Abstract
Toll-like receptor 4 (TLR4) is involved in activation of the innate immune response in a large number of different diseases. Despite numerous studies, the role of separate domains of TLR4 in the regulation of receptor activation is poorly understood. Replacement of the TLR4 ectodomain with LPS-binding proteins MD-2 or CD14 resulted in a robust ligand-independent constitutive activation comparable with the maximal stimulation of the receptor with LPS. The same effect was achieved by the replacement of the ectodomain with a monomeric fluorescent protein or a 24-kDa gyrase B fragment. This demonstrates an intrinsic dimerization propensity of the transmembrane and cytoplasmic domains of TLR4 and reveals a previously unknown function of the ectodomain in inhibiting spontaneous receptor dimerization. Constitutive activation was abolished by the replacement of the ectodomain by a bulkier protein ovalbumin. N-terminal deletion variants of TLR4 revealed that the smallest segment of the ectodomain that already prevents constitutive activity comprises only 90 residues (542 to 631) of the total 608 residues. We conclude that TLR4 represents a receptor with a low threshold of activation that can be rapidly activated by the release of inhibition exerted by its ectodomain. This is important for the sensitivity of TLR4 to activation by different agonists. The TLR4 ectodomain has multiple roles in enabling ligand regulated activation, providing proper localization while serving as an inhibitor to prevent spontaneous, ligand-independent dimerization.
Keywords: Fusion Protein, Innate Immunity, Lipopolysaccharide (LPS), Toll IL-1 Receptor (TIR) Domain, Toll-like Receptors (TLR)
Introduction
Pattern recognition receptors are proteins expressed by cells of the innate immune system and act as a first line of host defense against invading microorganisms, recognizing diverse but highly conserved structural pathogen-associated molecular patterns of bacteria, fungi, and viruses (1). Activation of pattern recognition receptors by their respective ligands triggers an immediate immune response, characterized by proinflammatory cytokine production (1, 2). Toll-like receptors (TLRs)2 are a family of pattern recognition receptors central to the vertebrate innate immune response. Activation of TLRs links innate and adaptive immunity through proinflammatory signaling and induction of costimulatory molecules on immune cells (3, 4). However, activation of TLRs has to be a tightly regulated process. TLRs have to be able to mount an immediate immune response upon binding of the agonist, whereas in the absence of the ligand it is imperative that they remain in an inactive state to prevent unwanted activation that may lead to excessive inflammation and autoimmune disease (5). This is especially important for TLR4, the cellular receptor for LPS, a molecular signature of the outer cell membrane of Gram-negative bacteria (6). Activation of TLR4 by LPS affects the regulation of more than 1000 genes (7). Additionally, TLR4 signaling has been implicated in sterile inflammation through activation of the receptor by endogenous ligands (8–14).
TLR4 is a type I transmembrane glycoprotein. The ectodomain of TLR4 is composed of 22 repeats of a leucine-rich repeat module, adopting a solenoidal conformation (15, 16). A single transmembrane helix connects the ectodomain to the Toll/IL-1R (TIR) cytoplasmic domain, responsible for intracellular signal propagation upon receptor activation by LPS (2, 17). TLR4 is unique among the TLRs, being able to trigger two distinct intracellular signal transduction pathways. The MyD88-dependent signaling pathway culminates primarily in proinflammatory cytokine production (18), whereas the TRIF-dependent pathway leads to the production of type I interferons (19).
TLR4 differs from other TLRs in another aspect also, as it requires a coreceptor molecule myeloid differentiation protein-2 (MD-2) for the activation by its agonist LPS (20). MD-2 is a secreted glycoprotein that binds to the N terminus of the ectodomain of TLR4 (21) and is the actual LPS-binding protein of the receptor complex and indispensable for signaling (20, 22, 23). Another molecule, cluster of differentiation 14 (CD14), facilitates signaling by TLR4 (24, 25). CD14 is, like TLR4, a LRR module-containing molecule with overall similar horseshoe-shaped conformation (26). It exists either as a glycosylphosphatidylinositol-anchored molecule at the cell surface or as a soluble molecule circulating in serum (27). CD14 serves as a lipotransferase, transferring LPS monomers to MD-2, and is nonetheless indispensable for physiological responses to low concentrations of LPS (28).
Despite the overwhelming data on the TLR4-mediated signaling cascade (29) as well as the solved crystal structure of the ectodomain of TLR4 in complex with MD-2 and its agonistic and antagonistic ligands, LPS and lipid IVa, respectively (15, 16), the mechanisms governing receptor activation and its dynamics remain unclear. Structural information is available for the soluble domains. However, the membrane proximity introduces important structural constraints and the issue of interdomain flexibility. In this study, we have set out to investigate the contributions of separate domains of the LPS-sensing complex to receptor activation and subsequent signal transduction by creating various fusion proteins as well as dissecting TLR4 into separate functional domains. We show that although the transmembrane and cytoplasmic TIR domain of TLR4 contribute the major interaction energy for receptor dimerization, the ectodomain plays an important role in providing controlled responsiveness to LPS, primarily by inhibiting spontaneous dimerization of the transmembrane and cytoplasmic domains, thereby preventing constitutive, ligand-independent receptor signaling.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
The HEK293 cell line was provided by Dr. J. Chow (Eisai Research Institute, Andover, MA). Expression plasmids containing sequences of TLR4, TLR2, MD-2, and CD14, as well as the pELAM-1 firefly luciferase plasmid, were a gift from Dr. C. Kirschning (Technical University of Munich, Germany). A Renilla luciferase phRL-TK plasmid was from Promega. A clone for mCherry and a TLR4-YFP expression plasmid were provided by Dr. T. Espevik (Norwegian Institute of Science and Technology, Norway), and a clone for mCerulean was provided by Dr. D. Piston (Vanderbilt University, Nashville, TN). A clone for OVA was purchased from Addgene. A Renilla luciferase plasmid phRL-TK was from Promega. Smooth LPS (from Salmonella abortus equi) was kindly provided by Dr. K. Brandenburg (Research Center Borstel, Germany) and FITC-LPS was a gift from Dr. K. Triantafilou (University of Sussex, UK). DMEM with GlutaMAX-I was from Invitrogen, FBS from BioWhittaker. All other reagents and chemicals were purchased from Sigma unless stated otherwise.
DNA Construct Preparation
N-terminally truncated versions of TLR4 used in this work were prepared by classical PCR amplification of a specific DNA segment of TLR4 and cloned into the pFLAG-CMV-3 expression vector (Sigma) to obtain the N-terminal preprotrypsin leader sequence and a FLAG tag. All chimeric DNA constructs were created by means of a PCR overlap extension technique and cloned into pFLAG-CMV-3, except for the TM-TIR-TLR4-CHERRY and TIR-TLR4-CERULEAN constructs that were inserted into the pcDNA3 vector (Invitrogen), with the TM-TIR-TLR4-CHERRY containing the endogenous TLR4 signal sequence and TIR-TLR4-CERULEAN being devoid of it. In the OVA-L9-TM-TLR4 construct, the nucleotide sequence of a nine-amino acid linker (L9) connecting ovalbumin with the transmembrane segment of TLR4 was incorporated into forward and reverse primers. Proofreading DNA polymerases Platinum Pfx or AccuPrime Pfx (both Invitrogen) were used in all reactions. All constructs were sequenced. Primer sequences are available upon request.
Luciferase Reporter Assay
HEK293 cells were seeded in 96-well Costar plates (Corning, NY) at 5 × 104 cells per well and incubated overnight in a humidified atmosphere (5% CO2) at 37 °C. The next day, cells were cotransfected with expression plasmids coding for specific constructs, along with NF-κB-dependent firefly luciferase (40 ng) and constitutive Renilla (5 ng) luciferase reporter plasmids using Lipofectamine 2000 (Invitrogen). After 20 h, the cells were either lysed or the medium was replaced and cells stimulated with LPS or coumermycin A1 (Promega) for a further 18 h before lysis. Cell lysis was performed in passive lysis buffer (Promega) and analyzed for reporter protein activities using a dual-luciferase reporter assay system on a Mithras LB940 luminometer. Relative luciferase activity was calculated by normalizing the firefly luciferase activity of each sample for constitutive Renilla luciferase activity measured within the same sample.
Immunoblotting
HEK293T cells were seeded in a 6-well plate (Techno Plastic Products, Switzerland) at a density of 3 × 105 cells/well. 48 h later, they were transfected with 2 μg of appropriate plasmid DNA using Lipofectamine 2000. 24 to 48 h after transfection, cells were lysed in lysis buffer (50 mm Tris-HCl (pH 8), 1 mm EDTA (pH 8), 1 mm EGTA (pH 8), 150 mm NaCl, 1% Triton X-100, 1% Na-DOC, 10% glycerol, 1 mm Na3VO4, 50 mm NaF, 1 mm β-glycerophosphate) containing a mixture of protease inhibitors (Roche). Protein samples mixed with Laemmli sample buffer containing dithiothreitol were resolved on SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was washed with Tris-buffered saline (pH 7.5) containing 0.01% Tween 20 (TBST), blocked to reduce nonspecific background with 5% dried nonfat milk in TBST for 1 h at 25 °C, and incubated with primary rabbit anti-FLAG polyclonal antibody (Sigma) in the blocking solution for 1 h. After washing with TBST, the blot was incubated with goat anti-rabbit horseradish peroxidase-labeled antibody (Abcam) for 45 min at 25 °C in the blocking solution and washed extensively with TBST. Blots were developed using the Pierce Super Signal Pico substrate system.
Cell Surface Expression of TLR4 Chimeric Receptors and Deletion Variants
Cell surface expression of TLR4 chimeric and deletion variants was assessed by flow cytometry. Briefly, HEK293T cells were prepared as described for Western blotting and collected 24 h or 48 h after transfection in PBS containing 3% FBS (FACS buffer). Cells were resuspended in 100 μl of FACS buffer containing 8 μg/ml polyclonal primary rabbit anti-FLAG antibody (Sigma) and incubated on ice for 30 min. Afterward, cells were washed three times in FACS buffer, resuspended, and incubated for another 30 min in the dark with 2 μg/ml DyeMer 488/615-labeled goat anti-rabbit or phycoerythrin (PE)-labeled goat anti-rabbit antibody (both Molecular Probes). The cells were then washed three more times and finally resuspended in 500 μl of FACS buffer. Flow cytometry analysis was performed on an EPICS ALTRA flow cytometer (Beckman Coulter). In each sample, at least 30,000 cells were analyzed.
Binding of FITC-LPS
Binding of FITC-LPS was assayed using flow cytometry. HEK293T cells expressing transfected DNA constructs were prepared as described for Western blotting and collected 24 h or 48 h after transfection in fresh serum-free medium. Cells were incubated with FITC-LPS at a concentration of 2 μg/ml in the dark at 37 °C After 20 min of incubation, cells were extensively washed in PBS, finally resuspended in FACS buffer, and immediately analyzed on an EPICS ALTRA flow cytometer (Beckman Coulter).
Confocal Microscopy
HEK293 cells were seeded at a density of 5 × 104 cells per well in an 8-well microscope chamber slides (Ibidi, DE). 24 h later, they were transfected with a total amount of 160 ng of plasmid DNA using Lipofectamine 2000. Cells were analyzed live 24 h to 48 h after transfection on a Leica TCS5 confocal microscope using a 63× magnification. Where needed, cell membranes were stained before imaging with Dil (1,1′ dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) or SynaptoRed (both Invitrogen) according to the manufacturer's instructions.
RESULTS
The TLR4 Ectodomain Has Additional Roles Besides Providing a Scaffold for the Coreceptor and Ligand Binding
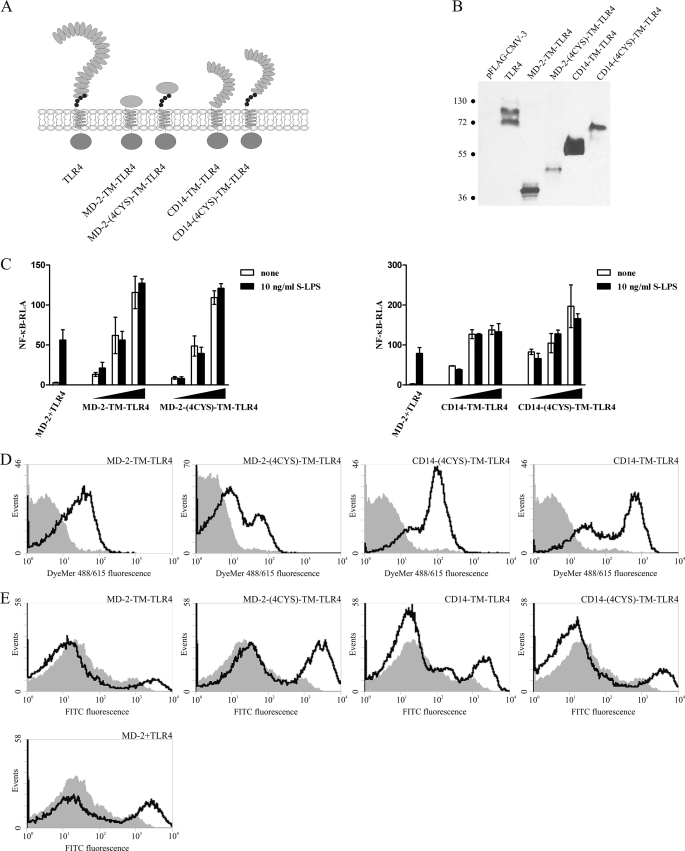
To investigate additional roles of the TLR4 ectodomain, we have examined the effect of replacement of the ectodomain of TLR4 with either of the two LPS-binding proteins, MD-2 or CD14, generating MD-2-TM-TLR4 and CD14-TM-TLR4 chimeric constructs (Fig. 1A). Additionally, two other constructs were prepared, MD-2-(4CYS)-TM-TLR4 and CD14-(4CYS)-TLR4, which retained the membrane proximal segment of the TLR4 ectodomain (Fig. 1A). This segment, conserved in all TLRs as well as other extracellular leucine-rich repeat proteins, contains two disulfide bridges and has been previously shown to play an important structural role, conferring conformational rigidity to the C-terminal segment of the ectodomain (2, 30).
FIGURE 1.
Replacement of the ectodomain of TLR4 with LPS-binding proteins leads to constitutive activation. A, schematic representation of the MD-2-TM-TLR4, MD-2-(4CYS)-TM-TLR4, CD14-TM-TLR4, and CD14-(4CYS)-TM-TLR4 chimeric constructs. The conserved cysteine residues of the TLR4 juxtamembrane region are represented as small black circles juxtaposed to the membrane. B, constitutive activation of the NF-κB promoter by the MD-2- and CD14-based TLR4 chimeric constructs. HEK293 cells were transiently transfected with reporter plasmids as well as increasing amounts (1 ng, 5 ng, and 10 ng) of plasmid DNA encoding the MD-2-TM-TLR4, MD-2-(4CYS)-TM-TLR4, CD14-TM-TLR4, or CD14-(4CYS)-TM-TLR4 constructs or with MD-2- and TLR4-encoding plasmids (1 ng each). After 18 h of stimulation with LPS, luciferase activity (RLA) was measured in the cell lysates. C, expression levels of chimeric constructs in HEK293T cells. HEK293T cells were transiently transfected with 2 μg of MD-2-TM-TLR4, MD-2-(4CYS)-TM-TLR4, CD14-TM-TLR4, CD14-(4CYS)-TM-TLR4, or TLR4 plasmid DNA. 48 h post-transfection, cell lysates were prepared, and 30 μg of total proteins were loaded onto an SDS-PAGE gel and blotted using anti-FLAG antibody. Molecular weight marker sizes are given in kDa. D, MD-2- and CD14- based TLR4 chimeric constructs are expressed at the cell surface. HEK293T cells were transiently transfected with 2 μg of MD-2-TM-TLR4, MD-2-(4CYS)-TM-TLR4, CD14-TM-TLR4, or CD14-(4CYS)-TM-TLR4 plasmid DNA. Cell surface expression levels of chimeric constructs were determined by flow cytometric analysis 48 h post-transfection. Histograms of mock-transfected cells (filled areas) or cells transfected with the appropriate DNA (solid lines) are shown. E, MD-2- and CD14-based TLR4 chimeric constructs bind LPS. HEK293T cells were transiently transfected with 2 μg of MD-2-TM-TLR4-, MD-2-(4CYS)-TM-TLR4-, CD14-TM-TLR4-, or CD14-(4CYS)-TM-TLR4-encoding plasmids or with TLR4 and MD-2 plasmid DNA (1 μg each). 48 h post-transfection, binding of FITC-LPS by the receptors was assessed by flow cytometric analysis. Histograms of mock-transfected cells treated with LPS (filled areas) or transfected cells treated with LPS (solid lines) are shown.
The MD-2-TM-TLR4 and CD14-TM-TLR4 fusion constructs exhibited strong constitutive activation that was entirely LPS-independent (Fig. 1B). The inclusion of the conserved juxtamembrane cysteine-rich stretch of the TLR4 ectodomain did not make a difference, as the MD-2-(4CYS)-TM-TLR4 and CD14-(4CYS)-TM-TLR4 constructs were as active as MD-2-TM-TLR4 and CD14-TM-TLR4 (Fig. 1B). The observed effect of constitutive activation of these chimeric constructs was not due to misfolding, as all proved to be functional. Chimeric proteins were expressed in the cells (Fig. 1C) and at the cell surface (D), and the latter were capable of binding LPS (E). Furthermore, they all signaled through the same signaling pathways as the native receptor pair MD-2/TLR4 because their signaling could effectively be inhibited by dominant negative versions of the downstream adapter proteins MyD88 and TRIF (data not shown).
These results indicate that the ectodomain of TLR4 does not merely serve as a scaffold for MD-2 binding and interaction with CD14 but rather seems to play an important role in enabling the controlled responsiveness of the receptor complex to LPS. Furthermore, at least in the case of the CD14- and MD-2-TLR4 chimeric constructs, the inclusion of the leucine-rich repeat C-cap of the juxtamembrane part of the ectodomain of TLR4 does not prevent the constitutive activity.
Replacement of the TLR4 Ectodomain by a Monomeric Domain Retains the Constitutive Activity
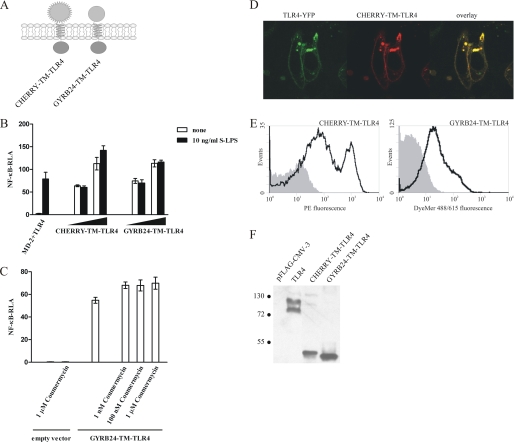
The prerequisite for TLR-induced cell activation is association of cytosolic TIR domains, as only dimeric TIRs provide the scaffold for the recruitment of cytosolic adapters and assembly of signalosome, which is a crucial step in the initiation of intracellular signal propagation (31). Thus, as MD-2- and CD14-based TLR4 chimeras are constitutively active, they must be present in the cell in the form of dimeric or oligomeric clusters. As MD-2 is known to form dimers and higher-order multimers (32, 33), and CD14 was in fact crystallized as a dimer (26), a dimeric or oligomeric state of the chimeric receptors could lead to the oligomerization of the ectodomains of these constructs, bringing the TIR domains close together. This is supported by the previous report on the addition of the CD4 dimerization domain to the transmembrane and cytoplasmic domains of TLR4, which also resulted in constitutive activity of CD4-TLR4 chimeras (17). To rule out the effect of intrinsic dimerization of ectodomain replacements, MD-2 and CD14 were substituted by the monomeric protein mCherry (34) or the 24-kDa N-terminal fragment of Escherichia coli gyrase B (GYRB24), which has previously been used as a fusion partner in coumermycin-induced dimerization (35, 36). The CHERRY-TM-TLR4 and GYRB24-TM-TLR4 constructs are schematically represented in Fig. 2A.
FIGURE 2.
Replacement of the TLR4 ectodomain by monomeric protein domains leads to the constitutive activation. A, schematic representation of the CHERRY-TM-TLR4 and GYRB24-TM-TLR4 chimeras. B, the CHERRY-TM-TLR4 and GYRB24-TM-TLR4 chimeric constructs constitutively activate target promoters. HEK293 cells were transiently transfected with reporter plasmids as well as increasing amounts (1 ng and 10 ng) of plasmid DNA encoding the CHERRY-TM-TLR4 or GYRB24-TM-TLR4 expression constructs or with MD-2- and TLR4-encoding plasmids (1 ng each). After 18 h of stimulation with LPS, luciferase activity (RLA) was measured in the cell lysates. C, addition of coumermycin A1 to GYRB24-TM-TLR4-expressing cells does not enhance cell activation. HEK293 cells were transiently transfected with reporter plasmids as well as with 5 ng of plasmid DNA encoding the GYRB24-TM-TLR4 construct or an empty vector. After 18 h of stimulation with different amounts of coumermycin A1, luciferase activity was measured in the cell lysates. D and E, the CHERRY-TM-TLR4 and GYRB24-TM-TLR4 chimeric constructs are expressed at the cell surface. D, HEK293 cells were transiently cotransfected with a plasmid encoding TLR4-YFP and a plasmid encoding the CHERRY-TM-TLR4 chimera (80 ng each). The expression profiles of both constructs was compared using a confocal microscope 36 h post-transfection. E, HEK293T cells were transiently transfected with 2 μg of CHERRY-TM-TLR4 or GYRB24-TM-TLR4 plasmid DNA. Cell surface expression levels of the constructs were determined by flow cytometric analysis 48 h after transfection. Histograms of mock-transfected cells (filled areas) or cells transfected with the appropriate DNA (solid lines) are shown. F, expression of chimeric constructs in HEK293T cells. HEK293T cells were transiently transfected with 2 μg of MD-2-TM-TLR4, MD-2-(4CYS)-TM-TLR4, CD14-TM-TLR4, CD14-(4CYS)-TM-TLR4, or TLR4 plasmid DNA. 48 h post-transfection, cell lysates were prepared, and 30 μg of total proteins were loaded onto an SDS-PAGE gel and blotted using anti-FLAG antibody. Molecular weight marker sizes are given in kDa.
Despite fusion of monomeric partners to the transmembrane and cytoplasmic domains of the TLR4, CHERRY-TM-TLR4, as well as GYRB24-TM-TLR4 constructs induced constitutive signaling in HEK293 cells (Fig. 2B). Addition of a gyrase dimerization-inducing agent, coumermycin A1 (35), to the GYRB-TM-TLR4-expressing cells did not significantly increase the cellular activation (Fig. 2C), indicating that dimers or higher-order oligomers of the chimeras must have already been formed even in the absence of coumermycin A1. CHERRY-TM-TLR4 clearly colocalized with TLR4-YFP (Fig. 2D) and was, like GYRB24-TM-TLR4, present at the cell surface (E). CHERRY-TM-TLR4, GYRB24-TM-TLR4, and TLR4 were expressed at similar levels in HEK293T cells (Fig. 2F).
These results indicate that the constitutive activity of the chimeric TLR4 receptor is a consequence of an intrinsic dimerization propensity of the transmembrane and/or cytoplasmic domain of TLR4 and not a consequence of the extracellular fusion partner-forced dimerization of these domains.
The Large Extracellular Fusion Partner to the Transmembrane and Cytoplasmic Domains of TLR4 Prevents the Constitutive Activation
As all TLRs have similar ectodomains in terms of their size, shape, and structure. The replacement of the TLR4 ectodomain with that of TLR2 would be expected to result in an inactive chimeric receptor. As expected, the TLR2-TM-TLR4 construct (Fig. 3A) behaved similarly to WT TLR4 in terms of basal cell activation in HEK293 cells (Fig. 3B) as well as in terms of its expression profile (C and D). A degree of basal activity, similar to that of wild-type TLR4, has previously been demonstrated also for the TLR9-TLR4 fusion (37), in agreement with our results. Thus, the ectodomains of at least TLRs 2 and 9, fused to the transmembrane and intracellular domains of TLR4, can functionally replace the TLR4 ectodomain in terms of sustaining only the basal level of cell activation.
FIGURE 3.
Replacement of the TLR4 ectodomain by the ectodomain of another TLR or a large monomeric protein prevent constitutive activation. A, schematic representation of the TLR2-TM-TLR4 and OVA-L9-TM-TLR4 chimeric constructs. The conserved cysteine residues of the TLR4 juxtamembrane region are represented as small black circles. B, proper choice of the extracellular fusion partner to the transmembrane and cytoplasmic domains of TLR4 can inhibit constitutive activation of cells. HEK293 cells were transiently transfected with reporter plasmids as well as increasing amounts (1 ng, 10 ng, and 50 ng) of plasmid DNA encoding TLR4 or the OVA-L9-TM-TM-TLR4 construct or with MD-2- and TLR4-encoding plasmids (5 ng each). After 18 h of stimulation with LPS, luciferase activity (RLA) was measured in the cell lysates. C, expression of chimeric constructs in HEK293T cells. HEK293T cells were transiently transfected with 2 μg of MD-2-TM-TLR4, MD-2-(4CYS)-TM-TLR4, CD14-TM-TLR4, CD14-(4CYS)-TM-TLR4, or TLR4 plasmid DNA. 48 h post-transfection, cell lysates were prepared, and 30 μg of total proteins were loaded onto an SDS-PAGE gel and blotted using anti-FLAG antibody. Molecular weight marker sizes are given in kDa. D, the inactive TLR4-based chimeric constructs TLR2-TM-TLR4 and OVA-L9-TM-TLR4 are expressed at the cell surface. HEK293T cells were transiently transfected with 2 μg of TLR4, TLR2-TM-TLR4, or OVA-L9-TM-TLR4 plasmid DNA. Cell surface expression levels of receptors were determined by flow cytometric analysis 48 h post-transfection. Histograms of mock-transfected cells (filled areas) or cells transfected with the appropriate DNA (solid lines) are shown.
Next, we expected that in chimeric receptors with fusion partners not adopting a TLR-like structure, inhibition of transmembrane and cytoplasmic domain dimerization could be achieved by a large size or appropriate shape of the fusion partner, exerting steric hindrance to dimerization. This was confirmed by the ovalbumin-based chimera (Fig. 3A) that was inactive even at a high level of expression (B) despite being localized at the cell plasma membrane (D). Ovalbumin is a 45-kDa ellipsoidally shaped monomeric protein (38, 39). Thus, the size, shape, and/or aggregation tendency of the fusion partner to the transmembrane and intracellular domains of TLR4 determine the activity of the chimeric protein.
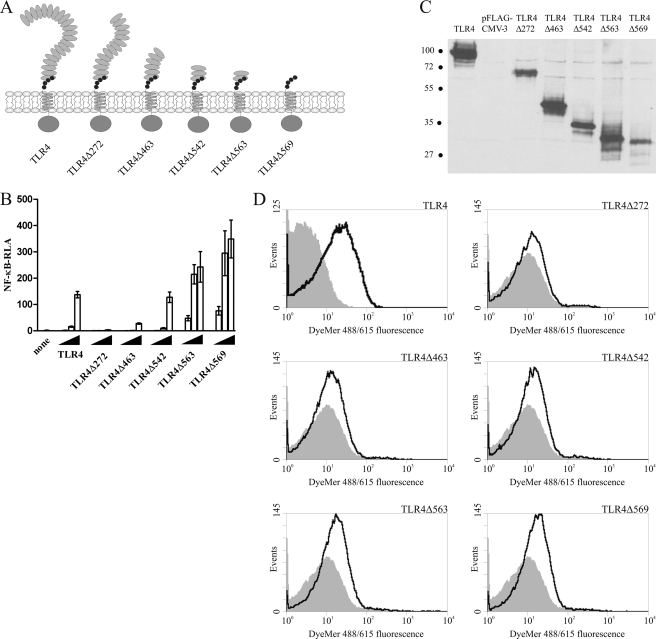
Mapping the Region of the TLR4 Ectodomain Responsible for the Inhibition of the Constitutive Activity
To allocate the minimal region in the TLR4 ectodomain responsible for exerting the inhibitory effect on spontaneous dimerization of the cytoplasmic domains, we prepared constructs with sequential N-terminal deletions of the TLR4 ectodomain (Fig. 4A). Deletions of 272, 463, or even 542 N-terminal amino acid residues, retaining 41, 19, or 10 kDa of the ectodomain, respectively, did not suffice to impart constitutive signaling upon the receptors (Fig. 4B), although they were expressed at the cell surface (C and D). Only the TLR4Δ563 and TLR4Δ569 variants with ∼8 and 7 kDa ectodomains, respectively, strongly activated cells (Fig. 4B). This positions the threshold inhibitory part of the TLR4 molecule between amino acid residues 563 and 542, which means that only 90 amino acid residues of the TLR4 ectodomain are sufficient to inhibit spontaneous dimerization of the cytoplasmic domains of the receptor. These results indicate that in the wild-type receptor, specific structural features of the ectodomain of TLR4 rather than solely its size are responsible for exerting inhibition on the spontaneous dimerization of the receptor cytoplasmic domains.
FIGURE 4.
Deletion mapping of the region in the TLR4 ectodomain domain responsible for inhibition of constitutive activation. A, schematic representation of the deletion variants of TLR4 as compared with the full-length TLR4. Deletion variants were made by sequential N-terminal deletions of the ectodomain of TLR4. The conserved cysteine residues of the TLR4 juxtamembrane region are represented as small black circles. B, comparison of basal activity of N-terminal TLR4 deletions. HEK293 cells were transiently transfected with reporter plasmids as well as increasing amounts (1 ng, 10 ng, and 50 ng) of plasmid DNA encoding TLR4 or the appropriate TLR4 N-terminal deletion variant. After 26 h, luciferase activity (RLA) was measured in the cell lysates. C and D, TLR4 N-terminal deletion variants are expressed in the cells and reach the cell surface. HEK293T cells were transiently transfected with 2 μg of appropriate plasmid DNA. 48 h after transfection, cells were either lysed and expression of deletion variants was determined by Western blot using anti-FLAG antibody or were analyzed by flow cytometry to determine the cell surface expression level of the constructs. Representative immunoblot analysis results (C) and histograms of mock-transfected cells (filled areas) or cells transfected with the TLR4 deletion variants (solid lines) (D) are shown. Molecular weight marker sizes are given in kDa.
TLR4 Signaling Depends on Stable Insertion of the Transmembrane and Cytoplasmic Domains into the Cell Membrane
Despite the intrinsic dimerization propensity of the transmembrane and cytoplasmic domain of TLR4, the deletion variant TM-TIR-TLR4 (i.e. TLR4Δ631) (Fig. 5A), lacking the entire ectodomain of TLR4, was unable to trigger cell activation in HEK293 cells even when overexpressed (Fig. 5B). The same lack of activity was observed for the TM-TIR-TLR4-CHERRY variant (Fig. 5B). In comparison, expression of the TLR4Δ569 deletion variant, encompassing only 63 amino acid residues of the juxtamembrane ectodomain of TLR4, induced robust cellular activation (Fig. 5B). Flow cytometry data confirmed localization of the TLR4Δ569 variant at the cell surface (Fig. 4D), whereas the TM-TIR-TLR4 and TM-TIR-TLR4 -CHERRY variants did not reach the plasma membrane, despite being expressed within the cell (Fig. 5, C and D). Similar results were obtained with the TIR-TLR4 and TIR-TLR4-CERULEAN variants, which were also unable to initiate signaling (Fig. 5, A, B, and D).
FIGURE 5.
TLR4 signaling requires stable insertion into the cell plasma membrane. A, schematic representation of the TM-TIR-TLR4 and TIR-TLR4 deletion constructs and their GFP-tagged variants. The TM-TIR-TLR4 and TIR-TLR4 deletion variants were modified by the addition of fluorescent proteins mCherry and Cerulean, respectively, to their C termini. B, the TM-TIR-TLR4 and TIR-TLR4 deletion constructs as well as their GFP-modified variants are unable to support cell activation. HEK293 cells were transiently transfected with reporter plasmids along with 100 ng of empty vector or plasmid DNA encoding the TIR-TLR4, TIR-TLR4-CERULEAN, TM-TIR-TLR4, or TM-TIR-TLR4-CHERRY variants or with 5 ng of plasmid DNA for the TLR4Δ569 deletion variant. 24 h after transfection, luciferase activity (RLA) was measured in the cell lysates. C, the TMCYTO-TLR4 deletion variant is not present at the cell surface despite being expressed in the cell. HEK293T cells were transiently transfected with 2 μg of TM-TIR-TLR4 plasmid DNA or an empty vector. 48 h after transfection, cells were either analyzed by flow cytometry to determine the cell surface expression level of the construct or were lysed, and expression of TM-TIR-TLR4 was determined by Western blot analysis using anti-FLAG antibody. A representative histogram of mock-transfected cells (filled areas) or cells transfected with TM-TIR-TLR4 DNA (solid lines) and immunoblot analysis results are shown. Molecular weight marker sizes are given in kDa. D, expression profile of the GFP-modified TM-TIR-TLR4 and TIR-TLR4 deletion variants. HEK293 cells were transiently transfected with plasmid DNA encoding the TM-TIR-TLR4-CHERRY or TIR-TLR4-CERULEAN constructs (100 ng each). 24 h after transfection, cell membranes were stained with appropriate dyes, and expression profiles of both constructs were examined by confocal microscopy.
These observations suggest that signaling from the receptor has to be initiated at the cell surface membrane and that the transmembrane domain of TLR4 alone is unable to provide stable insertion of the TM-TIR-TLR4 variant into the cell plasma membrane. Judging by the cell surface expression profile of signaling-competent deletion and chimeric TLR4 variants, a small ectodomain is sufficient for stable anchoring to the plasma membrane.
The Transmembrane and/or Cytoplasmic Domains of TLR4 Can Modulate MD-2/TLR4 Signaling by Sequestering Downstream Adapter Proteins
Despite their lack of constitutive activity, the TM-TIR-TLR4 and TIR-TLR4 deletion variants were functional because they inhibited LPS-dependent MD-2/TLR4 signaling as well as activation of cells by the constitutively active TLR chimeras in a concentration-dependent manner (Fig. 6A). This was not accomplished by modulating the level of the signaling-competent cell-surface expressed receptor as demonstrated by CD14-TM-TLR4 and TMCYTO-TLR4 coexpression (Fig. 6B) but was due to the sequestration of downstream adapter proteins, as deletion variants also inhibited cell activation induced by MyD88-overexpression (C). This can be explained by the tendency of the TLR4 TIR domains to dimerize, thereby creating a scaffold for binding of MyD88. Inhibition results from the fact that signaling can only be initiated from the cell surface, whereas the TM-TIR-TLR4 and TIR-TLR4 variants do not anchor into the cell membrane.
FIGURE 6.
The transmembrane and/or cytoplasmic domains of TLR4 can modulate MD-2/TLR4 signaling by sequestering downstream adapter proteins. A, the TM-TIR-TLR4 and TIR-TLR4 deletion variants inhibit MD-2/TLR4 and constitutive signaling. HEK293 cells were transiently cotransfected with reporter plasmids and plasmid DNA encoding MD-2 and TLR4 (1 ng each) as well as increasing amounts (10 ng, 50 ng, and 100 ng) of the TM-TIR-TLR4 or TIR-TLR4 expression plasmids. After 18 h of stimulation with LPS, luciferase activity (RLA) was measured in the cell lysates. Alternatively, HEK293 cells were transiently cotransfected with reporter plasmids, 120 ng of TM-TIR-TLR4 expression plasmid, and 1 ng of the MD-2-TM-TLR4-, CD14-TM-TLR4-, CHERRY-TM-TLR4-, or TLR4Δ569-encoding plasmids. Luciferase activity was measured in the cell lysates 26 h after transfection. B, inhibitory effect of the TMCYTO-TLR4 deletion variant on constitutive cell activation is not achieved through receptor down-regulation from the cell surface. HEK293T cells were transiently transfected with 500 ng of the CD14-TM-TLR4 and 1500 ng of TM-TIR-TLR4 expression plasmids. Cell surface expression levels of CD14-TM-TLR4 were determined by flow cytometric analysis 48 h after transfection. Histograms of TM-TIR-TLR4-transfected cells (filled area) or cells transfected with either CD14-TM-TLR4 plasmid alone (solid line) or the CD14-TM-TLR4 and TM-TIR-TLR4 plasmids (dotted line) are shown. C, the TM-TIR-TLR4 and TIR-TLR4 deletion variants inhibit signaling induced by overexpression of MyD88. HEK293 cells were transiently transfected with reporter plasmids, a plasmid encoding MyD88 (5 ng), as well as plasmid DNA encoding either the TM-TIR-TLR4 or TIR-TLR4 deletion variants (100 ng). After 36 h, luciferase activity was measured in the cell lysates.
DISCUSSION
TLR4 is probably the most studied among the TLRs, as it is unique in many aspects. TLR4 is the only known TLR to require a coreceptor molecule for ligand recognition, as the response to LPS is entirely dependent on the presence of MD-2 (20, 22, 23). TLR4 is also the only TLR with the ability to signal through two distinct adaptor proteins, MyD88 and TRIF, inducing either inflammatory cytokine secretion or IFN-β production, respectively (18, 19). Furthermore, in addition to LPS, many other exogenous (40–50) as well as some endogenous (8–14) TLR4 agonists have been proposed.
We have investigated the additional roles of the TLR4 ectodomain in receptor activation and subsequent signal transduction by its replacement and deletions. We have demonstrated that the TLR4 ectodomain on one hand provides stable association of the transmembrane and cytoplasmic domains with the plasma membrane, wherefrom signaling can be initiated, whereas on the other hand, it inhibits their spontaneous, ligand-independent dimerization. Strong dimerization of TIR domains is a unique feature of TLR4 among all the TLRs, as demonstrated by Zhang et al. (51) on an example of TLR chimeras with integrins α and β as the fusion partners to the transmembrane and cytoplasmic domains of separate TLRs. Additionally, we have found that in contrast to TLR4, the replacement of the ectodomain of TLR3 by a small polypeptide domain does not lead to receptor activation (unpublished results)3. Lee et al. (52) used the reconstitution of β-lactamase fragments at the C termini of the TLR4 TIR domains to monitor dimerization. However, in that case, dimerization detected by a β-lactamase complementation did not lead to substantial activation. Our results explain these observations, as we have shown that the biological activity of the dimeric cytoplasmic domains of TLR4 depends on their membrane localization. Although their stable anchoring into the plasma membrane imparts induction of signaling, their cytoplasmic localization on the other hand leads to inhibition of WT TLR4 signaling.
Although the transmembrane and cytoplasmic domains of TLRs define their subcellular targeting (37, 53), they alone are insufficient to provide stable anchoring to the plasma membrane and require the presence of an ectodomain. Ectodomains can be very diverse protein domains, as all of the chimeric TLR4 molecules used in this work localized to the plasma membrane, irrespective of the extracellular fusion partner used. The lack of constitutive activity of the OVA-L9-TM-TLR4 construct cannot be attributed solely to the size of OVA, as chimeras with integrins or CD4, both larger than OVA, exhibited constitutive activity in previous experiments (17, 51, 54). However, CD4 is an elongated molecule, and its diameter around their rotational axis at the C-terminal fusion site to the TLR4 TM segment is smaller than that of ovalbumin. Additionally, CD4 and integrins promote dimerization (51, 55, 56), which could contribute to constitutive signaling. On the other hand, a monomeric fusion partner alone does not necessarily prevent the constitutive activity, as the CHERRY-TM-TLR4 chimera still constitutively activated the cells despite the monomeric nature of mCherry. Thus, a number of factors, among them particularly size, shape, and aggregation propensity combined, seem to provide the inhibitory action to the transmembrane and cytoplasmic domain dimerization in the TLR4 chimeras.
Because the cytoplasmic linker between the transmembrane and TIR domain is relatively long and could be extended without affecting receptor activation (57), we conclude that the transmembrane segment must also contribute to the intrinsic receptor dimerization propensity. This is in agreement with the data of Weber et al. (58), who demonstrated increased basal activity of the chimeric Drosophila Toll (dToll), where the transmembrane and cytoplasmic segments of the native protein have been replaced for those of TLR4, as compared with the dToll chimera, where only the cytoplasmic domain of dToll was replaced for that of TLR4.
In contrast to ovalbumin, in the wild-type receptor the size of the ectodomain does not play the main role in terms of preventing spontaneous receptor activation. This is evidenced by N-terminal deletions of substantial parts of the TLR4 ectodomain, and the TLR4Δ542 deletion variant fails to render the receptor constitutively active despite the relatively small size of the remaining ectodomain. Therefore, we conclude that the specific structural features of the C-terminal part of the ectodomain prevent dimerization. This somewhat contrasts the findings of Weber et al. (58) for dToll, reporting that the juxtamembrane segment is critical for stable, high-affinity receptor-receptor interactions and thus activation or derepression of the downstream signaling pathway. This is similar to the results of Hu et al. (59), who proposed a conserved juxtamembrane cysteine-rich motif as a self-inhibitory module to dToll activation. The C-terminal parts of the TLR4 and dToll ectodomains contain a conserved stretch of cysteine residues that in dToll is involved in the formation of two intramolecular disulfide bonds, maintaining receptor rigidity, and is required for the controlled responsiveness to the ligand. Mutation of either one of these cysteines results in constitutive activity of the receptor (30, 59). However, we were unable to create the constitutively active variant of TLR4Δ542 by mutating the cysteine 583 to alanine. Furthermore, the TLR4Δ569 variant, despite retaining all four conserved juxtamembrane cysteines, was constitutively active. Therefore, it seems that in the case of TLR4, the mechanism of inhibition of receptor dimerization is not restricted specifically to the cysteine residues nor to the cysteine-rich domain alone and thus differs from that of dToll.
Thus, long-range interactions must govern the inhibitory activity of the TLR4 ectodomains to TM-TIR domain dimerization. Electrostatic repulsion because of the pattern of charged residues may be involved, although the net charge of this segment is only slightly negative. Nevertheless, we have shown previously that insertion of a peptide linker between the TLR4 ectodomain and the transmembrane domain decreases receptor activation, which could be due to the interaction of the ectodomain with the membrane surface (57). It has been discovered recently that the TLR4 ectodomain is cross-linked by nickel ions mediated by nonconserved histidine residues, contributing to the nickel allergy (60). In this case, the TLR4 ectodomain cross-linking with nickel ions is sufficient to provide the stabilizing interactions to overcome the inhibition exerted by the ectodomains and allow interaction between the transmembrane and cytosolic domains of TLR4.
The ability of the TM-TIR-TLR4 and TIR-TLR4 deletion variants to suppress signaling is in agreement with the results of Du et al. (61), demonstrating the effect on a mouse macrophage cell line. Although the TM-TIR-TLR4 variant is capable of interacting with the WT TLR4, as demonstrated by means of an in vivo β-lactamase complementation assay (52), this mechanism cannot be responsible for the observed inhibitory effect. Interaction of TM-TIR-TLR4 with the WT TLR4 at the cell surface would be expected to trigger or potentiate signaling rather than inhibit it, as a dimeric interaction surface at the plasma membrane would be created, capable of binding the adapter proteins, thus initiating the downstream signal propagation. However, the β-lactamase complementation assay was unable to distinguishing between the cell surface-expressed TLR4 and the cytosolic pool. Therefore, inhibition of signaling can be attributed to the engagement of the adapter molecules. This is supported by the inhibition of cell activation induced by overexpression of MyD88 with the TLR4 deletion variants. The TM-TIR-TLR4 and TIR-TLR4 variants thus sequester adapter proteins.
Constitutive TLR4 induced-cell activation is 100–500 times more potent than the activation exerted by constitutive dimeric versions of other TLRs (54). The role of the ectodomain of TLR4 in preventing dimerization of the TIR domains is therefore essential in maintaining the receptor in an inactive state prior to the recognition of the ligand. Activation of TLR4 can therefore be viewed as a release from inhibition rather than ligand-induced dimerization. A similar concept has previously been proposed by Brunn et al. (62), suggesting that under quiescent conditions, TLR4 is kept in an inactive state by the intact extracellular matrix, whereas the release of proteases, as it occurs in tissue injury, degrades the extracellular matrix, simultaneously relieving the constraint on TLR4 function as well as generating endogenous ligands, thus allowing them to stimulate the receptor. We have shown that truncation of the TLR4 ectodomain from its N terminus ultimately leads to constitutively active receptor variants. It is therefore tempting to speculate that activation of TLR4 could also be achieved directly by cleavage of the TLR4 ectodomain by an as yet unidentified protease(s) released in the inflammatory milieu or by an infecting microorganism. The proteolytic cleavage of the ectodomains of TLR7 and TLR9 has already been demonstrated to play a role in receptor activation (63, 64). Based on the reports of the mediation of the proinflammatory effects of some proteases through TLR4 (65–67), the role of proteolytic cleavage of the TLR4 ectodomain for receptor activation remains to be thoroughly examined. We could, however, not detect cell activation of TLR4-transfected HEK293 cells by elastase, which has been reported previously (67).
This work was supported by grants from the Slovenian Research Agency.
J. Lonzarić and R. Jerala, unpublished results.
- TLR
- Toll-like receptor
- IL-1R
- interleukin-1 receptor
- TIR
- Toll/IL-1R
- MyD88
- myeloid differentiation primary response gene (88)
- TRIF
- Toll/IL-1R domain-containing adapter-inducing interferon β
- MD-2
- myeloid differentiation protein 2
- CD14
- cluster of differentiation 14
- OVA
- ovalbumin
- GYRB24
- 24 kDa N-terminal fragment of Escherichia coli gyrase B
- RLA
- relative luciferase activity.
REFERENCES
- 1. Takeda K., Akira S. (2005) Int. Immunol. 17, 1–14 [DOI] [PubMed] [Google Scholar]
- 2. Gay N. J., Gangloff M. (2008) Handb. Exp. Pharmacol. 181–200 [DOI] [PubMed] [Google Scholar]
- 3. Iwasaki A., Medzhitov R. (2004) Nat. Immunol. 5, 987–995 [DOI] [PubMed] [Google Scholar]
- 4. Hoebe K., Janssen E., Beutler B. (2004) Nat. Immunol. 5, 971–974 [DOI] [PubMed] [Google Scholar]
- 5. Akashi-Takamura S., Miyake K. (2006) J. Infect. Chemother. 12, 233–240 [DOI] [PubMed] [Google Scholar]
- 6. Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 7. Amit I., Garber M., Chevrier N., Leite A. P., Donner Y., Eisenhaure T., Guttman M., Grenier J. K., Li W., Zuk O., Schubert L. A., Birditt B., Shay T., Goren A., Zhang X., Smith Z., Deering R., McDonald R. C., Cabili M., Bernstein B. E., Rinn J. L., Meissner A., Root D. E., Hacohen N., Regev A. (2009) Science 326, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taylor K. R., Trowbridge J. M., Rudisill J. A., Termeer C. C., Simon J. C., Gallo R. L. (2004) J. Biol. Chem. 279, 17079–17084 [DOI] [PubMed] [Google Scholar]
- 9. Termeer C., Benedix F., Sleeman J., Fieber C., Voith U., Ahrens T., Miyake K., Freudenberg M., Galanos C., Simon J. C. (2002) J. Exp. Med. 195, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smiley S. T., King J. A., Hancock W. W. (2001) J. Immunol. 167, 2887–2894 [DOI] [PubMed] [Google Scholar]
- 11. Okamura Y., Watari M., Jerud E. S., Young D. W., Ishizaka S. T., Rose J., Chow J. C., Strauss J. F., 3rd. (2001) J. Biol. Chem. 276, 10229–10233 [DOI] [PubMed] [Google Scholar]
- 12. Biragyn A., Ruffini P. A., Leifer C. A., Klyushnenkova E., Shakhov A., Chertov O., Shirakawa A. K., Farber J. M., Segal D. M., Oppenheim J. J., Kwak L. W. (2002) Science 298, 1025–1029 [DOI] [PubMed] [Google Scholar]
- 13. Park J. S., Gamboni-Robertson F., He Q., Svetkauskaite D., Kim J. Y., Strassheim D., Sohn J. W., Yamada S., Maruyama I., Banerjee A., Ishizaka A., Abraham E. (2006) Am. J. Physiol. Cell Physiol. 290, C917–924 [DOI] [PubMed] [Google Scholar]
- 14. Miller Y. I., Viriyakosol S., Binder C. J., Feramisco J. R., Kirkland T. N., Witztum J. L. (2003) J. Biol. Chem. 278, 1561–1568 [DOI] [PubMed] [Google Scholar]
- 15. Kim H. M., Park B. S., Kim J. I., Kim S. E., Lee J., Oh S. C., Enkhbayar P., Matsushima N., Lee H., Yoo O. J., Lee J. O. (2007) Cell 130, 906–917 [DOI] [PubMed] [Google Scholar]
- 16. Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., Lee J. O. (2009) Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
- 17. Medzhitov R., Preston-Hurlburt P., Janeway C. A., Jr. (1997) Nature 388, 394–397 [DOI] [PubMed] [Google Scholar]
- 18. Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C. A., Jr. (1998) Mol. Cell 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 19. Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., Akira S. (2002) J. Immunol. 169, 6668–6672 [DOI] [PubMed] [Google Scholar]
- 20. Shimazu R., Akashi S., Ogata H., Nagai Y., Fukudome K., Miyake K., Kimoto M. (1999) J. Exp. Med. 189, 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fujimoto T., Yamazaki S., Eto-Kimura A., Takeshige K., Muta T. (2004) J. Biol. Chem. 279, 47431–47437 [DOI] [PubMed] [Google Scholar]
- 22. Viriyakosol S., Tobias P. S., Kitchens R. L., Kirkland T. N. (2001) J. Biol. Chem. 276, 38044–38051 [DOI] [PubMed] [Google Scholar]
- 23. Nagai Y., Akashi S., Nagafuku M., Ogata M., Iwakura Y., Akira S., Kitamura T., Kosugi A., Kimoto M., Miyake K. (2002) Nat. Immunol. 3, 667–672 [DOI] [PubMed] [Google Scholar]
- 24. Lee J. D., Kato K., Tobias P. S., Kirkland T. N., Ulevitch R. J. (1992) J. Exp. Med. 175, 1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright S. D., Ramos R. A., Tobias P. S., Ulevitch R. J., Mathison J. C. (1990) Science 249, 1431–1433 [DOI] [PubMed] [Google Scholar]
- 26. Kim J. I., Lee C. J., Jin M. S., Lee C. H., Paik S. G., Lee H., Lee J. O. (2005) J. Biol. Chem. 280, 11347–11351 [DOI] [PubMed] [Google Scholar]
- 27. Haziot A., Chen S., Ferrero E., Low M. G., Silber R., Goyert S. M. (1988) J. Immunol. 141, 547–552 [PubMed] [Google Scholar]
- 28. Prohinar P., Re F., Widstrom R., Zhang D., Teghanemt A., Weiss J. P., Gioannini T. L. (2007) J. Biol. Chem. 282, 1010–1017 [DOI] [PubMed] [Google Scholar]
- 29. Kawai T., Akira S. (2006) Cell Death Differ. 13, 816–825 [DOI] [PubMed] [Google Scholar]
- 30. Schneider D. S., Hudson K. L., Lin T. Y., Anderson K. V. (1991) Genes Dev. 5, 797–807 [DOI] [PubMed] [Google Scholar]
- 31. Núñez Miguel R., Wong J., Westoll J. F., Brooks H. J., O'Neill L. A., Gay N. J., Bryant C. E., Monie T. P. (2007) PLoS ONE 2, e788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mullen G. E., Kennedy M. N., Visintin A., Mazzoni A., Leifer C. A., Davies D. R., Segal D. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3919–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Visintin A., Mazzoni A., Spitzer J. A., Segal D. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12156–12161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 35. Gilbert E. J., Maxwell A. (1994) Mol. Microbiol. 12, 365–373 [DOI] [PubMed] [Google Scholar]
- 36. Farrar M. A., Olson S. H., Perlmutter R. M. (2000) Methods Enzymol. 327, 421–429 [DOI] [PubMed] [Google Scholar]
- 37. Barton G. M., Kagan J. C., Medzhitov R. (2006) Nat. Immunol. 7, 49–56 [DOI] [PubMed] [Google Scholar]
- 38. Veerman C., de Schiffart G., Sagis L. M., van der Linden E. (2003) Int. J. Biol. Macromol. 33, 121–127 [DOI] [PubMed] [Google Scholar]
- 39. Nisbet A. D., Saundry R. H., Moir A. J., Fothergill L. A., Fothergill J. E. (1981) Eur. J. Biochem. 115, 335–345 [DOI] [PubMed] [Google Scholar]
- 40. Kawasaki K., Gomi K., Kawai Y., Shiozaki M., Nishijima M. (2003) J. Endotoxin Res. 9, 301–307 [DOI] [PubMed] [Google Scholar]
- 41. Gomi K., Kawasaki K., Kawai Y., Shiozaki M., Nishijima M. (2002) J. Immunol. 168, 2939–2943 [DOI] [PubMed] [Google Scholar]
- 42. Debierre-Grockiego F., Campos M. A., Azzouz N., Schmidt J., Bieker U., Resende M. G., Mansur D. S., Weingart R., Schmidt R. R., Golenbock D. T., Gazzinelli R. T., Schwarz R. T. (2007) J. Immunol. 179, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 43. Haynes L. M., Moore D. D., Kurt-Jones E. A., Finberg R. W., Anderson L. J., Tripp R. A. (2001) J. Virol. 75, 10730–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kurt-Jones E. A., Popova L., Kwinn L., Haynes L. M., Jones L. P., Tripp R. A., Walsh E. E., Freeman M. W., Golenbock D. T., Anderson L. J., Finberg R. W. (2000) Nat. Immunol. 1, 398–401 [DOI] [PubMed] [Google Scholar]
- 45. Rassa J. C., Meyers J. L., Zhang Y., Kudaravalli R., Ross S. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2281–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ashkar A. A., Mossman K. L., Coombes B. K., Gyles C. L., Mackenzie R. (2008) PLoS Pathog. 4, e1000233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mossman K. L., Mian M. F., Lauzon N. M., Gyles C. L., Lichty B., Mackenzie R., Gill N., Ashkar A. A. (2008) J. Immunol. 181, 6702–6706 [DOI] [PubMed] [Google Scholar]
- 48. Sasu S., LaVerda D., Qureshi N., Golenbock D. T., Beasley D. (2001) Circ. Res. 89, 244–250 [DOI] [PubMed] [Google Scholar]
- 49. Bulut Y., Shimada K., Wong M. H., Chen S., Gray P., Alsabeh R., Doherty T. M., Crother T. R., Arditi M. (2009) Infect. Immun. 77, 2683–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trompette A., Divanovic S., Visintin A., Blanchard C., Hegde R. S., Madan R., Thorne P. S., Wills-Karp M., Gioannini T. L., Weiss J. P., Karp C. L. (2009) Nature 457, 585–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang H., Tay P. N., Cao W., Li W., Lu J. (2002) FEBS Lett. 532, 171–176 [DOI] [PubMed] [Google Scholar]
- 52. Lee H. K., Dunzendorfer S., Tobias P. S. (2004) J. Biol. Chem. 279, 10564–10574 [DOI] [PubMed] [Google Scholar]
- 53. Nishiya T., DeFranco A. L. (2004) J. Biol. Chem. 279, 19008–19017 [DOI] [PubMed] [Google Scholar]
- 54. Hasan U. A., Dollet S., Vlach J. (2004) Biochem. Biophys. Res. Commun. 321, 124–131 [DOI] [PubMed] [Google Scholar]
- 55. Koskinen R., Lamminmäki U., Tregaskes C. A., Salomonsen J., Young J. R., Vainio O. (1999) J. Immunol. 162, 4115–4121 [PubMed] [Google Scholar]
- 56. Wu H., Kwong P. D., Hendrickson W. A. (1997) Nature 387, 527–530 [DOI] [PubMed] [Google Scholar]
- 57. Treeby M., Vasl J., Ota P., Friedrich J., Jerala R. (2009) Biochem. Biophys. Res. Commun. 381, 65–69 [DOI] [PubMed] [Google Scholar]
- 58. Weber A. N., Moncrieffe M. C., Gangloff M., Imler J. L., Gay N. J. (2005) J. Biol. Chem. 280, 22793–22799 [DOI] [PubMed] [Google Scholar]
- 59. Hu X., Yagi Y., Tanji T., Zhou S., Ip Y. T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9369–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schmidt M., Raghavan B., Müller V., Vogl T., Fejer G., Tchaptchet S., Keck S., Kalis C., Nielsen P. J., Galanos C., Roth J., Skerra A., Martin S. F., Freudenberg M. A., Goebeler M. (2010) Nat. Immunol. 11, 814–819 [DOI] [PubMed] [Google Scholar]
- 61. Du X., Poltorak A., Silva M., Beutler B. (1999) Blood Cells Mol. Dis. 25, 328–338 [DOI] [PubMed] [Google Scholar]
- 62. Brunn G. J., Bungum M. K., Johnson G. B., Platt J. L. (2005) FASEB J. 19, 872–874 [DOI] [PubMed] [Google Scholar]
- 63. Park B., Brinkmann M. M., Spooner E., Lee C. C., Kim Y. M., Ploegh H. L. (2008) Nat. Immunol. 9, 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sepulveda F. E., Maschalidi S., Colisson R., Heslop L., Ghirelli C., Sakka E., Lennon-Duménil A. M., Amigorena S., Cabanie L., Manoury B. (2009) Immunity 31, 737–748 [DOI] [PubMed] [Google Scholar]
- 65. Hietaranta A., Mustonen H., Puolakkainen P., Haapiainen R., Kemppainen E. (2004) Biochem. Biophys. Res. Commun. 323, 192–196 [DOI] [PubMed] [Google Scholar]
- 66. Walsh D. E., Greene C. M., Carroll T. P., Taggart C. C., Gallagher P. M., O'Neill S. J., McElvaney N. G. (2001) J. Biol. Chem. 276, 35494–35499 [DOI] [PubMed] [Google Scholar]
- 67. Devaney J. M., Greene C. M., Taggart C. C., Carroll T. P., O'Neill S. J., McElvaney N. G. (2003) FEBS Lett. 544, 129–132 [DOI] [PubMed] [Google Scholar]