Abstract
The ATP-sensitive potassium (KATP) channel regulates pancreatic β-cell function by linking metabolic status to electrical activity. Syntaxin-1A (Syn-1A), a SNARE protein mediating exocytotic fusion, binds and inhibits the KATP channel via the nucleotide-binding folds (NBFs) of its sulfonylurea receptor-1 (SUR1) regulatory subunit. In this study, we elucidated the precise regions within the NBFs required for Syn-1A-mediated KATP inhibition, using in vitro binding assays, whole cell patch clamp and FRET assay. Specifically, NBF1 and NBF2 were each divided into three subregions, Walker A (WA), signature sequence linker, and Walker B (WB), to make GST fusion proteins. In vitro binding assays revealed that Syn-1A associates with WA and WB regions of both NBFs. Patch clamp recordings on INS-1 and primary rat β-cells showed that Syn-1A-mediated channel inhibition was reversed by co-addition of NBF1-WB (not NBF1-WA), NBF2-WA, and NBF2-WB. The findings were corroborated by FRET studies showing that these truncates disrupted Syn-1A interactions with full-length SUR1. To further identify the binding sites, series single-site mutations were made in the Walker motifs of the NBFs. Only NBF1-WA (K719M) or NBF2-WA (K1385M) mutant no longer bound to Syn-1A; K1385M failed to disrupt Syn-1A-mediated inhibition of KATP channels. These data suggest that NBF1-WA (Lys-719) and NBF2-WA (Lys-1385) are critical for Syn-1A-KATP channel interaction. Taken together, Syn-1A intimately and functionally associates with the SUR1-NBF1/2 dimer via direct interactions with WA motifs and sites adjacent to WB motifs of NBF1 and NBF2 but transduces its inhibitory actions on KATP channel activity via some but not all of these NBF domains.
Keywords: ATP, Ion Channels, Membrane Biophysics, Microscopic Imaging, Pancreatic Islet, ATP-sensitive Potassium Channel, Nucleotide-binding Folds, Pancreatic Islet Beta-Cell, Sulfonylurea Receptor-1, Syntaxin-1A
Introduction
The ATP-sensitive potassium (KATP)4 channel is a key regulator of the pancreatic β-cell, effectively linking metabolic status to electrical activity (1). Under resting metabolic conditions, the KATP channel is primarily responsible for maintaining the resting membrane potential. Upon an increase in cytosolic ATP, as a result of increased glucose uptake and metabolism, the KATP channel closes, causing the membrane to depolarize. This consequently activates voltage-gated Ca2+ channels and ultimately triggers insulin exocytosis (2). The KATP channel is an octameric complex composed of four repeats of two distinct subunits: the central pore-forming Kir6.2 and the regulatory SUR1 (3, 4). As the main regulatory subunit, SUR1 is the target site for various important pharmaceuticals such as sulfonylureas and K+ channel openers (5–8). Structurally, it is a three-transmembrane domain protein with two major cytosolic domains located between the second and third transmembrane domains and at the C terminus, collectively named the nucleotide-binding folds (NBFs) (9). Each NBF (see Fig. 1A) contains three characteristic motifs: Walker A (WA), linker or signature sequence, and Walker B (WB). It is primarily through these motifs that the NBFs are able to coordinate molecules of ATP, a process essential to proper nucleotide regulation of the channel (9–11).
FIGURE 1.
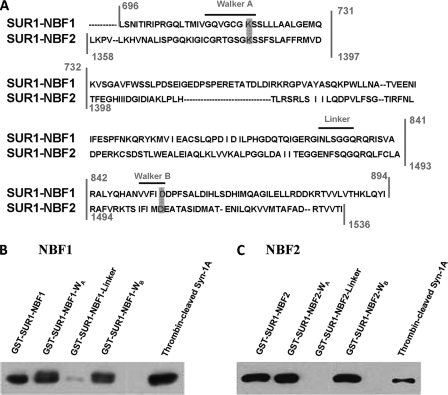
Syntaxin-1A binds SUR1 at WA and WB regions of NBF1 and NBF2. A, SUR1-NBF1 and -NBF2 sequence alignment. The vertical lines indicate the truncation segments that we generated (see “Materials and Methods”) for each NBF, with each truncate containing WA, linker, or WB motifs. Horizontal dashes within sequences indicate a lack of residue for the respective NBF. Characteristic motifs are labeled above the sequence with horizontal lines. Notice that there is one motif in each truncated region, for a total of three truncated sections per NBF. The gray boxes indicate the specific sites we chose to mutate (see Fig. 5). B, binding of Syn-1A to SUR1-NBF1-WA and -WB domains. GST-SUR1-NBF1 (as a positive control), GST-SUR1-NBF-WA, -WB, and -linker (all bound to glutathione agarose beads, 400 pmol of protein each) were used to pull down thrombin-cleaved Syn-1A (300 μg of protein). Thrombin-cleaved Syn-1A (15 μg of protein) was used as a positive control. The precipitated proteins were separated on 15% SDS-PAGE, and the protein of interest was probed with mouse anti-Syn-1A antibody. C, binding of Syn-1A to SUR1-NBF2-WA and -WB domains. The same binding assay was performed as described in B, but WT GST-SUR1-NBF2 and truncates were used to replace WT GST-SUR1-NBF1 and truncates.
It has been well studied that the SNARE proteins form a complex necessary for driving the fusion of plasma and vesicle membranes during exocytosis (for reviews, see Refs. 12–14). In the context of the pancreatic islet β-cell and insulin exocytosis, we have previously shown that Syntaxin-1A (Syn-1A), a core member of this SNARE complex, appears to play multiple roles during this secretory process, effectively regulating various channels involved in both the triggering and cessation of membrane fusion (15). To date, Syn-1A has been demonstrated to regulate β-cell voltage-gated K+ channels (16, 17), voltage-gated Ca2+ channels (18), and more recently KATP channels (19, 20). Syn-1A perhaps possesses a conserved binding pocket in all of these molecules; or alternatively, Syn-1A could be a structurally flexible molecule capable of assuming many conformations that render specificity to respective ion channels in the local neighborhood (microdomain) of the plasma membrane. Syn-1A is indeed distributed along the entire plasma membrane of neurons and neuroendocrine cells (including β-cells) to be involved in not only the exocytotic apparatus per se (i.e. synapse) but also ion channels (K+) located more distantly from exocytotic sites (15). For β-cell KATP channels, we had shown that Syn-1A binds SUR1 at both NBF1 and NBF2 and not the Kir6.2 subunit (19, 20). Furthermore, it appears that at least some Syn-1A is endogenously bound to the β-cell KATP channels, because botulinum neurotoxin C1 proteolytic cleavage of endogenous Syn-1A increased KATP channel activity (19, 21). Moreover, physiologic changes in cytosolic ATP concentrations in β-cells could profoundly modulate Syn-1A interactions with KATP channels presumably acting on Walker motifs (21). These NBFs are large with complex functional domains; thus this work was directed at examining what putative domains within SUR1-NBF1 and -NBF2 bind Syn-1A to transduce Syn-1A inhibitory actions on insulin-secreting β-cell KATP channels.
MATERIALS AND METHODS
Cell Culture
The INS-1 (832/13) cell line was a gift from Dr. Christopher Newgard (Duke University Medical Centre, Durham, NC). The cells were grown at 37 °C in 5% CO2 in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 2 mm l-glutamine, 10 mm HEPES, 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mm sodium pyruvate, and 50 μm 2-mercaptoethanol. One day prior to whole cell voltage clamp recordings, the cells were trypsinized and plated onto glass coverslips.
For binding studies, we used HEK293 cells infected with recombinant adenoviruses containing wild type rat Kir6.2 with a tetracycline-inhibited transactivator, and wild type hamster SUR1 expressed in a tetracycline-inhibited transactivator-regulated gene (both gifts from Dr. S. L. Shyng, Oregon Health & Science University, Portland, OR) (22). The transfected HEK cells were cultured at 37 °C in 5% CO2 in DMEM high glucose 1× with l-glutamine and sodium pyruvate (Invitrogen), supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. For overexpression of Syn-1A, the HEK cells were transfected with pcDNA3-Syn-1A using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
For FRET imaging studies, HEK293 cells were co-transfected with Syn-1A-mCherry and SUR1-EGFP (gift from Dr. C. G. Nichols, Washington University, School of Medicine, St. Louis, MO) plus Kir6.2 (gift from Dr. S. Seino, Chiba University, Chiba, Japan) by using Lipofectamine 2000 (Invitrogen). The cells were examined 2 days post-transfection.
Pancreatic Islet β-Cell Isolation
Islets were isolated from male Wistar rats by collagenase digestion as described previously 0.015% trypsin (17). Dispersed single β-cells for electrophysiological recordings were obtained by islet treatment with 0.015% trypsin in Ca2+- and Mg2+-free phosphate-buffered saline. The cells were plated on glass coverslips and cultured in 2.8 mm glucose (with 7.5% fetal calf serum, 0.25% sodium, and 100 g/ml streptomycin) prior to recordings.
Recombinant GST Fusion Proteins
The pGEX 4T-1-Syn-1A plasmid was a gift from W. Trimble (The Hospital for Sick Children, Toronto, Canada). The constructs (see Fig. 1A) expressing truncated SUR1-NBF1 and -NBF2 containing WA (NBF1, amino acids 696–731; NBF2, amino acids 1358–1397), linker (NBF1, amino acids 732–841; NBF2, 1398–1493), or WB (NBF1, amino acids 842–894; NBF2, amino acids 1494–1536), respectively, were generated using the PCR method and hamster SUR1 as template (a gift from Dr. Lydia Aguilar-Bryan, Pacific Northwest Diabetes Research Institute, Seattle, WA). Mutants on WA and WB motifs in these truncated NBF1 (K719M and D854N, respectively) and NBF2 (K1385M and D1506N, respectively) regions were also generated (see Fig. 5A) using QuikChange® site-directed mutagenesis according to the manufacturer's instructions (Stratagene, La Jolla, CA). All of the constructs were confirmed by sequencing (ACGT Corp., Toronto, Canada). The resulting PCR products were subsequently cloned into the pGEX 4T-1 vector for expression as GST fusion proteins. GST fusion protein expression and purification were performed following the manufacturer's instructions (Amersham Biosciences). Syn-1A was obtained by cleavage of GST-Syn-1A immobilized on agarose beads using thrombin (Sigma).
FIGURE 5.
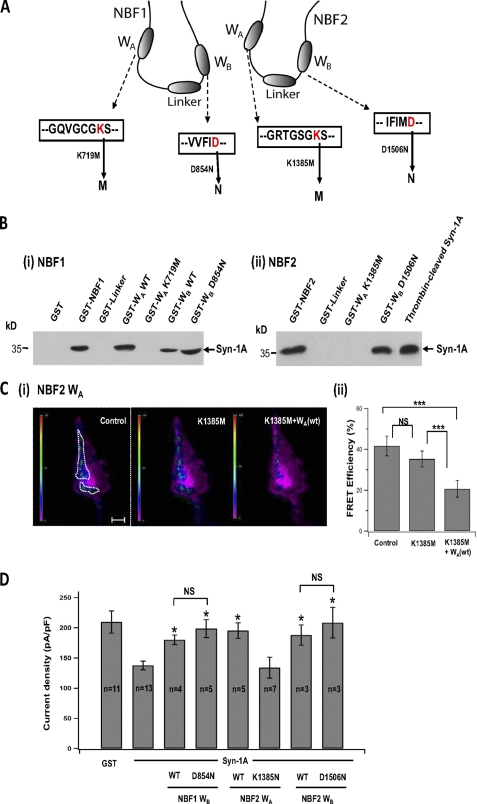
Assessment of whether Syn-1A binding and KATP channel signal transduction is through WA and WB motifs per se in NBF1 and NBF2 of SUR1. A, anatomy of SUR1-NBF1 and -NBF2, and their respective WA and WB motifs. Conserved lysine residues in WA motifs were substituted with methionine, generating NBF1-WA (K719M) and NBF2-WA (K1385M) truncated proteins. Conserved aspartic acid residues were substituted with asparagines, generating NBF1-WB (D854N) and NBF2-WB (D1506N) truncated proteins. B, binding of Syn-1A to Walker motifs in NBF1 and NBF2 (as described for Fig. 1). Panel (i), NBF-1: WA motif mutant NBF1-WA (K719M) could not bind Syn-1A, whereas WB motif mutant NBF1-WB (D854N) remained able to bind Syn-1A. Full-length NBF1, WT NBF1-WA, and WT NBF1-WB are positive controls. Panel (ii), NBF-2: WA motif mutant NBF1-WA (K1385M) could not bind Syn-1A, whereas WB motif mutant NBF2-WB (D1506N) remained able to bind Syn-1A. Full-length NBF2 is positive control. C, NBF2 WA motif mutant NBF2-WA (K1385M) could not disrupt Syn-1A interaction with SUR1 in live cells by FRET assay (as described in Fig. 4). Panel (i), representative recordings of FRET signal on the plasma membrane of the same HEK cell before and after addition of 1 μm GST-NBF2-WA (K1385M) followed by WT NBF2-WA. The scale bar indicates 5 μm. The vertical scale bar indicates FRET efficiency in pseudocolor. Panel (ii), summary of FRET efficiency change in seven experiments. The bar graph (means ± S.E.). ***, indicates p < 0.001. FRET studies shown in supplemental Fig. S3 demonstrated that WB motif mutants including NBF1-WB (D854N) and NBF2-WB (D1506N), like their corresponding wild type proteins, disrupted Syn-1A interactions with SUR1. D, NBF2-WA (K1385M), unlike its wild type protein, could not block Syn-1A inhibition of KATP channel activity in INS-1 cells (as in Fig. 2; cells were held at a resting potential of −70 mV and stimulated to −110 mV to elicit whole cell inward currents). WB motif mutants NBF1-WB (D854N) and NBF2-WB (D1506N), like their WT proteins, remained able to block Syn-1A inhibition of KATP channels. Positive (Syn-1A alone) and negative controls (GST) are shown. The bar graph shows the means ± S.E. *, significant difference when compared with Syn-1A alone (p < 0.05). Representative traces are in supplemental Fig. S2.
In Vitro Binding Assays
In vitro binding assays were performed as described previously (19). Briefly, the GST fusion proteins were bound to glutathione-agarose beads and incubated with thrombin-cleaved Syn-1A (10 μg) in 250 μl of binding buffer (25 mm HEPES, pH 7.4, 50 mm NaCl, 1.2 mm MgCl2, 0.1% gelatin, 0.1% Triton X-100, 0.1% bovine serum albumin, 0.2% β-mercaptoethanol) at 4 °C for 2 h with constant agitation. The beads were then washed twice with washing buffer (20 mm HEPES, pH 7.4, 150 mm KOAC, 1 mm EDTA, 1.2 mm MgCl2, 5% glycerol, 0.1% Triton X-100). The precipitated proteins were separated on 15% SDS-PAGE, transferred to nitrocellulose membrane, and identified with specific antibody against Syn-1A (1:2000; Sigma). For binding assays using overexpressed Syn-1A cell lysate, the transfected HEK293 cells were washed with ice-cold PBS and harvested in lysis buffer (15 mm MES plus 15 mm HEPES, 100 mm KCl, 1.5% Triton X-100, 2 μm pepstatin A, 1 μg/ml leupeptin, and 10 μg/ml aprotinin). The cells were sonicated on ice (30 s/1 ml) and then left on ice for 30 min before removal of insoluble material by centrifugation at 55,000 × g at 4 °C for 40 min. GST fusion proteins bound to glutathione agarose beads (400 pmol each) were each individually incubated with cell lysate extract (300 μg) at 4 °C for 2 h. The samples were then washed with lysis buffer, separated on 15% SDS-PAGE, and probed for Syn-1A by Western blot analysis.
Electrophysiology
Whole cell patch clamping recordings (ruptured) were made from INS-1 and β-cells using an EPC-9 amplifier and Pulse software (HEKA Electronik, Lambrecht, Germany). Pipettes were pulled from 1.5-mm borosilicate glass capillary tubes and were fire-polished to a tip resistance of 3–5 MΩ. Pipettes were filled with intracellular solution containing 140 mm KCl, 1 mm MgCl2, 1 mm EGTA, and 10 mm HEPES (pH 7.3 adjusted with KOH). External bath solution contained 138 mm NaCl, 5.6 mm KCl, 1.2 mm MgCl2, 2.6 mm CaCl2, and 5 mm HEPES (pH 7.3 with NaOH). Recombinant proteins were premixed with intracellular pipette solution (1 μm or as indicated) and delivered to cell cytosol through the patch pipette after whole cell configuration was made. Current was evoked by a series of 250-ms step voltage protocols of −110 mV stimulating potential from a holding potential of −70 mV. Maximum current was verified by the recording of 10 consecutive pulses with no further increase after a suspected maximum to ensure no larger current was observed. Tolbutamide (0.3 mm) was added to the bath solution following maximum current to verify the current identity. Current density (pA/pF), as a means of normalization across different cells, was calculated by dividing the current over cell membrane capacitance. The data are presented as the means ± S.E. All of the recordings were performed at room temperature.
FRET Imaging
FRET imaging was performed as we recently reported (21). Transfected cells were plated on autoclaved glass coverslips overnight and then imaged in intracellular buffer containing 20 mm HEPES, 5 mm NaCl, 140 mm potassium gluconate, and 1 MgCl2, and pre-equilibrated with 95:5 O2:CO2, pH 7.4) at 37 °C using an Nikon TE-2001U inverted microscope fitted with an argon laser unit (480 ± 10 nm; Spectra-Physics), a helium-neon laser units (545 ± 10 nm; Melles Griot), and a Quantum 512SC charge-coupled device camera (Photometrics, Tucson, AZ) controlled by NIS software (Nikon). For FRET measurements, a Plan apo ×60 oil immersion objective (1.65 NA), a Dual-View imaging system (Optical Insights, Tucson, AZ) containing a EGFP/mCherry dual band dichroic splitter (565dcxr), and an emission filter (HQ530/30 for EGFP and HQ630/50 for mCherry) were combined together to allow simultaneous two-channel monitoring of emission fluorescence. The images were acquired at 900-ms exposure time. The cells were permeabilized by incubation in intracellular buffer containing digitonin (50 μm) for 5 min at 37 °C and subsequently maintained in intracellular buffer. For FRET image analysis, EGFP fused with SUR1 was used as FRET donor and mCherry fused with Syn-1A as FRET acceptor. Four images including donor excitation/donor emission (Dd), donor excitation/acceptor emission (Da), acceptor excitation/acceptor emission (Aa), and acceptor excitation/donor emission (Ad) were acquired in the same condition in each FRET experiment. In addition, donor-only sample and acceptor-only sample were acquired as part of the FRET protocol before each experiment for calculation of bleedthrough. FRET efficiency was used to indicate the interaction of the two proteins, calculated as FRETcorr = FRETraw − (CoB × DdFRET) − (CoA × AaFRET)). FRET efficiency% = (FRETcorr/DdFRET)*100%. CoB means the amount of donor bleedthrough in the absence of an acceptor, and CoA means the amount of acceptor bleedthrough in the absence of a donor. To examine only the molecular interactions of Syn-1A and SUR1 on the surface of the plasma membrane and avoid contamination from intracellular FRET signals, we used total internal reflection fluorescence microscopy to optically isolate the plasma membrane, enabling high spatial image resolution of the protein interactions on entire surfaces of the plasma membrane. This advantage of total internal reflection fluorescence microscopy is attributed to the exponentially decaying evanescent wave that enables illumination of only the fluorescent signal within a thin layer (<200 nm) at and beneath the plasma membrane and thus significantly reduces out-of-focus background fluorescence. For statistical analysis of FRET efficiency, we draw regions of interest around the entire area of the plasma surface expressing any FRET signal (in blue, green, and red; see pseudocolor bar) as indicated and do not include the purple areas where there is no FRET signal. For example, see Fig. 4A(i). From these regions of interest, we calculated the average FRET efficiency and then tracked the change of FRET efficiency in the same region of interest under the indicated different conditions.
FIGURE 4.
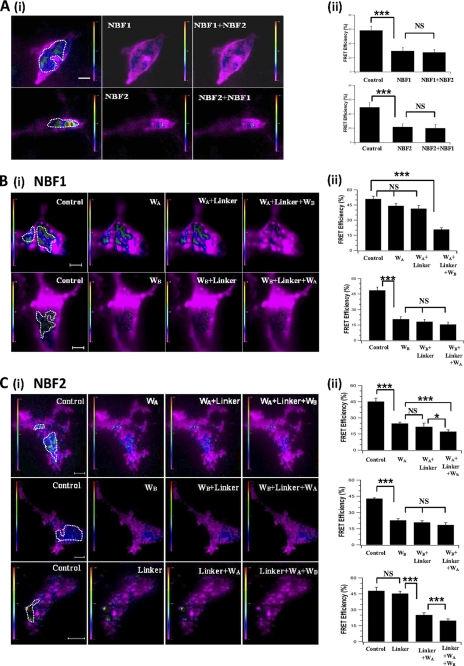
NBF1 and NBF2 truncated proteins containing Walker domains can disrupt Syn-1A interaction with SUR1 in live cells. A, NBF1 and NBF2 can inhibit Syn-1A interaction with SUR1. Panel (i), representative recording of FRET signals on the plasma membrane of HEK cell expressing Syn-1A-mCherry, SUR1-EGFP, and Kir6.2 before and after addition of 1 μm truncated NBF1/NBF2 proteins to the permeabilized cells in the reverse sequences (NBF1 then NBF2; or NBF2 then NBF1). The excitation wavelength was 488 nm, and fluorescence emission was measured at 605–655 nm. The scale bar indicates 5 μm. The vertical scale bar indicates FRET efficiency in pseudocolor. Panel (ii), summary of FRET efficiency before and after addition of different NBF truncations. The bar graph shows the means ± S.E., n = 15. ***, p < 0.001. B, WB but not WA domain of NBF1 can inhibit the interaction between Syn-1A and SUR1. Panel (i), representative recordings of the FRET signal on the plasma membrane of one same HEK cell before and after addition of 1 μm NBF1 truncations in different sequence (WA, linker then WB; or WB, linker then WA). Panel (ii), bar graph (means ± S.E.), n = 5. ***, p < 0.001. C, both WA and WB domains of NBF2 can inhibit the interaction between Syn-1A and SUR1. Panel (i), representative recordings of the FRET efficiency on the plasma membrane of one same HEK cell before and after addition of 1 μm NBF2 truncations in different sequence (WA, linker, and then WB; or WB, linker, and then WA; or linker, WA, and then WB). Panel (ii), corresponding bar graph (means ± S.E.), n = 7. ***, p < 0.001; *, p < 0.05.
Data Analysis
The data are presented as the means ± S.E. Statistical analysis of electrophysiology and FRET data were carried out using SigmaStat v3.1 (Systat Software Inc.). Differences between experimental groups were evaluated using one-way analysis of variance followed by Tukey post hoc test with p < 0.05 considered as statistically significant.
RESULTS
Syntaxin-1A Binds SUR1 at WA and WB Domains of Both NBF1 and NBF2
We reported previously that NBF1 and NBF2 of SUR1 appear to encode the binding domains for Syn-1A to inhibit insulin secreting islet β-cell KATP channels (19, 20). Therefore, we postulated that Syn-1A must require specific putative subdomains within each NBFs to coordinate protein assembly and transduce channel regulation onto the pore-forming Kir6.2 subunit. To further identify the putative regions in each NBF critical for this interaction, NBF truncations were constructed and expressed as GST fusion proteins. Specifically, each NBF was divided into three individual sections (Fig. 1A), each containing a characteristic motif: WA (NBF1, amino acids 696–731; NBF2, amino acids 1358–1397), linker (NBF1, amino acids 732–841; NBF2, amino acids 1398–1493), and WB (NBF1, amino acids 842–894; NBF2, amino acids 1494–1536). Each set of the three truncated domains proteins would encompass the full-length region of NBF1 (amino acids 696–894) and NBF2 (amino acids 1358–1536). When applied to in vitro GST-binding assays, the truncated fragments containing NBF1-WA, NBF1-WB, NBF2-WA, and NBF2-WB bound to agarose beads were all individually able to pull down thrombin-cleaved recombinant Syn-1A (Fig. 1, B and C). Similar results were obtained when using cell lysate of HEK293 cells overexpressing Syn-1A (data not shown), that these NBF fragments pulled down expressed Syn-1A. In contrast, NBF1-linker and NBF2-linker containing fragments were unable to pull down Syn-1A. Full-length NBF1 and NBF2 were included as positive controls, each of which bound to Syn-1A as previously reported (19).
Syntaxin-1A Inhibition of β-Cell KATP Channel Activity at NBF1 Is through WB but Not WA Domain
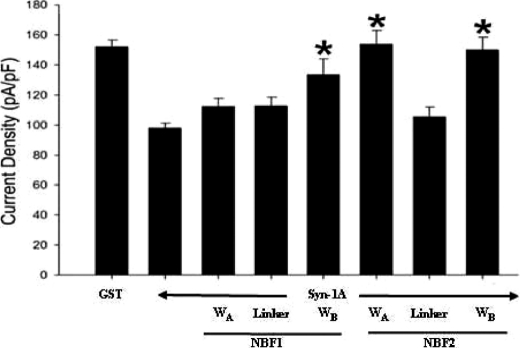
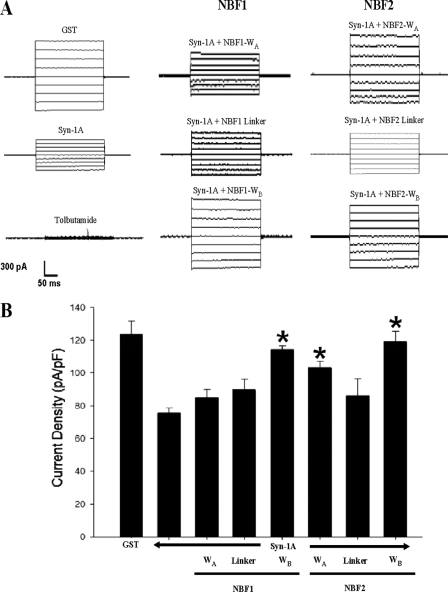
We next examined whether the domains on NBF1 that bind Syn-1A would transduce Syn-1A inhibitory actions on KATP channels. This was examined through whole cell patch clamp analysis on the rat insulinoma INS-1 cell line (23). As per our previous study (19), recombinant proteins were introduced into the cell interior via patch pipette during dialysis, which also allowed for washout of cytosolic ATP and thus maximization of KATP channel current. We first compared the effect of GST-syn-1A (positive control) with GST-only (negative control) on the current to confirm the inhibitory effect of Syn-1A on KATP, as previously reported (19). As shown in Fig. 2, the current density (pA/pF) recorded from GST-Syn-1A (1 μm) dialyzed INS-1 cells (97.9 ± 3.3 pA/pF; n = 20) was 36% less than that from GST (1 μm) control cells (151.9 ± 4.6 pA/pF; n = 19). Having established the base level of the Syn-1A-induced current inhibition, we then proceeded to investigate combinations of GST-Syn-1A with single truncated proteins. GST-Syn-1A was premixed with a NBF truncate in equal concentrations (1 μm) in the pipette solution before loading and establishment of whole cell configuration. This allowed GST-Syn-1A to prebind the recombinant NBF truncate protein first (assuming interaction), thus blocking the ability of Syn-1A to functionally interact with endogenous SUR1. Co-dialysis of GST-Syn-1A with NBF1-WA (112.2 ± 5.6 pA/pF; n = 15) or NBF1-linker (112.6 ± 5.9 pA/pF; n = 10) yielded current densities similar to that of the Syn-1A positive control (p > 0.05), indicative of the fact that neither NBF1-WA nor linker could interrupt Syn-1A action on KATP channel activity. In contrast, the NBF1-WB significantly reduced Syn-1A blockade of KATP channels (133.5 ± 10.7 pA/pF; n = 9; p < 0.05) (Fig. 2). Because Syn-1A inhibition remained despite co-dialysis with WA or the linker, we dialyzed these NBF truncates into the cells alone to ensure that they were not able to inhibit current activity. Indeed, when dialyzed alone, both NBF1-WA (150 ± 4.9 pA/pF; n = 7) and NBF1-linker (148.7 ± 5.7 pA/pF; n = 8) yielded current densities similar to GST control cells.
FIGURE 2.
Syntaxin-1A inhibits KATP channel activity in INS-1 cells via Walker regions of NBF1 and NBF2 of SUR1. The bar graph (means ± S.E.) of the effects of syntaxin-1A (1 μm) co-dialyzed with truncated NBF proteins (1 μm) containing NBF1-WA (n = 16), NBF1-linker (n = 10), NBF1-WB (n = 9), NBF2-WA (n = 10), NBF2-linker (n = 10), and NBF2-WB (n = 10) regions alone with KATP currents normalized to cell capacitance (pA/pF). Positive (Syn-1A, n = 26) and negative controls (GST, n = 19) are shown. The cells were held at a resting potential of −70 mV and stimulated to −110 mV to elicit inward currents. *, significant difference when compared with Syn-1A-positive control (p < 0.05).
Because cell lines, although excellent surrogate models, may not completely represent the true physiological model, we proceeded to confirm our results on freshly isolated rat pancreatic islet β-cells (Fig. 3A). Similar to that seen in INS-1 cells, GST-Syn-1A reduced current activity by 39% (75.5 ± 3.2 pA/pF; n = 6) when compared with GST control currents (123.7 ± 8.2 pA/pF; n = 6, p < 0.05) (left traces in Fig. 3A and summary in Fig. 3B). The addition of NBF1-WB (middle bottom trace) significantly reversed Syn-1A-mediated inhibition with resulting current activity of 114.2 ± 2.5 pA/pF (n = 5, p < 0.05), whereas NBF1-WA (middle top trace, 84.5 ± 5.2 pA/pF; n = 5) or NBF1-linker (middle trace, 89.4 ± 6.7 pA/pF; n = 5) did not affect the Syn-1A block.
FIGURE 3.
Syntaxin-1A inhibits rat pancreatic islet β-cell KATP channel activity via Walker regions of NBF1 and NBF2 of SUR1. A, representative whole cell currents after dialysis of indicated proteins. Tolbutamide (0.3 mm) was perfused through the extracellular solution. B, bar graph (mean ± S.E.) of the effects of syntaxin-1A (1 μm) co-dialyzed with truncated NBF proteins (1 μm) containing NBF1-WA (n = 5), NBF1-linker (n = 5), NBF1-WB (n = 5), NBF2-WA (n = 5), NBF2-linker (n = 5), and NBF2-WB (n = 5) regions alone with KATP currents normalized to cell capacitance (pA/pF). Positive (Syn-1A, n = 6) and negative controls (GST, n = 6) are shown. The cells were held at a resting potential of −70 mV and stimulated to −110 mV to elicit inward currents. *, significant difference when compared with the Syn-1A positive control (p < 0.05).
Taken together, our data showed that although Syn-1A bound to both NBF1-WA and -WB domains in SUR1 (Fig. 1B), its binding of WB domain transduces Syn-1A inhibition of KATP channel activity. The WA domain of NBF1 thus appears to play a more important role in stabilizing the binding of Syn-1A without actually transducing Syn-1A actions.
Syntaxin-1A Inhibits KATP Channel Activity at NBF2 through WA and WB Domains
We next examined the functional roles of NBF2 domains in Syn-1A-mediated blockade effect on KATP channels, using the similar approaches for NBF1. In INS-1 cells, co-dialysis of GST-Syn-1A with NBF2-WA (153.5 ± 9.6 pA/pF; n = 10) and NBF2-WB (149.7 ± 8.8 pA/pF; n = 10) yielded currents significantly different from the Syn-1A but not the GST control cells (Fig. 2). Similar to NBF1, NBF2-linker (105.3 ± 6.6 pA/pF; n = 10) was unable to significantly suppress Syn-1A-mediated channel inhibition. Once again, to ensure that the NBF2-linker truncate was unable to inhibit the channel alone, we dialyzed it into cells alone and yielded currents similar to GST control cells (151.1 ± 4.8 pA/pF; n = 8).
Findings from the INS-1 cell line were again verified in primary rat islet β-cells (Fig. 3A, right traces), which yielded results identical to those of INS-1 cells. NBF2-WA (103.2 ± 3.7 pA/pF; n = 5) and NBF2-WB (118.8 ± 6.7 pA/pF; n = 5) were both able to significantly reverse Syn-1A-mediated inhibition but not NBF2-linker (86.1 ± 10.2 pA/pF; n = 5) (Fig. 3B). As with NBF1, the NBF2-linker truncate was not predicted to have a functional effect on Syn-1A inhibition of channel activity based on GST binding assay results. Therefore, for NBF2, it appears that Syn-1A interaction with both WA and WB regions are required for functional inhibition of the KATP channel. Based on binding and functional data, Syn-1A is able to coordinate at NBF1/NBF2 via the WA and WB domains; however, it would appear that only three of four of these Walker domains are critical for transducing channel inhibition.
In Vivo Syntaxin-1A Complex Formation with Full-length SUR1
SUR1 exists in highly complex intramolecular interactions, including the formation of the NBF1 and NBF2 dimer, and intermolecular interactions with the Kir6.2 subunit. It is possible that Syn-1A intimately binds to the NBF1/2 dimer along with the existing SUR1/Kir6.2 complex and thus regulates the KATP channel properties. We thus explored this possibility to test whether each of these small Syn-1A-binding NBF subdomains is sufficient to disrupt this intimate association of this tri-protein complex in vivo. Here, we employed FRET analysis in living cells expressing these three interacting proteins in their physiological configuration and targeted to their native compartment, the plasma membrane, as we had recently reported (21). Specifically, HEK293 cells expressed with Kir6.2 subunit were co-transfected with SUR1 tagged with EGFP as a donor and Syn-1A tagged with mCherry as an acceptor (see “Materials and Methods”).
NBF1 or NBF2 Alone Is Sufficient to Disrupt Syn-1A-SUR1 Interactions
To examine the FRET signal between SUR1-EGFP and Syn-1A-mCherry co-expressed with Kir6.2 in HEK293 cells, a 488-nm laser was applied to excite EGFP, and mCherry signal (FRET) was recorded with 605–655-nm band pass filter. The FRET efficiency, described as the mean value of [FRETcorr/DdFRET], indicates this very close proximity between SUR1 and Syn-1A and thus their presumed interactions (Fig. 4A). Under control conditions (HEK cells permeabilized with 50 μm digitonin in intracellular buffer for 5 min), FRET efficiency was 58.20 ± 3.41 (n = 15) (Fig. 4A, panels (i) and (ii)). When 1 μm NBF1 truncate was applied to the same HEK cell for 3 min (Fig. 4A, panel (i), top images), we observed a dramatic decrease in FRET signal (purple indicating no FRET) precisely at the same high FRET area (compare the top images in Fig. 4A, panel (i)), indicating that NBF1 interrupted the protein interaction at the cell membrane. The FRET efficiency decreased by 52.2% from 58.20 ± 3.41 to 27.8 ± 2.82 (Fig. 4A, panel (ii), top graph), before and after application of NBF1, respectively. When 1 μm NBF1 and 1 μm NBF2 were added together, the FRET signal recorded in the same cell did not show further decrease; FRET efficiency was 25.3 ± 1.88 (p > 0.05). These findings indicate that NBF1 can disrupt Syn-1A interactions with SUR1 independent of NBF2. We next examined the role of NBF2 (Fig. 4A, panel (i), bottom images) by adding the truncated proteins in opposite sequence: NBF2 and then NBF1, and a similar phenomenon was observed; FRET efficiency (50.3 ± 3.95; Fig. 4A, panel (ii), bottom graph) was reduced by 56.1% after 1 μm NBF2 truncate was added (22.1 ± 1.82, n = 15). The addition of NBF1 to NBF2 did not show further disruption of protein interaction (FRET efficiency: 21.4 ± 2.52, n = 15; p > 0.05).
For NBF1, WB but Not WA Domain Could Disrupt Syn-1A-SUR1 Interactions
To further identify the functional domains in NBF that are responsible for Syn-1A-SUR1 interaction, we investigated the role of single truncated proteins from sectioned regions of NBF1 and NBF2. First, we tested the effect of WA, linker, and WB domains of NBF1 (Fig. 4B). Sequential application of 1 μm WA and linker domains into the cell did not significantly decrease FRET efficiency (Fig. 4B, panel (i), top images; analysis in panel (ii), top graph: WA: 44.14 ± 2.29; WA+ linker: 41.37 ± 3.06; control: 50.97 ± 2.75, n = 5; p > 0.05). Subsequent addition of WB domain reduced the FRET to 20.69 ± 1.72 (n = 5, p < 0.001). This indicated that the WB domain of NBF1 disrupted Syn-1A interaction with SUR1. To confirm this, we added the WB domain to the permeabilized HEK cells first (Fig. 4B, panel (i), bottom images), resulting in a dramatic decrease of FRET efficiency (20.67 ± 2.45; Fig. 4B, panel (ii), bottom graph) from the control (48.59 ± 3.1; n = 5, p < 0.001). No further effect was observed upon sequential addition of linker (WB + linker: 18.19 ± 2.32) and WA (WB + linker + WA: 15.49 ± 2.2, n = 5, p > 0.05). Because the WA domain binds to Syn-1A (Fig. 1B), the inability of WA to disrupt Syn-1A-SUR1 interactions and Syn-1A blockade of KATP channel (Figs. 2 and 3) might be due to low concentrations of WA domain (1 μm) used in these experiments. We thus performed dose-response assays of WA domain of up to 10 μm for FRET (supplemental Fig. S1A) and also tested the highest concentration of WA domain (10 μm) in the patch clamp study (INS-1, supplemental Fig. S1B). We found that the high concentrations of WA did not cause significant disruption of either the FRET signal or reversal of Syn-1A inhibition of KATP channels.
For NBF2, Either WA or WB Could Disrupt Syn-1A-SUR1 Interactions
Similar experiments were performed to test the function of NBF2 subdomains (Fig. 4C). First, 1 μm WA domain applied into the cell (Fig. 4C, panel (i), upper images) caused a 45.1% decrease of FRET efficiency (Fig. 4C, panel (ii), top graph; WA: 24.79 ± 1.32; control: 45.18 ± 3.21; n = 6, p < 0.001). Subsequent addition of linker did not further disrupt FRET efficiency (WA + linker: 21.71 ± 3.4; n = 6, p > 0.05). Following the linker domain, the addition of WB domain resulted in a further decrease in FRET (WA+ linker + WB: 17.22 ± 0.98 versus WA+ linker: 21.71 ± 3.4, n = 6, p < 0.05). To ensure that prebinding of one region did not influence binding of additional regions, we performed the same experiment in reverse sequence: WB, linker then WA (Fig. 4C, panel (i), middle images, and panel (ii), middle graph). A decrease of FRET efficiency of 47% was recorded upon addition of WB alone (WB: 22.87 ± 1.6; control: 42.77 ± 0.75; n = 6, p < 0.001), but no further significant decrease was recorded upon subsequent addition of linker (WB + linker: 20.92 ± 1.6) and WA (WB + linker + WA: 18.5 ± 1.3; n = 6, p > 0.05). To confirm the lack of linker disruption in NBF2, we added the linker first in a final combination (Fig. 4C, panel (i), bottom images, and panel (ii), bottom graph). Predictably, no effect on the FRET efficiency was observed (linker: 45.3 ± 2.2; control: 47.9 ± 3.4; n = 6, p > 0.05). Subsequent addition of WA domain resulted in a significant decrease in FRET (linker + WA: 25.1 ± 2.1; n = 6, p < 0.001). A further slight but significant decrease was observed when WB domain was subsequently added (linker + WA+ WB: 19.77 ± 1.7; n = 6, p < 0.01). It therefore appears that for NBF2, WA, and WB domains seem to interact intimately with each other and with Syn-1A such that either one was sufficient to disrupt Syn-1A interactions with SUR1.
Assessment of Whether Syn-1A Binding and KATP Channel Signal Transduction Is through WA and WB Motifs per se in NBF1 and NBF2 of SUR1
Our studies above showed that large truncated segments of NBFs containing Walker motifs bind and/or transduce Syn-1A inhibition of KATP channels. The next step was to examine whether it is the Walker motifs per se in these NBF domain proteins that affect these actions of Syn-1A. We thus introduced single mutations into conserved sites within WA (lysine) and WB (aspartic acid) in each of these NBF subdomains (Fig. 5A) previously reported. The mutations were made by substitutions of lysine in WA and aspartic acid in WB with methionine and asparagine, respectively, and the consequent constructs are GST-NBF1-WA (K719M) (24), GST-NBF1-WB (D854N) (24, 25), GST-NBF2-WA (K1385M) (26), and GST-NBF2-WB (D1506N) (27). Of note, these Walker motif mutations had been reported to be major binding and transduction sites for magnesium and adenine nucleotides (24–27).
The WA motif mutations in NBF1-WA (K719M) (Fig. 5B, panel (i)) and NBF2-WA (K1385M) (Fig. 5B, panel (ii)) did not bind Syn-1A in contrast to its respective wild type proteins (Fig. 1B). Because NBF1-WA (wt) domain did not disrupt Syn-1A/SUR1 interactions on our FRET or patch clamp studies (Figs. 2–4 and supplemental Fig. S1), we did not further examine NBF1-WA (K719M). Nonetheless, these results demonstrate that the WA motif per se in NBF1 is a binding site for Syn-1A. Remarkably, GST-NBF2-WA (K1385M) did not disrupt Syn-1A-SUR1 interactions in the FRET assay (Fig. 5C, panels (i) and (ii): NBF2-WA (K1385M): 35.28 ± 3.9; control: 41.7 ± 4.8, n = 7; p > 0.05), and further addition of the wild type NBF2-WA domain would do so (20.65 ± 2.6, n = 7, p < 0.001). Consistently, GST-NBF2-WA (K1385M) co-dialyzed with Syn-1A did not block Syn-1A inhibition of KATP channel in INS-1 cells (NBF2-WA (K1385M) + Syn-1A: 134.1 ± 17.2 pA/pF (n = 7) versus Syn-1A alone: 137.8 ± 7.2 pA/pF; n = 13) in contrast to WT NBF2-WA (195.4 ± 12.9 pA/pF, n = 5), which blocked Syn-1A inhibition. Representative traces of these experiments are in supplemental Fig. S2. These results indicate that the WA motif in NBF2 is both a binding and transduction site for Syn-1A in inhibiting β-cell KATP channels.
WB motif mutations in NBF1-WB (D854N) and NBF2-WB (D1506N) remained able to bind Syn-1A (Fig. 5B). Both NBF1-WB (D854N) and NBF2-WB (D1506N) were also as effective as their wild type proteins in blocking Syn-1A inhibition of INS-1 β-cell KATP channels (Fig. 5D and representative traces in supplemental Fig. S2) and in disrupting Syn-1A interactions with full-length SUR1 in the FRET assay (supplemental Fig. S3). These results indicate that the binding and transduction domains within the WB regions of NBF1 and NBF2 are actually not at the WB motifs per se but rather at domains adjacent to the WB motifs.
DISCUSSION
In this work, we employed complementary strategies (protein binding assays, patch clamp electrophysiology, and FRET imaging analysis) to gain further insight into how Syn-1A may coordinate with the NBFs of SUR1 to mediate inhibition of islet β-cell KATP channels. Collectively, our results indicate that Syn-1A favors the WA and WB regions of NBF1 and NBF2 because suppression of channel activity can be transduced through three of four regions: NBF1-WB, NBF2-WA, and NBF2-WB. Within these regions in NBF1 and NBF2, Syn-1A binds the WA motifs per se but binds to as yet undefined domains adjacent to the WB motifs.
Our binding data predicted that the linker regions of both NBF1 and NBF2 would not functionally inhibit Syn-1A, because no physical interaction was observed between the linkers and Syn-1A. Indeed, this prediction was verified through whole cell dialysis of the NBF1-linker and NBF2-linker into both INS-1 and primary rat pancreatic islet β-cells, as well as FRET microscopy of transfected HEK cells. We have found that Syn-1A binds to NBF1-WA motif but peculiarly would not reverse Syn-1A inhibition of KATP channel, indicating that this a putative Syn-1A binding site but that would not transduce Syn-1A actions on KATP channel. The fact that NBF1-WA domain would not disrupt Syn-1A interaction with SUR1 indicates that other domains in NBF1 and NBF2 are sufficient to stabilize binding to Syn-1A. In contrast to NBF1-WA, NBF2-WA motif is the putative binding domain that could both bind and transduce Syn-1A inhibition on KATP channels. Although both WB regions of NBF1 and NBF2 could bind and transduce Syn-1A inhibition KATP channel, the binding sites are likely to be adjacent to the Walker motifs and not the motifs per se. These binding sites in NBF1 and NBF2 could bind and relay the signal or transduce conformational changes required to induce channel closure.
The level of syntaxin-1A expression is uneven along the plasma membrane: higher at the exocytotic sites and lower at nonexocytotic sites. Regulation of KATP channels is nonexocytotic; thus it is likely that recombinant Syn-1A applied into the cell even at a low concentration has inhibitory effects on KATP channels. Although Syn-1A is distributed all along the entire plasma membrane, the KATP channels may not be all colocalized and bound to endogenous Syn-1A, although some are bound; as our previous work shows, BoNT/C1 cleavage of endogenous Syn-1A could increase KATP channel activity (19, 21).
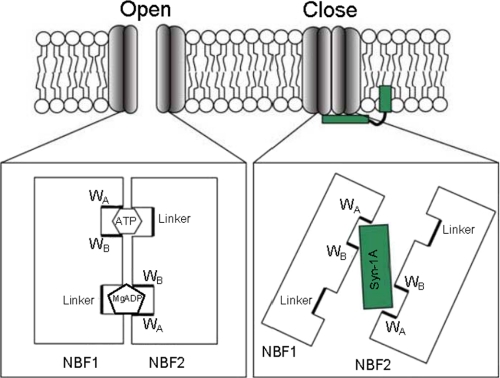
Based on our data, we asked: If Syn-1A transduces its action through the NBFs, how can nucleotide binding activate the channel while Syn-1A inhibits it? In an attempt to answer this, we propose a mechanism by which Syn-1A would serve to stabilize the NBFs in an open dimer conformation, opposite that of the closed dimer required for coordination of ATP. NBF dimerization has been strongly supported as a conserved mechanism across various ABC family members (9, 28–30). It is thought that ATP binding and subsequent hydrolysis provides the regulatory powerstroke required to elicit conformational change in the ABC protein. For example, the CFTR Cl− channel activates upon ATP binding to the NBF dimer but is thought to close upon hydrolysis, dimer disruption, and subsequent release of the nucleotide (30). Stabilization of ATP requires tight packing of the NBFs, to which we refer as the closed dimer conformation. In contrast, electron microscopy has revealed that in the absence of ATP, the NBFs of KATP channels appear much less intimate with each other (31). In fact, the NBFs are spread in such a manner that they appear to interact more with neighboring NBFs of adjacent SUR subunits. We believe that this open dimer conformation of the NBFs may be how and where Syn-1A clamps onto the channel (Fig. 6). It should be noted that the KATP channel in the electron microscopy study was assumed to be in a closed state because PIP2, primarily responsible for endogenous KATP activation, was excluded from the solutions used for purification and structural measurement (31). Based on protein sizes obtained from the Misura et al. study of the three-dimensional structure and crystallization of Syn-1A (32), the putative Syn-1A H3 interacting domain (∼9 nm in length) would have clear access to the NBFs (∼5 nm in height), even when tethered to the plasma membrane (Fig. 6). This would allow Syn-1A to effectively “clamp” onto the channel. Although a previous report indicates the HABC domain does not bind to SUR1 (20), initial H3 attachment may facilitate HABC binding to further clamp the channel. Our results provide important pieces to this model by identifying the interacting NBF domains, including the WA motifs per se in NBF1 and NBF2.
FIGURE 6.
Modeling syntaxin-1A interaction with specific Walker domains of NBF1 and NBF2. A tight NBF-1/2 dimer supports the open conformation of the KATP channel, where NBF1 binds to ATP, whereas NBF2 binds to MgADP (33). During the closed conformation, syntaxin-1A is sandwiched inside the NBF-1/2 dimer, interacting only with Walker domain regions. Various Walker and linker motifs are labeled, as well as NBFs and syntaxin-1A.
The FRET signal in our assay is generated between two fluorescent proteins: the GFP and mCherry pair of the interacting proteins in living β-cells. The GFP-mCherry pair generates the FRET signal when the pair molecules are less than 10 nm apart and thus extremely sensitive in picking up very small intermolecular disruptions. Our data showed that Walker domains (NBF1-WB, NBF2-WA, and NBF2-WB), but not the linkers, sufficed to inhibit the FRET signals generated between EGFP-SUR1 subunit and mCherry-Syn-1A, indicating that a tight interaction of Syn-1A to SUR1 is mediated through the Walker domains. These findings further support the model that the NBF-1/2 dimer constrains the open conformation and snugly binds/interacts intimately with Syn-1A, leading to a close conformation (Fig. 6). A slight disruption within a putative Walker motif can sufficiently reduce this fit as to disrupt the FRET signal; thus it is likely that these two Walker domains of the same NBF may prefer a conformation in which they rely on each other for stable binding to Syn-1A. However, the linker regions are not required for stabilization of the NBF1/2 dimer to Syn-1A binding.
Overall, this work has provided valuable insight into the structure-function relationship between Syn-1A and the KATP channel. Although further work is necessary to reveal additional residues of interaction located on both the NBFs and Syn-1A, we have established a strong starting point for these prospective studies. The foreseeable crystallization of the KATP structure in the future will be essential to modeling how Syn-1A fits into the channel and, more importantly, how this channel truly works.
Supplementary Material
This work was supported by Canadian Institutes for Health Research Grant MOP 69083 and Heart and Stroke Foundation of Ontario Grant T-6064 (to H. Y. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- KATP channel
- ATP-sensitive potassium channel
- SUR1
- sulfonylurea receptor-1
- NBF
- nucleotide binding fold
- Syn-1A
- syntaxin-1A
- WA
- Walker A
- WB
- Walker B
- MES
- 2-morpholino-ethanesulfonic acid
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Ashcroft F. M., Harrison D. E., Ashcroft S. J. (1984) Nature 312, 446–448 [DOI] [PubMed] [Google Scholar]
- 2. Ashcroft F. M., Gribble F. M. (1999) Diabetologia 42, 903–919 [DOI] [PubMed] [Google Scholar]
- 3. Inagaki N., Gonoi T., Clement J. P., 4th, Namba N., Inazawa J., Gonzalez G., Aguilar-Bryan L., Seino S., Bryan J. (1995) Science 270, 1166–1170 [DOI] [PubMed] [Google Scholar]
- 4. Shyng S., Nichols C. G. (1997) J. Gen. Physiol. 110, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ashford M. L., Sturgess N. C., Cook D. L., Hales C. N. (1986) Adv. Exp. Med. Biol. 211, 69–76 [DOI] [PubMed] [Google Scholar]
- 6. Niki I., Nicks J. L., Ashcroft S. J. (1990) Biochem. J. 268, 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwanstecher M., Sieverding C., Dörschner H., Gross I., Aguilar-Bryan L., Schwanstecher C., Bryan J. (1998) EMBO J. 17, 5529–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uhde I., Toman A., Gross I., Schwanstecher C., Schwanstecher M. (1999) J. Biol. Chem. 274, 28079–28082 [DOI] [PubMed] [Google Scholar]
- 9. Campbell J. D., Sansom M. S., Ashcroft F. M. (2003) EMBO Rep. 4, 1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nichols C. G., Shyng S. L., Nestorowicz A., Glaser B., Clement J. P., 4th, Gonzalez G., Aguilar-Bryan L., Permutt M. A., Bryan J. (1996) Science 272, 1785–1787 [DOI] [PubMed] [Google Scholar]
- 11. Matsuo M., Kimura Y., Ueda K. (2005) J. Mol. Cell Cardio. 38, 907–916 [DOI] [PubMed] [Google Scholar]
- 12. Sørensen J. B. (2005) Trends Neurosci. 28, 453–455 [DOI] [PubMed] [Google Scholar]
- 13. Rizo J., Rosenmund C. (2008) Nat. Struct. Mol. Biol. 15, 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lang T., Jahn R. (2008) Handbook Exp. Pharmacol. 184, 107–127 [DOI] [PubMed] [Google Scholar]
- 15. Leung Y. M., Kwan E. P., Ng B., Kang Y., Gaisano H. Y. (2007) Endocr. Rev. 28, 653–663 [DOI] [PubMed] [Google Scholar]
- 16. Michaelevski I., Chikvashvili D., Tsuk S., Fili O., Lohse M. J., Singer-Lahat D., Lotan I. (2002) J. Biol. Chem. 277, 34909–34917 [DOI] [PubMed] [Google Scholar]
- 17. Leung Y. M., Kang Y., Gao X., Xia F., Xie H., Sheu L., Tsuk S., Lotan I., Tsushima R. G., Gaisano H. Y. (2003) J. Biol. Chem. 278, 17532–17538 [DOI] [PubMed] [Google Scholar]
- 18. Bezprozvanny I., Scheller R. H., Tsien R. W. (1995) Nature 378, 623–626 [DOI] [PubMed] [Google Scholar]
- 19. Pasyk E. A., Kang Y., Huang X., Cui N., Sheu L., Gaisano H. Y. (2004) J. Biol. Chem. 279, 4234–4240 [DOI] [PubMed] [Google Scholar]
- 20. Cui N., Kang Y., He Y., Leung Y. M., Xie H., Pasyk E. A., Gao X., Sheu L., Hansen J. B., Wahl P., Tsushima R. G., Gaisano H. Y. (2004) J. Biol. Chem. 279, 53259–53265 [DOI] [PubMed] [Google Scholar]
- 21. Kang Y., Zhang Y., Liang T., Leung Y. M., Ng B., Xie H., Chang N., Chan J., Shyng S. L., Tsushima R. G., Gaisano H. Y. (2011) J. Biol. Chem. 286, 5876–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin Y. W., Bushman J. D., Yan F. F., Haidar S., MacMullen C., Ganguly A., Stanley C. A., Shyng S. L. (2008) J. Biol. Chem. 283, 9146–9156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hohmeier H. E., Mulder H., Chen G., Henkel-Rieger R., Prentki M., Newgard C. B. (2000) Diabetes 49, 424–430 [DOI] [PubMed] [Google Scholar]
- 24. Ueda K., Inagaki N., Seino S. (1997) J. Biol. Chem. 272, 22983–22986 [DOI] [PubMed] [Google Scholar]
- 25. Matsuo M., Kioka N., Amachi T., Ueda K. (1999) J. Biol. Chem. 274, 37479–37482 [DOI] [PubMed] [Google Scholar]
- 26. de Wet H., Mikhailov M. V., Fotinou C., Dreger M., Craig T. J., Vénien-Bryan C., Ashcroft F. M. (2007) FEBS J. 274, 3532–3544 [DOI] [PubMed] [Google Scholar]
- 27. Gribble F. M., Tucker S. J., Haug T., Ashcroft F. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7185–7190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J., Lu G., Lin J., Davidson A. L., Quiocho F. A. (2003) Mol. Cell 12, 651–661 [DOI] [PubMed] [Google Scholar]
- 29. Higgins C. F., Linton K. J. (2004) Nat Struct Mol Biol 11, 918–926 [DOI] [PubMed] [Google Scholar]
- 30. Vergani P., Lockless S. W., Nairn A. C., Gadsby D. C. (2005) Nature 433, 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mikhailov M. V., Campbell J. D., de Wet H., Shimomura K., Zadek B., Collins R. F., Sansom M. S., Ford R. C., Ashcroft F. M. (2005) EMBO J. 24, 4166–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Misura K. M., Scheller R. H., Weis W. I. (2000) Nature 404, 355–362 [DOI] [PubMed] [Google Scholar]
- 33. Ueda K., Komine J., Matsuo M., Seino S., Amachi T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.