR from MFG membranes (5). These data are the primary basis for the current hypothesis that ADPH directly associates with BTN and XOR to achieve CLD secretion (2, 11). However, direct support for this mechanism of CLD secretion is lacking, and alternative mechanisms have been proposed (11, 14). Thus, additional studies are required to establish the details by which mammary epithelial cells envelop and secrete CLD.
Understanding of the possible physiological functions of ADPH has been advanced by studies demonstrating that it is composed of discrete structural and functional domains (15–18). Cell culture studies have shown that the CLD binding function of ADPH is located within its N-terminal half (15, 17). In addition, sequences located in the N terminus of ADPH within a region of homology to perilipin and TIP47 referred to as the PAT-1 domain (3) have been shown to be essential for CLD stabilization and accumulation and to encode determinants of its proteasomal degradation (16). ADPH and TIP47 share over 40% similarity in their amino acid sequence. The crystallographic structure of the C-terminal portion of TIP47 has been solved and has been shown to form a four-helix bundle motif (18). At present, the importance of this structure to the physiological properties of TIP47 has not been established. Based on its sequence similarity to TIP47, the C-terminal half of ADPH is predicted to form a four-helix bundle (18); however, there are currently no experimental data to validate this prediction.
In this study, we provide evidence that the C-terminal portion of ADPH is important for CLD secretion by mammary epithelial cells. In addition, we developed a homology model of the ADPH C-terminal domain that supports the presence of a four-helix bundle and provides direct biochemical evidence that the C-terminal portion of ADPH encodes an independently folding helical structure. For the first time, we show that the ADPH C-terminal portion directly binds liposomes in vitro and that the lipid-membrane association is mediated by electrostatic interactions. Expression of the C-terminal domain in HEK 293 cells further shows that the ADPH four-helix bundle motif localizes at the plasma membrane. These results suggest that ADPH contains a structured, independently folding C-terminal domain that interacts with lipid membranes to regulate CLD secretion.
EXPERIMENTAL PROCEDURES
Antibodies
Rabbit polyclonal antibodies specific to the C-terminal 15 amino acids (anti-ADPHcterm) or the N-terminal 25 amino acids (anti-ADPHnterm) of mouse ADPH were generated as described previously (19). Antibodies to GFP were obtained from BD Biosciences Clontech (Madison, WI). 10-nm gold-conjugated goat anti-rabbit IgG was obtained from Amersham Biosciences. Mouse monoclonal VSV-G antibody was obtained from Roche Applied Science. Rabbit polyclonal antibodies to human ezrin were obtained from Abcam (Cambridge, MA). Alexa594-conjugated phalloidin, Alexa488, and Alexa594 antibodies were obtained from Invitrogen. Rabbit polyclonal antibodies to the human transferrin receptor were a kind gift from Professor Paul Seligman (University of Colorado, Denver, CO). Alexa 488- and 594-wheat germ agglutinin were obtained from Invitrogen. DAPI and Hoechst 33342 were purchased from Sigma-Aldrich and Anaspec (Freemont, CA), respectively.
Animals
CD-1 mice (Charles River, Inc., Wilmington, MA) were maintained as breeding colonies at the United States Department of Agriculture-approved Center for Comparative Medicine at the University of Colorado Anschutz Medical Campus and housed individually. Pregnancy was timed by the observation of vaginal plugs after mating. The first day of pregnancy is taken as the day of vaginal plug detection. Parturition occurs on approximately day 19 of gestation and is also designated day 1 of lactation. Mammary tissue was removed from animals euthanized by carbon dioxide and cervical dislocation and frozen in liquid nitrogen or processed for immunofluorescence microscopy (8, 19). Animal procedures were approved by the University of Colorado Anschutz Medical Campus Institutional Animal Care and Use Committee.
Plasmids
Plasmids containing cDNA encoding ADPH(fl)-VSV and ADPH(1–220)-VSV have been described (15). The portion of this plasmid encoding ADPH(fl)-VSV or ADPH(1–220)-VSV was excised and ligated into pEGFP-C3 to generate GFP-ADPH(fl)-VSV and GFP-ADPH(1–220)-VSV plasmids, respectively. cDNA encoding the murine ADPH(172–425) sequence was PCR-amplified from ADPH(fl)-VSV and ligated into the pGEX-4T1 bacterial expression vector. ADPH(170–425) was generated with a C-terminal VSV-G epitope tag (YTDIEMNRLGK). The ADPH(170–425)-VSV sequence was PCR-amplified from a full-length, murine ADPH-VSV plasmid (16) and ligated into a pcDNA3.1 Zeo+ vector. All plasmid sequences were confirmed by DNA sequencing.
Adenoviral Transduction and Tissue Processing
Adenovirus expressing GFP (AdGFP) was constructed as described previously (20). The adenoviruses encoding GFP-ADPH(fl)-VSV (AdGFP-ADPH(fl)-VSV) or GFP-ADPH(1–220)-VSV (AdGFP-ADPH(1–220)-VSV) were constructed by cloning the respective coding sequences in frame with the GFP coding sequence into the plasmid pShuttleCMV (21). Viruses were grown and purified as described previously (22). CsCl gradient-purified viruses were dialyzed into storage buffer containing 50% (v/v) glycerol as a cryoprotectant (23). Mammary epithelial cells were transduced on pregnancy day 17 with the indicated adenovirus particles (24). Transduced mammary glands were excised on lactation day 2 and processed for immunofluorescence microscopy (8). Mammary gland sections were incubated with Alexa 594- or Alexa 488-labeled wheat germ agglutinin to detect luminal borders and with DAPI to detect nuclei (8). The sections were imaged by confocal fluorescence microscopy using an Olympus iX81 microscope equipped with a DSU spinning disc and Slidebook software (Intelligent Imaging Innovations, Inc., Denver, CO). All fluorescent images were digitally deconvolved using the No Neighbors algorithm (Slidebook), converted to TIFF files, and processed using Photoshop (Adobe Systems Inc., Mountain View, CA).
CLD Quantitation
Average CLD diameters were determined by analysis of individual CLD coated with GFP-ADPH(fl)-VSV, GFP-ADPH(1–220)-VSV, or endogenous ADPH in immunostained sections containing 60–80 randomly chosen alveoli at ×600 magnification as described previously (8). Five sections each from three different mice were used for quantitation. All values are means ± S.E. Statistical significance was determined using Student's t test.
Cell Lines
Stable cell lines expressing GFP-ADPH(fl)-VSV, GFP-ADPH(1–220)-VSV, or ADPH(170–425)-VSV were generated by transfecting HEK 293 cells (ATCC, Manassas, VA) with plasmids containing their respective cDNA (16). Cell lines were cultured in the presence of oleic acid, processed for immunofluorescence analysis, and imaged by confocal microscopy as described (16).
Immunoelectron Microscopy
Cells were processed for immunoelectron microscopy using a modified Tokuyasu method (25). Briefly, pelleted cells were fixed overnight at 4 °C in PBS-buffered 4% paraformaldehyde containing 5% sucrose and 100 mm HEPES and infiltrated with PBS containing 2.1 m sucrose over ∼10 h, with repeated solution changes. Fixed cells were transferred to an aluminum cryosectioning stub (Ted Pella, Inc., Redding, CA) and immediately frozen in liquid nitrogen. Semithin (90 nm) cryosections were cut at −110 °C with an UltraCut UCT/FCS cryomicrotome (Leica), using a diamond knife (Diatome) and transferred to a Formvar-coated, carbon-coated, glow-discharged 100-mesh copper-rhodium electron microscopy grid. Following blocking of nonspecific antibody binding sites with 10% calf serum in PBS, the sections were labeled by sequential incubation with antibodies to GFP and colloidal gold-conjugated secondary antibodies (Ted Pella Inc., Redding, CA) and then negatively stained and embedded with 1% uranyl acetate, 1% methylcellulose in distilled water. Samples were viewed in a Tecnai TF20 electron microscope (FEI) operating at 200 KeV, and images were collected digitally.
Expression and Purification of Recombinant ADPH(172-425)
ADPH(172–425) was expressed as a GST fusion protein in Escherichia coli BL21 (DE3) strain (Invitrogen). The GST fusion protein was purified by chromatography on glutathione-Sepharose 4B, Q-Sepharose, and Superdex75 (GE Healthcare). Purified GST-ADPH(172–425) was collected, rebound to glutathione-Sepharose, and digested with 50 units of thrombin (Sigma-Aldrich) to remove the GST tag. The purity of eluted protein was determined by SDS-PAGE and silver staining and immunoblot analysis using anti-ADPHcterm antibodies (19). The purified protein was verified to correspond to the ADPH 172–425 fragment by mass spectrometry.
Circular Dichroism Spectroscopy
Circular dichroism (CD) measurements were collected with a Jasco J-815 spectropolarimeter (Jasco Analytical Instruments, Easton, MD). Six accumulations of scans ranging from 195 to 250 nm were measured at 4 °C with 0.27 mg/ml ADPH(172–425) in 10 mm potassium phosphate, pH 7.2, 100 mm potassium chloride or in 50% trifluoroethanol (Sigma-Aldrich). The molar ellipticity ([θ]) in degrees cm2 dmol−1 was calculated from the equation, [θ] = θ·MRW/10lc as described (26), where MRW represents mean residue weight, l is path length in cm, and c is concentration in mg/ml. The α-helical content of ADPH(172–425) was calculated from the equation, % α-helix = (−[θ]222 + 3000)/39,000 (27).
Proteolytic Mapping
Purified ADPH(172–425) was digested with sequencing grade trypsin (Promega, Madison, WI), EndoGluC (New England BioLabs, Inc., Ipswich, MA), or chymotrypsin (Sigma-Aldrich). Trypsin digestion was carried out in 20 mm Tris, pH 8.0, 50 mm NaCl, 2 mm MgCl2, 2 mm DTT with 35 μm ADPH(172–425) in a 1:862 mass ratio of trypsin/ADPH. EndoGluC digestion was carried out in 50 mm Tris, pH 8.0, 0.5 mm Glu-Glu buffer (New England BioLabs, Inc., Ipswich, MA) at a 1:674 mass ratio. Chymotrypsin digestion was carried out in 100 mm Tris, pH 7.8, 10 mm CaCl2 buffer in a 1:800 mass ratio. For urea stability experiments, ADPH(172–425) was incubated in 0.5, 1, 2, and 5 m urea overnight at 22 °C, diluted to 0.8 m urea, and incubated with trypsin at a 1:862 mass ratio. Digestion products were separated by SDS-PAGE on 16% gels and visualized by silver staining or immunoblot analysis using anti-ADPHcterm antibodies. Proteolytic fragment identities were determined by mass spectrometry using a Q-Tof2 mass spectrometer (Waters Corp., Milford, MA) following peptide separation by liquid chromatography separation on a Vydac C18 column. Data analysis was performed using MassLynx 4.1 software (Waters Corp.).
Liposome Preparation and Binding Assay
Liposomes were generated according to Lee et al. (28) with minor modifications. Briefly, a solution of 2 mm 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (Sigma-Aldrich) and 2 mm 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS) (Avanti Polar Lipids, Alabaster, AL) in chloroform/methanol/water (65:25:4) were dried under vacuum and resuspended in 20 mm Tris, pH 7.2, 150 mm NaCl, 1 mm DTT. The liposomes were frozen in liquid nitrogen and thawed at 42 °C for three cycles and extruded through a 1.0-μm polycarbonate membrane (GE Healthcare) to produce uniform 1.0-μm vesicles. Liposomes were collected by centrifugation and resuspended in the specified buffers and incubated with 15 μg of purified ADPH(172–425) for 60 min at 22 °C. ADPH-bound liposomes were collected by centrifugation at 16,000 × g for 15 min, resuspended in 100 μl of buffer, and analyzed by anti-ADPHcterm immunoblotting following SDS-PAGE.
Homology Modeling
All computation-based modeling was performed using Discovery Studio (version 2.5, Accelrys Inc. (San Diego, CA)). An NCBI BLAST search cross-referenced with the RCSB Protein Data Bank identified known crystal structures to be used as structural templates for the generation of the mouse ADPH (Ser172–Glu425) model. The structure of TIP47 (Protein Data Bank code 1SZI) (18) for residues Phe189–Lys263 and Glu302–Glu409 (identities 28%, positives 47%) and apolipoprotein A-I (Protein Data Bank code 2A01) (29) for residues Ser172–Tyr188 (identities 22%, positives 41%) were used to generate the ADPH model. The sequences of the proteins were aligned, and five homology models were created. The residues in the models were corrected for physiological pH, and loop refinement was performed on residues His266–Cys300. The most energy-favored model was retained for further consideration. The model was refined further using CHARMm (30) and subjected to energy minimization (conjugate gradient, 1000 iterations) at a convergence of 0.001 kcal/mol using a Generalized Born implicit solvent model (31). In the initial minimization, the protein backbone atoms were fixed, followed by a final minimization where all atoms were unfixed and restraints were removed.
ADPH with a Lipid Bilayer Modeling
The Add Membrane and Orientate Molecule protocol within Discovery Studio was used to examine the potential interaction of ADPH(172–425) with a phosphatidylcholine-based lipid bilayer. The membrane was defined as a low dielectric planar slab, ADPH(172–425) was treated as a rigid body, and the models were corrected for physiological pH. No NaCl was introduced into the system. The protocol used a stepwise search algorithm for the optimal orientation of ADPH(172–425) relative to the membrane. The Generalized Born implicit membrane CHARMm module was used to calculate the polar contribution to solvation energy, and the non-polar contribution to solvation energy was approximated with a solvent-accessible surface area-dependent term, which is calculated with a uniform surface tension coefficient with the same value applied to all atoms (32, 33). The optimal interaction of ADPH(172–425) with the membrane corresponded to the orientation with the minimum solvation energy.
RESULTS
Adenoviral Expression of GFP-ADPH(1–220) in Mammary Epithelial Cells
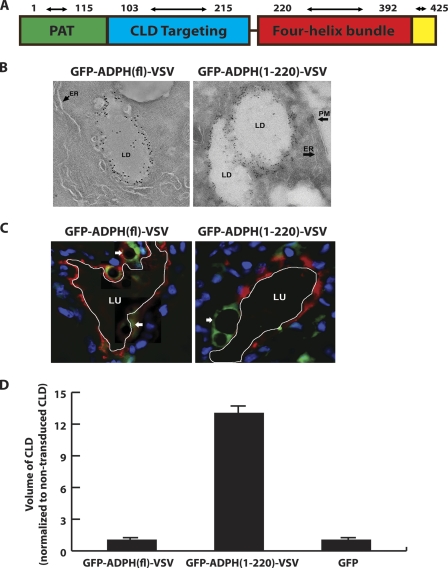
Initial structure-function studies indicating that CLD binding and stabilization functions of ADPH are located within its N-terminal half (Fig. 1A) (15–17) have led to speculation that the C-terminal portion of ADPH may contribute to CLD secretion by mediating interactions with elements of the plasma membrane (11). To test this hypothesis, we investigated secretion of CLD coated with exogenously expressed truncated ADPH lacking the C-terminal region, ADPH(1–220). To simplify detection of exogenously expressed forms of ADPH in vivo, we generated GFP fusion proteins of VSV epitope-tagged forms of full-length ADPH (GFP-ADPH(fl)-VSV) and ADPH(1–220) (GFP-ADPH(1–220)-VSV). Immunofluorescence (data not shown) and tomographic analyses of immunogold-stained cells stably expressing GFP-ADPH(fl)-VSV or GFP-ADPH(1–220)-VSV (Fig. 1B) demonstrate that these constructs correctly localize to the CLD surface and verify that the C-terminal region is not required for the CLD binding function of ADPH (15, 17). We did not observe significant differences in the density of gold particles on the surface of CLD or in the size of CLD coated with the GFP-ADPH(fl)-VSV or GFP-ADPH(1–220)-VSV (Fig. 1).
FIGURE 1.
Removal of the C-terminal portion of ADPH impairs CLD secretion. A, proposed domain organization of ADPH. B, tomographic sections of cells expressing GFP-ADPH(fl)-VSV or GFP-ADPH(1–220)-VSV following reaction with antibodies to GFP and staining with 10-nm gold-conjugated secondary antibodies. The large arrows indicate localization of immunogold-stained GFP-ADPH(fl)-VSV or GFP-ADPH(1–220)-VSV on the CLD (LD) surface. The arrowheads indicate the location of smooth endoplasmic reticulum (ER) and plasma membrane (PM). C, confocal fluorescence images of mammary gland alveoli from mice at lactation day 2 following transduction on pregnancy day 17 with AdGFP-ADPH(1–220)-VSV or AdGFP-ADPH(fl)-VSV. The arrows indicate CLD coated with GFP-ADPH(1–220)-VSV or GFP-ADPH(fl)-VSV (green). The small arrow indicates the localization of clusters of small CLD coated with GFP-ADPH(fl)-VSV (green). The apical border of each alveolus was identified by staining with Alexa 594-labeled wheat germ agglutinin and is outlined in white. Scale bars, 50 μm. D, relative size of CLD coated with GFP-ADPH(fl)-VSV, GFP-ADPH(1–220)-VSV, or endogenous ADPH in mammary epithelial cells transduced with AdGFP-ADPH(fl)-VSV, AdGFP-ADPH(1–220)-VSV, or AdGFP, respectively. All values are normalized to CLD size in non-transduced cells. n = 3 mice/group, 150 alveoli/group. +, p < 0.05.
We next tested the importance of the ADPH C-terminal region in CLD secretion by determining the relative abilities of CLD coated with GFP-ADPH(fl)-VSV or GFP-ADPH(1–220)-VSV to undergo secretion by mammary glands of intact mice using CLD size on lactation day 2 as an index of secretion (8, 34). CLD size increases during pregnancy, reaching a maximum between pregnancy days 17 and 18. Following parturition (at about pregnancy day 19) and the onset of lactation, there is a dramatic decrease in CLD size with the commencement of their secretion (24). GFP-ADPH(fl)-VSV, GFP-ADPH(1–220)-VSV, and GFP were expressed in epithelial cells of intact mammary alveoli prior to the onset of lactation by transduction with adenovirus encoding the respective construct on pregnancy day 17. Consistent with the immunogold staining of cultured cells, GFP-ADPH(fl)-VSV and GFP-ADPH(1–220)-VSV selectively localize to the CLD in mammary epithelial cells in vivo (Fig. 1C). Most of the CLD coated with GFP-ADPH(fl)-VSV appeared to be less than 2 μm in diameter (Fig. 1C, arrowhead), although a few larger CLD were detected (Fig. 1C, arrow) in some cells. The average size of GFP-ADPH(fl)-VSV-coated CLD was comparable with that of CLD coated with endogenous ADPH (Fig. 1D). In contrast, CLD coated with GFP-ADPH(1–220)-VSV were larger diameter structures (Fig. 1C). Because CLD size can vary with the extent of secretory activity of a given alveolus, we compared the relative sizes of CLD coated with endogenous ADPH, GFP-ADPH(fl)-VSV, or GFP-ADPH(1–220)-VSV in multiple randomly selected sections from three independent transduction experiments. As shown in Fig. 1D, we found that CLD coated with GFP-ADPH(1–220)-VSV were significantly larger than those coated with either GFP-ADPH(fl)-VSV or endogenous ADPH, whereas CLD coated by GFP-ADPH(fl)-VSV were similar in size to those coated with endogenous ADPH in both non-transduced and AdGFP-transduced cells. Together, these results indicate that truncated ADPH lacking the C-terminal region is able to bind to CLD; however, secretion of CLD coated with the mutant protein appears to be impaired relative to that of CLD coated with endogenous ADPH or exogenously expressed GFP-ADPH(fl)-VSV.
Homology Modeling of the ADPH C-terminal Region
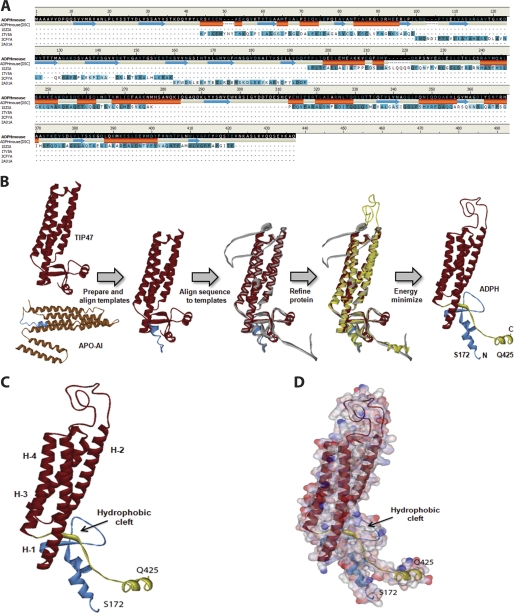
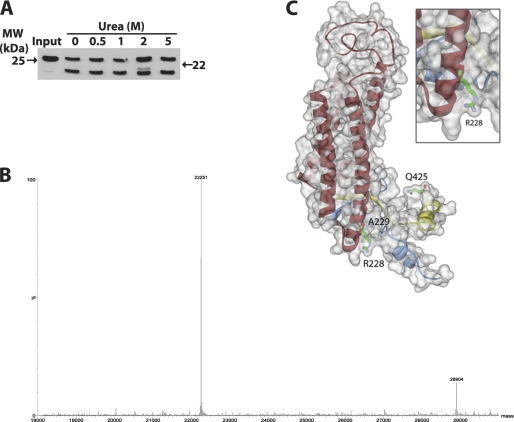
The concept that the C-terminal portion of ADPH consists of a four-helix bundle structure is based on evidence showing similarity between the primary sequence of the C-terminal region of ADPH and that of TIP47, whose crystal structure has been solved (18). To develop a more complete picture of structural properties of the ADPH C-terminal region, we constructed a homology model of this region using the known three-dimensional structures of murine TIP47 and human apolipoprotein A-I as templates (Fig. 2B). The resulting model had a DOPE score of −34,957.7 and revealed an overall α-helical structure forming a deep hydrophobic cleft and four-helix bundle, as predicted by previous sequence alignments (Fig. 2C) (18). The hydrophobic cleft is ∼9.5 Å deep with an α/β fold at the amino end and short stretches of α-helices at the carboxyl end (residues 393–425) that contribute to formation of the cleft (Fig. 2C). Adjacent to the cleft are four amphipathic α-helices that adopt a helical bundle motif. Each helix varies in length, with 37, 29, 22, and 26 amino acids, respectively, ranging from 7 to 10 helical turns (Table 1). Similar to the helical bundle motif of apoE, there appear to be hydrogen bonding interactions within each helix and a lack of disulfide bridges (35). Fig. 2D illustrates the surface charge distribution of the domain and highlights the amphipathic nature of the bundle. An interesting feature of the ADPH C-terminal domain is the presence of a 40-residue loop between helices 1 and 2. An unstructured loop of this size may also be present in the TIP47 C terminus but is excluded from the crystal structure (18). Computational modeling thus supports the structural similarity between ADPH and TIP47 C termini and the presence of a putative four-helix bundle motif in ADPH.
FIGURE 2.
Homology model of murine ADPH(172–425). A, alignment of mADPH sequence with the template sequences of human TIP47 (Protein Data Bank code 1SZI, chain A) and apolipoprotein A-I (Protein Data Bank code 2A01, chain A). Conserved amino acid residues between mADPH sequence and the structural templates are highlighted in dark green, structurally similar residues are highlighted in light green, and residues that have no match are not highlighted. A predicted secondary structure based on the amino acid sequence is displayed (helices are shown in orange, and sheets are shown in blue). B, the sequences of TIP47 and apolipoprotein A-I were aligned, and five homology models were created. The residues in the models were corrected for physiological pH and loop refinement. The model was refined further using CHARMm and subjected to energy minimization using a Generalized Born implicit solvent model. C, the homology model of murine ADPH(172–425), with residues Ser172–Glu219 colored blue (prehelix region), Ser220–Thr394 colored red (four-helix bundle), and Pro395–Gln425 colored yellow. The location of the putative hydrophobic cleft is indicated. D, surface map colored by electrostatic potential overlain on the four-helix bundle structure of ADPH(172–425). Negatively charged residues are indicated in red, and positively charged residues are indicated in blue.
TABLE 1.
Organization of helices in ADPH(172–425) homology model
| Residues | Number of residues | Length of helix | Turns/helix | Net charge | |
|---|---|---|---|---|---|
| Å | |||||
| Helix 1 | 223–260 | 37 | 54.218 | 10 | +1 |
| Helix 2 | 301–330 | 29 | 44.124 | 9 | 0 |
| Helix 3 | 335–357 | 22 | 33.460 | 7 | 0 |
| Helix 4 | 366–392 | 26 | 38.912 | 8 | −1 |
Recombinant ADPH(172–425) Forms a Stable Helical Structure
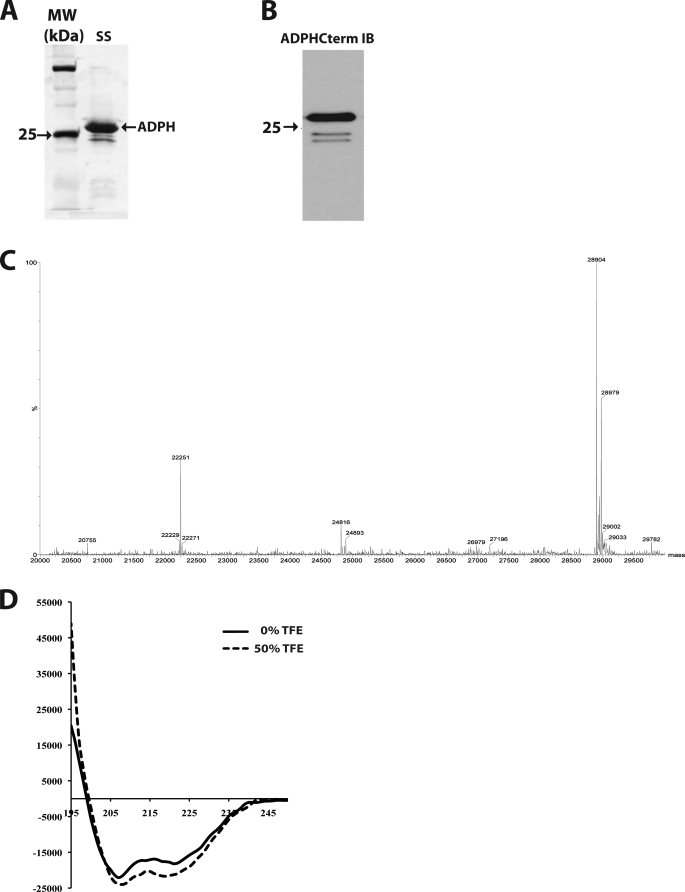
The homology modeling results suggest that the ADPH C-terminal region may be an independently folding structure. To directly test this prediction, we expressed ADPH(172–425) in E. coli as a GST fusion protein. After thrombin cleavage and removal of the N-terminal GST tag, silver stain analysis shows that purified recombinant ADPH(172–425) is the overwhelmingly predominant protein species (Fig. 3A). Immunoblotting with anti-ADPHcterm antibodies shows that the minor contaminants in our preparation contain an intact C terminus and thus appear to correspond to related cleavage fragments (Fig. 3B). LC/MS mass spectrometry (Fig. 3C) revealed that the molecular mass of the recombinant protein is 28,904 Da, which is consistent with the theoretical mass of 28,903 Da for residues 172–425 with a Gly-Ser N-terminal linker.
FIGURE 3.
Expression and purification of recombinant ADPH(172–425) from E. coli. A, silver stain of purified ADPH(172–425). Lane 1, molecular weight markers; lane 2, purified ADPH(172–425). The arrows indicate the positions of the 75, 25, and 15 kDa markers. B, immunoblot of purified ADPH(172–425) with anti-ADPHcterm antibodies. The arrow indicates the position of the 25 kDa marker. C, LC/MS analysis of purified recombinant ADPH(172–425). D, far-UV CD spectrum of purified ADPH(172–425) in the absence (solid line) and presence of 50% trifluoroethanol (TFE) (dashed line), indicating helical contents of 53 and 61% respectively.
To determine if the recombinant protein follows the helical predictions of our homology model, a far-UV CD spectrum was measured. Fig. 3D demonstrates the helical nature of the protein by the depressions at 208 and 222 nm, typical of α-helical proteins. The calculated helical content of ADPH(172–425) in aqueous solution was 53%. Its degree of helicity increased to 61% after incubation in 50% trifluoroethanol, suggesting that unstructured regions of the recombinant protein may have additional helix formation potential in a membrane-like environment (36). These initial experiments support our homology model and demonstrate that ADPH(172–425) is an independently folding structure with significant α-helical nature.
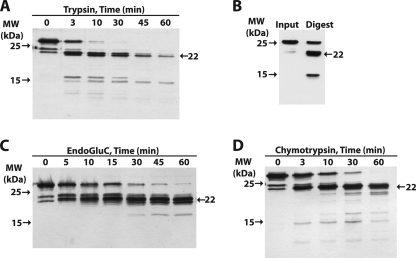
Evidence of ADPH(172–425) tertiary structure was demonstrated by limited proteolysis with trypsin, chymotrypsin, and EndoGluC. Brief trypsin proteolysis resulted in peptides between 10 and 15 kDa and a larger fragment near 22 kDa, whereas a 1-h digestion led to two fragments, one of ∼22 kDa and one of ∼15 kDa (Fig. 4A). Anti-ADPHcterm immunoblotting (Fig. 4B) shows that both the 22- and 15-kDa proteolysis fragments retain the 15 terminal residues of ADPH and thus have an intact C terminus. The EndoGluC and chymotrypsin digests also generated stable 22-kDa fragments (Fig. 4, C and D), which retained intact C termini (data not shown).
FIGURE 4.
Proteolytic mapping of ADPH(172–425). A, time dependence of trypsin cleavage of ADPH(172–425) analyzed by SDS-PAGE and silver staining. Size markers are indicated to the left. The fragment at 22 kDa (arrow on right) appears to be relatively resistant to trypsin digestion. B, immunoblot analysis of trypsin-digested ADPH(172–425) with anti-ADPHcterm showing that the 22- and 15-kDa fragments contain intact C termini. Time courses of EndoGluC (C) and chymotrypsin (D) cleavage of ADPH(172–425) analyzed by SDS-PAGE and silver staining. The presence of the stable 22-kDa fragment is indicated.
The structural stability of the ADPH C-terminal region was assessed by trypsin proteolysis following incubation in urea. Purified ADPH(172–425) was incubated in 0.5, 1, 2, or 5 m urea prior to trypsin proteolysis in 0.8 m urea. Fig. 5A shows that in each condition, trypsin digestion generated the 22-kDa fragment to roughly the same extent, suggesting that this fragment is relatively resistant to proteolysis. Mass spectrometric analysis revealed the mass of the 22-kDa fragment to be 22,251 Da (Fig. 5B), which corresponds to the calculated mass of residues 229–425 (22,251 Da) and cleavage at Arg228 located at the base of helix 1 of the four-helix bundle motif (Fig. 5C). Accordingly, our data indicate that the region bounded by helix 1 of the four-helix bundle to the C-terminal end of ADPH (Fig. 5C) forms a stable structure that is resistant to urea denaturation and proteolysis.
FIGURE 5.
Urea stability of ADPH(172–425). A, immunoblot analysis of the trypsin cleavage pattern of ADPH(172–425) in 0.8 m urea following overnight incubation in the indicated concentrations of urea. Anti-ADPHcterm antibodies were used for immunodetection. B, LC/MS identification of a 22 kDa proteolysis band after deconvolution. C, ADPH homology model of the 22-kDa cleavage fragment from Ala229 to Gln425 generated by trypsin cleavage at Arg228.
ADPH(172–425) Directly Binds Membranes
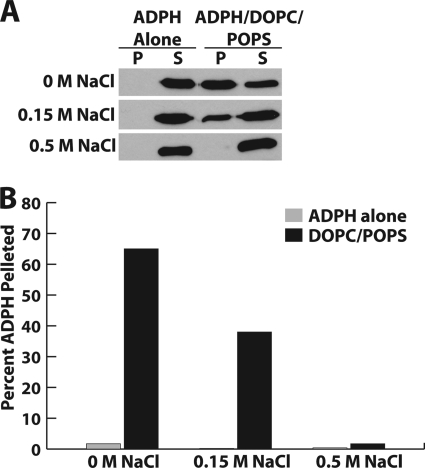
The four-helix bundle is a common lipid-binding motif among the exchangeable apolipoproteins (37). Our homology model highlights the amphipathic nature of the ADPH C terminus with charged residues at the surface of the protein and hydrophobic residues buried within the helical bundle. This organization is optimal for binding to charged phospholipid headgroups. Therefore, we sought to determine if ADPH(172–425) was able to directly bind liposomes composed of a DOPC/POPS. Under physiological salt conditions (150 mm), ADPH(172–425) was recovered primarily in the lipid-free fraction (Fig. 6, A and B). When ionic conditions were reduced to 0 m NaCl, we found that binding to DOPC/POPS liposomes was greatly enhanced. This result is explained by the presence of a net positive charge in helix 1 and a net negative charge in helix 4 allowing maximal electrostatic interactions with the negatively charged phosphoserine and positively charged choline. This observation was confirmed by the abolished interaction of ADPH(172–425) with DOPC/POPS liposomes in the presence of 0.5 m NaCl (Fig. 6B).
FIGURE 6.
ADPH(172–425) membrane interaction. A, immunoblot analysis of the binding reaction of ADPH(172–425) with DOPC/POPS liposomes showing negative correlation with NaCl concentration. B, quantitation of ADPH(172–425) in pelleted DOPC/POPS liposomes showing the percentage of total ADPH(172–425) found in the pellet. White bars, no liposome controls. Black bars, ADPH(172–425) liposome-bound. P, pellet; S, supernatant.
ADPH(170–425)-VSV Is Associated with the Plasma Membrane in HEK 293 Cells
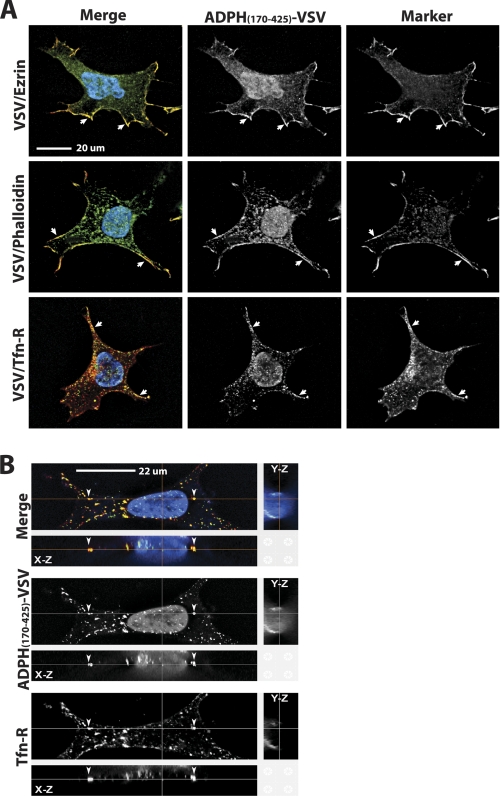
To determine if the ADPH C-terminal domain associates with cellular membranes, we next investigated its cellular localization by confocal immunofluorescence microscopy. Cell lines stably expressing the ADPH C-terminal domain were generated by transfecting HEK 293 cells with cDNA encoding ADPH(170–425) fused to a C-terminal VSV-G epitope tag to simplify immunodetection (16) (Fig. 7A). To reduce the possibility of mislocalization due to overexpression, we selected clones of these cells that expressed moderate amounts of ADPH(170–425)-VSV for immunolocalization analysis. Fig. 7A shows the localization of ADPH(170–425)-VSV relative to three plasma membrane markers: the transferrin receptor, ezrin, and phalloidin. In each case, we found significant overlap between ADPH(170–425)-VSV immunofluorescence and the immunofluorescence of the specific marker at the plasma membrane. In addition to membrane staining, we also detected punctate ADPH(170–425)-VSV immunostaining within the cytoplasm that appeared to correspond to vesicular compartments containing the transferrin receptor (Fig. 7B). Unlike VSV-tagged forms of full-length or ADPH(1–220), which exclusively localize to CLD (15, 16), we did not detect ADPH(170–425)-VSV immunostaining of CLD. Together, these results indicate that ADPH(170–425)-VSV interacts with internal membrane elements as well as the plasma membrane but does not associate with lipid components of CLD.
FIGURE 7.
Immunolocalization of ADPH(170–425)-VSV in HEK 293 cells. A, immunolocalization of ADPH(170–425)-VSV, ezrin, phalloidin, and the transferrin receptor (Tfn-R) in HEK 293 cells. ADPH(170–425)-VSV is detected by anti-VSV-G immunostaining (green). Endogenous ezrin and the transferrin receptor were detected by immunostaining with their respective antibodies (red). F-actin was stained with Alexa 594-labeled phalloidin (red). The panels show cells individually stained for ADPH(170–425)-VSV and either ezrin, transferrin receptor, or phalloidin and the corresponding merged images. Nuclei are detected by Hoechst staining (blue in the merged image). The arrows indicate sites of staining overlap between ADPH(170–425)-VSV and each membrane marker. B, representative Z-stack analysis of ADPH(170–425)-VSV (green) and transferrin receptor (red) immunostaining. The top panel in each group shows a stained cell in the x-y orientation. The x-z and y-z projections are shown above and to the right of the top panel, respectively. Individual staining patterns of ADPH(170–425)-VSV and transferrin receptor are shown along with the merged image for clarity. The arrowheads indicate transferrin receptor-positive vesicles that co-stain for ADPH(170–425)-VSV.
Modeling ADPH with a Lipid Bilayer
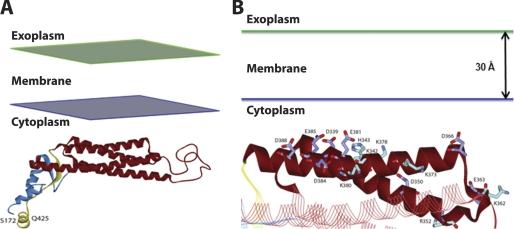
We next used the Add Membrane and Orientate Molecule protocol (33) to explore potential mechanisms for the interaction ADPH(172–425) with a lipid bilayer. This protocol initially attempts to position of ADPH(172–425) in an implicit membrane represented by a planar low dielectric slab to approximate the non-polar part of the lipid bilayer. However, the predicted optimal orientation of ADPH(172–425) was not transmembrane, and at the end of the search, the positive inside rule (38) was applied, and ADPH(172–425) was found to reside near the cytoplasmic membrane leaflet (Fig. 8A). Interestingly, helices 3 and 4 of ADPH(172–425) were preferred at the membrane interface with the acidic residues Asp366 and Asp339 of helix 3 and Glu381, Glu385, and Asp388 of helix 4 being concentrated in one region of the protein interface (Fig. 8B). This model supports our finding of an electrostatic dependent interaction between ADPH(172–425) and lipid membranes.
FIGURE 8.
Model of ADPH(172–425) lipid bilayer interactions. A, global view of the orientation of ADPH(172–425) with the lipid bilayer. B, the potential interface between helices 3 and 4 of ADPH(172–425) with the lipid bilayer. Carbon atoms of acidic residues are colored violet, and basic residues are colored cyan.
DISCUSSION
ADPH is predicted to be organized into functionally and structurally distinct domains (1, 15, 17). However, there has been a lack of direct evidence linking a specific structural feature of ADPH to a particular physiological function. Although sequences have been identified within the N-terminal region that function in targeting ADPH to CLD (15, 17) and that appear to modulate CLD accumulation (16), the structural features that mediate these functions remain undefined. Conversely, the C-terminal region of ADPH was predicted to encode a four-helix bundle motif homologous to the solved crystal structure of the TIP47 C terminus (18), but the function of this region was unclear. Our study shows for the first time that the ADPH C-terminal region encodes an independently folding, membrane-binding structure that appears to be important for milk lipid secretion. By demonstrating that the C-terminal region of ADPH folds independently of the N-terminal region and that it binds liposome membranes, we now have direct evidence that ADPH is composed of two independently functioning domains: an N-terminal domain that targets it to CLD and a C-terminal domain that mediates membrane binding. A similar functional organization of TIP47 has been proposed (39). Although direct evidence of the ability of the C-terminal region of TIP47 to bind membranes has not been demonstrated, it is likely that it also contains membrane binding elements based on the similarity to the ADPH C-terminal sequence and on observations that recombinant full-length TIP47 induces fragmentation of liposomes into discs (39). The apparent similarities in the structural properties and functional organizations of ADPH and TIP47 raise questions about whether these proteins serve similar physiological roles. Although TIP47 has been shown to compensate for ADPH in CLD accumulation in cultured cells (40), the extent to which the biological properties of these proteins overlap remains to be established.
The concept that ADPH possesses independently functioning CLD- and membrane-binding domains is further supported by our cell culture and in vivo adenovirus expression studies. Immunoelectron microscopy data presented in this study confirm earlier immunofluorescence studies showing that the N-terminal half of ADPH encodes a CLD binding function. The demonstration that the GFP-ADPH(1–220)-VSV fusion protein specifically localizes to the CLD surface also documents that the N-terminal region of ADPH can function independently of the C-terminal region to target other proteins to CLD. In agreement with previous studies of the effects of various ADPH truncation mutations on CLD properties (15, 17), we did not detect obvious differences in the localizations of GFP-ADPH(1–220)-VSV or GFP-ADPH(fl)-VSV on the CLD surface or in the size or number of CLD coated by these constructs (data not shown). Thus, the CLD binding properties of the N-terminal region of ADPH appear to be similar to those of full-length ADPH, although detailed quantitative measurements of their binding properties are required to validate this conclusion. In contrast, we detected significant differences in the size of CLD coated by GFP-ADPH(1–220)-VSV and GFP-ADPH(fl)-VSV in mammary epithelial cells of lactating mice. CLD size in the lactating mammary gland has been shown to increase when its secretion is impaired (12, 13, 34). Thus, the observation that CLD coated with GFP-ADPH(1–220)-VSV are markedly larger than those coated with GFP-ADPH(fl)-VSV or endogenous ADPH is consistent with the concept that loss of the C-terminal region of ADPH somehow interferes with CLD secretion. It is also conceivable that full-length and C-terminally truncated forms of ADPH could differentially affect CLD size through surface density effects or by influencing the ratio of surface to core lipids (41). However, such alternative explanations seem unlikely in light of observations that the CLD coated by GFP-ADPH(1–220)-VSV appear to be similar in size to those coated by GFP-ADPH(fl)-VSV (Fig. 1). Nevertheless, additional studies are needed to formally confirm that the ADPH C-terminal region is required for milk lipid secretion and to establish the mechanism underlying this process. However, coupled with demonstrations that the C-terminal region binds membrane bilayers in vitro and that it localizes to the plasma membrane in cultured cells, our mouse mammary gland data are consistent with the proposal that ADPH C-terminal region mediates interactions between CLD and the apical plasma membrane that lead to their secretion into milk (11).
The structural properties of the C-terminal region and the nature of its interaction with membranes were probed by homology modeling and by biochemical characterization of purified recombinant ADPH(172–425). The homology model reveals that this region has an α/β motif that forms a deep hydrophobic cleft that lies adjacent to the amphipathic four-helix bundle motif and that its overall structure is similar to that of the solved crystal structure of the C-terminal region of TIP47. Our biochemical studies document that ADPH(172–425) folds independently into a stable structure that is predominantly α-helical, supporting the structure predicted by three-dimensional modeling. Protease mapping demonstrates that the region corresponding to the four-helix bundle motif is resistant to digestion by a range of proteases with different specificities, suggesting that it remains highly structured in solution. Trypsin mapping of ADPH(172–425) exposed to up to 5 m urea further suggests that the four-helix bundle motif is resistant to urea denaturation.
The three-dimensional homology model predicts the presence of a large unstructured loop between helices 1 and 2. However, we failed to detect proteolytic fragments corresponding to cleavage within this region, suggesting that this loop is not exposed in solution. Similarly, we did not find significant evidence of proteolytic cleavage in the far C-terminal portion of ADPH(172–425) that lies beyond helix 4, indicating that this region is also not exposed in solution. In contrast, the N-terminal region preceding helix 1 appears to be highly susceptible to protease digestion. Interestingly, Arg228 at the base of helix 1 was identified as a primary trypsin-sensitive site. These observations, coupled with the three-dimensional model of ADPH(172–425), suggests that the region preceding the four-helix bundle exhibits significant flexibility in solution, which may be important in permitting the four-helix bundle domain to function independently of N-terminal regions involved in CLD binding. Collectively, our three-dimensional modeling and biochemical analyses indicate that the C-terminal region of ADPH is composed of a stable, rigidly structured four-helix bundle domain bounded by an inflexible protease-resistant region, on its C-terminal end, and by a flexible, protease-sensitive linker region on its N-terminal end.
Evidence from Robenek et al. (42, 43) and from Gao and Serrero (44) indicates that ADPH is capable of binding to the plasma membrane. Our cell culture data showing that stably expressed ADPH(170–425)-VSV co-localizes with three independent plasma membrane markers support these observations and provide direct evidence that the plasma membrane binding functions of ADPH are mediated by sequences within its C-terminal region. Evidence from liposome binding assays showing that ionic conditions influence the binding of ADPH(172–425) to DOPC/POPS liposomes further suggests that the membrane binding functions of the ADPH C terminus are mediated, in part, by electrostatic interactions. The pI of ADPH(172–425) is calculated to be 6.0; thus, at pH 7.2 its liposome interactions are expected to be driven by the negative charge on the protein and the positive choline group on these liposomes. Our modeling studies (Fig. 8) predict that helices 3 and 4 of the four-helix bundle motif are primary sites of membrane contact. Although deletion and mutational analyses of individual helices of the four-helix bundle are required to specify their individual contributions to membrane interactions, only helix 4 is predicted to have a net negative charge. Thus, this helix may be a primary determinant of membrane interaction.
In summary, we developed the first three-dimensional model of the ADPH C-terminal domain and identified this domain as an independently folding, α-helical structure. We show that this putative four-helix bundle motif directly associates with liposomes in vitro and that expression of ADPH lacking this domain impairs CLD secretion in vivo. Together, these results indicate that ADPH contains a third functional domain, a membrane-binding domain that is distinct from the PAT domain and CLD targeting region of the protein. In mammary epithelial cells of lactating animals, CLD undergo secretion by a novel apical membrane envelopment process, forming milk lipids (2). ADPH, along with BTN and XOR, is hypothesized to participate in CLD secretion by forming a complex that stabilizes CLD-plasma membrane interactions (1, 2); however, it is unclear what initiates interactions between CLD and plasma membrane elements. Our data provide evidence that CLD-plasma membrane interactions are mediated by the four-helix bundle domain of the ADPH C terminus. Proteins with helical bundles have been shown to induce changes in membrane composition resulting in formation of endocytic vesicles and redistribution of proteins and lipids within the plasma membrane (45). Although vesicle formation and CLD envelopment in milk lipid secretion are two distinct phenomena (46), binding of the ADPH four-helix bundle to the plasma membrane offers a tenable mechanism for initiating membrane changes that result in the recruitment of necessary factors, such as BTN and/or XOR, to sites of CLD secretion.
Acknowledgment
We thank E. Bales for excellent technical assistance and editing.
This work was supported, in whole or in part, by National Institutes of Health Grants 2RO1-HD045962 and PO1-HD38129 (to J. L. M.).
- MFG
- milk fat globule(s)
- CLD
- cytoplasmic lipid droplet(s)
- ADPH
- adipophilin
- BTN
- butyrophilin
- XOR
- xanthine oxidoreductase
- VSV
- vesicular stomatitis virus
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- EndoGluC
- endoproteinase GluC.
REFERENCES
- 1. McManaman J. L. (2009) Clin. Lipidol. 4, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mather I. H., Keenan T. W. (1998) J. Mammary Gland Biol. Neoplasia 3, 259–273 [DOI] [PubMed] [Google Scholar]
- 3. Miura S., Gan J. W., Brzostowski J., Parisi M. J., Schultz C. J., Londos C., Oliver B., Kimmel A. R. (2002) J. Biol. Chem. 277, 32253–32257 [DOI] [PubMed] [Google Scholar]
- 4. Heid H. W., Schnölzer M., Keenan T. W. (1996) Biochem. J. 320, 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McManaman J. L., Palmer C. A., Wright R. M., Neville M. C. (2002) J. Physiol. 545, 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reinhardt T. A., Lippolis J. D. (2008) J. Dairy Sci. 91, 2307–2318 [DOI] [PubMed] [Google Scholar]
- 7. Wu C. C., Howell K. E., Neville M. C., Yates J. R., 3rd, McManaman J. L. (2000) Electrophoresis 21, 3470–3482 [DOI] [PubMed] [Google Scholar]
- 8. Russell T. D., Palmer C. A., Orlicky D. J., Fischer A., Rudolph M. C., Neville M. C., McManaman J. L. (2007) J. Lipid. Res. 48, 1463–1475 [DOI] [PubMed] [Google Scholar]
- 9. Murphy D. J. (2001) Prog. Lipid. Res. 40, 325–438 [DOI] [PubMed] [Google Scholar]
- 10. Ducharme N. A., Bickel P. E. (2008) Endocrinology 149, 942–949 [DOI] [PubMed] [Google Scholar]
- 11. McManaman J. L., Russell T. D., Schaack J., Orlicky D. J., Robenek H. (2007) J. Mammary Gland Biol. Neoplasia 12, 259–268 [DOI] [PubMed] [Google Scholar]
- 12. Ogg S. L., Weldon A. K., Dobbie L., Smith A. J., Mather I. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10084–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vorbach C., Scriven A., Capecchi M. R. (2002) Genes Dev. 16, 3223–3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robenek H., Hofnagel O., Buers I., Lorkowski S., Schnoor M., Robenek M. J., Heid H., Troyer D., Severs N. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10385–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McManaman J. L., Zabaronick W., Schaack J., Orlicky D. J. (2003) J. Lipid. Res. 44, 668–673 [DOI] [PubMed] [Google Scholar]
- 16. Orlicky D. J., Degala G., Greenwood C., Bales E. S., Russell T. D., McManaman J. L. (2008) J. Cell Sci. 121, 2921–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Targett-Adams P., Chambers D., Gledhill S., Hope R. G., Coy J. F., Girod A., McLauchlan J. (2003) J. Biol. Chem. 278, 15998–16007 [DOI] [PubMed] [Google Scholar]
- 18. Hickenbottom S. J., Kimmel A. R., Londos C., Hurley J. H. (2004) Structure 12, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 19. Russell T. D., Palmer C. A., Orlicky D. J., Bales E. S., Chang B. H., Chan L., McManaman J. L. (2008) J. Lipid Res. 49, 206–216 [DOI] [PubMed] [Google Scholar]
- 20. Schaack J., Allen B., Orlicky D. J., Bennett M. L., Maxwell I. H., Smith R. L. (2001) Virology 291, 101–109 [DOI] [PubMed] [Google Scholar]
- 21. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Orlicky D. J., Schaack J. (2001) J. Lipid. Res. 42, 460–466 [PubMed] [Google Scholar]
- 23. Evans R. K., Nawrocki D. K., Isopi L. A., Williams D. M., Casimiro D. R., Chin S., Chen M., Zhu D. M., Shiver J. W., Volkin D. B. (2004) J. Pharm. Sci. 93, 2458–2475 [DOI] [PubMed] [Google Scholar]
- 24. Russell T. D., Fischer A., Beeman N. E., Freed E. F., Neville M. C., Schaack J. (2003) J. Virol. 77, 5801–5809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tokuyasu K. T. (1986) J. Microsc. 143, 139–149 [DOI] [PubMed] [Google Scholar]
- 26. Kirwan J. P., Hodges R. S. (2010) J. Struct. Biol. 170, 294–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chroni A., Pyrpassopoulos S., Thanassoulas A., Nounesis G., Zannis V. I., Stratikos E. (2008) Biochemistry 47, 9071–9080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S. A., Eyeson R., Cheever M. L., Geng J., Verkhusha V. V., Burd C., Overduin M., Kutateladze T. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ajees A. A., Anantharamaiah G. M., Mishra V. K., Hussain M. M., Murthy H. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2126–2131 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Brooks B. R., Brooks C. L., 3rd, Mackerell A. D., Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Kuczera K., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor B., Venable R. M., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009) J. Comput. Chem. 30, 1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feig M., Brooks C. L., 3rd (2004) Curr. Opin. Struct. Biol. 14, 217–224 [DOI] [PubMed] [Google Scholar]
- 32. Im W., Lee M. S., Brooks C. L., 3rd (2003) J. Comput. Chem. 24, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 33. Spassov V. Z., Yan L., Szalma S. (2002) J. Phys. Chem. B 106, 8726–8738 [Google Scholar]
- 34. McManaman J. L., Palmer C. A., Anderson S., Schwertfeger K., Neville M. C. (2004) Adv. Exp. Med. Biol. 554, 263–279 [DOI] [PubMed] [Google Scholar]
- 35. Hatters D. M., Peters-Libeu C. A., Weisgraber K. H. (2006) Trends Biochem. Sci. 31, 445–454 [DOI] [PubMed] [Google Scholar]
- 36. Narayanaswami V., Maiorano J. N., Dhanasekaran P., Ryan R. O., Phillips M. C., Lund-Katz S., Davidson W. S. (2004) J. Biol. Chem. 279, 14273–14279 [DOI] [PubMed] [Google Scholar]
- 37. Narayanaswami V., Kiss R. S., Weers P. M. (2010) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 155, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. von Heijne G. (1992) J. Mol. Biol. 225, 487–494 [DOI] [PubMed] [Google Scholar]
- 39. Bulankina A. V., Deggerich A., Wenzel D., Mutenda K., Wittmann J. G., Rudolph M. G., Burger K. N., Höning S. (2009) J. Cell Biol. 185, 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sztalryd C., Bell M., Lu X., Mertz P., Hickenbottom S., Chang B. H., Chan L., Kimmel A. R., Londos C. (2006) J. Biol. Chem. 281, 34341–34348 [DOI] [PubMed] [Google Scholar]
- 41. Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., Farese R. V. (2008) Nature 453, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robenek H., Hofnagel O., Buers I., Robenek M. J., Troyer D., Severs N. J. (2006) J. Cell Sci. 119, 4215–4224 [DOI] [PubMed] [Google Scholar]
- 43. Robenek H., Robenek M. J., Troyer D. (2005) J. Lipid Res. 46, 1331–1338 [DOI] [PubMed] [Google Scholar]
- 44. Gao J., Serrero G. (1999) J. Biol. Chem. 274, 16825–16830 [DOI] [PubMed] [Google Scholar]
- 45. McMahon H. T., Gallop J. L. (2005) Nature 438, 590–596 [DOI] [PubMed] [Google Scholar]
- 46. Murphy D. J., Vance J. (1999) Trends Biochem. Sci. 24, 109–115 [DOI] [PubMed] [Google Scholar]