Abstract
Mouse and human prothrombin (ProT) active site specifically labeled with d-Phe-Pro-Arg-CH2Cl (FPR-ProT) inhibited tissue factor-initiated thrombin generation in platelet-rich and platelet-poor mouse and human plasmas. FPR-prethrombin 1 (Pre 1), fragment 1 (F1), fragment 1.2 (F1.2), and FPR-thrombin produced no significant inhibition, demonstrating the requirement for all three ProT domains. Kinetics of inhibition of ProT activation by the inactive ProTS195A mutant were compatible with competitive inhibition as an alternate nonproductive substrate, although FPR-ProT deviated from this mechanism, implicating a more complex process. FPR-ProT exhibited ∼10-fold more potent anticoagulant activity compared with ProTS195A as a result of conformational changes in the ProT catalytic domain that induce a more proteinase-like conformation upon FPR labeling. Unlike ProT and ProTS195A, the pathway of FPR-ProT cleavage by prothrombinase was redirected from meizothrombin toward formation of the FPR-prethrombin 2 (Pre 2)·F1.2 inhibitory intermediate. Localization of ProT labeled with Alexa Fluor® 660 tethered through FPR-CH2Cl ([AF660]FPR-ProT) during laser-induced thrombus formation in vivo in murine arterioles was examined in real time wide-field and confocal fluorescence microscopy. [AF660]FPR-ProT bound rapidly to the vessel wall at the site of injury, preceding platelet accumulation, and subsequently to the thrombus proximal, but not distal, to the vessel wall. [AF660]FPR-ProT inhibited thrombus growth, whereas [AF660]FPR-Pre 1, lacking the F1 membrane-binding domain did not bind or inhibit. Labeled F1.2 localized similarly to [AF660]FPR-ProT, indicating binding to phosphatidylserine-rich membranes, but did not inhibit thrombosis. The studies provide new insight into the mechanism of ProT activation in vivo and in vitro, and the properties of a unique exosite-directed prothrombinase inhibitor.
Keywords: Coagulation Factors, Enzyme Kinetics, Enzyme Mechanisms, Fluorescence, Protein-Protein Interactions, Proteolytic Enzymes, Prothrombin, Serine Protease, Thrombosis
Introduction
The final serine proteinase zymogen activation step of the blood coagulation pathway is the proteolytic activation of prothrombin (ProT)4 to thrombin by prothrombinase. Prothrombinase consists of the proteinase, factor Xa (FXa), bound to its protein cofactor, factor Va (FVa), assembled in the presence of calcium on phospholipid membranes containing phosphatidylserine (PS) (1, 2). The lipid membrane provides a nidus for localization and assembly of the complex, which is accompanied by a ∼300,000-fold increased catalytic efficiency of ProT activation. The rate increase is due to a 100-fold decrease in apparent Km associated with membrane binding of FXa and ProT and a 3000-fold increase in apparent kcat that is a unique function of FVa (1–3). Specific recognition of ProT as a substrate of prothrombinase is mediated by exosite interactions with the FXa·FVa·membrane complex (1, 4–9). ProT substrate recognition by the FXa·FVa complex is dependent on the lower affinity precursor form of exosite I (proexosite I) on ProT (10–17).
Physiological membranes of several blood cell types including, platelets (18–22), endothelial cells (23–25), and mononuclear cells (21, 26) support ProT activation in vitro. Platelets have long been thought to be the primary cellular surface for prothrombinase assembly. Activation of platelets results in redistribution of membrane phospholipids with increased exposure of PS on the outer membrane surface, crucial for assembly of coagulation factor activating complexes (27). Assembly of prothrombinase on activated platelets results in an enhancement in the rate of thrombin formation similar to that obtained with optimal phospholipid vesicles (21, 28). Although the primary role of the endothelial cell layer lining blood vessels is to prevent clot formation and maintain normal flow through anticoagulant mechanisms, stimulation or injury of the endothelium can also provide a thrombogenic surface, which binds FXa (29, 30) and FVa (23), assembling prothrombinase (24, 31–33). The cellular location of ProT activation during thrombosis in vivo has not been established, but it has been widely anticipated to be activated platelet surfaces.
A model employing laser-induced focal damage to the vessel wall of cremaster arterioles in living mice has revealed several important aspects of the mechanism of thrombosis (34–36). In this model, both fibrin generation and initial platelet activation are thrombin-dependent and reflect tissue factor (TF)-initiated activation of the extrinsic coagulation pathway (37). This is in contrast to other mouse models of thrombosis, for example, the severe, ferric chloride-induced vessel wall injury, where the endothelium is denuded and initial platelet activation is induced by interaction of glycoprotein VI with exposed vessel wall collagen (37). Both vessel wall TF and microparticle borne TF participate in thrombus formation in the laser-injury model (38). Platelets accumulate at the site of injury; the platelet thrombus reaches maximal size between 90 and 120 s and then decreases. Fibrin accumulation begins essentially simultaneously with platelet deposition and is seen first at the vessel wall-platelet thrombus interface (35, 36, 39). Maximal fibrin deposition is achieved at about 100 s. Confocal microscopy studies confirm that fibrin accumulates primarily at the vessel wall-platelet interface and only partially colocalizes with the platelet thrombus (40). In Par4−/− mice, small platelet thrombi form rapidly but fail to grow and the platelets within this thrombus are activated very slowly. However, fibrin generation and deposition is normal in the Par4−/− mice (40). These results challenge the view that the activated platelet membrane is the primary site of prothrombinase assembly in vivo and suggest an important role for activated endothelium or microparticles. Recent studies support a role of the endothelium in thrombus formation by demonstrating that endothelial cells are rapidly activated by localized laser injury before platelet accumulation, and provide a procoagulant surface for prothrombinase-generated thrombin (41).
In the present studies, examination of the inhibition of thrombin generation by a novel active site-blocked ProT derivative in vitro in plasma and in model systems of purified proteins provided new insight into the mechanism of ProT activation and the exosite-mediated mechanism of inhibition of coagulation by active site-labeled ProT. Understanding these mechanisms allowed us to interpret in vivo wide-field and confocal microscopy experiments with active site fluorescently labeled ProT to determine the dynamics of formation of sites of ProT localization during thrombosis in the laser injury murine model.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6J mice (4–6 weeks old; 13–17 g) were obtained from the Jackson Laboratory. The Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee approved animal experiments.
Antibodies
Rat anti-mouse CD41 antibody was from BD Biosciences. Fab fragments from CD41 antibody were generated using an ImmunoPure Fab Preparation kit from Pierce Biotechnology. The anti-CD41 Fab fragments were conjugated with Alexa Fluor® 488-tetrafluorophenyl ester (Invitrogen).
Coagulation Proteins
Human ProT, FXa, and thrombin were purified, active site-titrated, and characterized as described previously (42–44). Human factor V (FV) and FVa were purified according the method of Dahlbäck (45) with modifications (46). Human prethrombin 1 (Pre 1), prethrombin 2 (Pre 2), fragment 1 (F1), and fragment 1.2 (F1.2) were purified from products of ProT cleavage as described (13, 43). Murine ProT (Hematologic Technologies) and human ProT and Pre 1 were inactivated with Nα-[(acetylthio)acetyl]-FPR-CH2Cl and labeled with Alexa Fluor 660 C2-maleimide (Invitrogen) using staphylocoagulase (47). ProT, Pre 1, and Pre 2 inactivated with FPR-CH2Cl were prepared similarly. Human F1.2 (Hematologic Technologies) was labeled with Alexa Fluor 660-tetrafluorophenyl ester according to the manufacturer's instructions (Invitrogen). CaCl2 (5 mm) was included in the reaction mixture to prevent labeling of the NH2 terminus necessary for maintaining the proper conformation of the Gla domain.
Protein Concentrations
Protein concentrations were determined by absorbance at 280 nm using the following absorption coefficients ((mg/ml)−1 cm−1) and molecular weights (48–50): ProT, ProTS195A, ProTR271Q, PtoTR320Q, and their FPR derivatives, 1.47, 71,600; Pre 1 and FPR-Pre 1, 1.78, 49,900; Pre 2 and FPR-Pre 2, 1.73, 37,000; and thrombin and FPR-thrombin, 1.74, 36,600. [5F]Hir-(54–65)(SO3−) was prepared as described (51). Thrombin and FXa concentrations were determined routinely using the chromogenic substrates d-Phe-l-pipecolyl-l-Arg-p-nitroanilide and Z-d-Arg-Gly-Arg-p-nitroanilide, respectively. Concentrations of fluorescently labeled proteins were determined by bicinchoninic acid assay (Pierce). Concentrations of ProT species in vivo in mice were calculated from the ratio of the dose in micrograms/g of mouse weight, divided by a blood volume to weight ratio of 0.078 ml/g (52) and expressed in a molar concentration to allow, for example, FPR-ProT, FPR-Pre 1, and FPR-thrombin to be compared on the same molar scale.
Thrombin Generation in Plasma
Human platelet-poor, mouse platelet-rich, platelet-poor, and human platelet-rich plasmas were prepared as described (53, 54). Human ProT-deficient plasma was purchased (George King Biomedical). Thrombin generation in plasma was monitored by calibrated automated thrombography (CAT), where the fluorescence enhancement accompanying hydrolysis of the low affinity fluorogenic substrate, Z-Gly-Gly-Arg-7-amido-4-methylcoumarin (Bachem), by thrombin was followed with time in a Fluoroskan Ascent® plate reader (Thermo Labsystems) (53, 55). The fluorescence changes in plasma with time were transformed into thrombin concentration by the use of a calibrated thrombin solution measured in parallel. Thrombin generation curves were analyzed to obtain the phenomenological parameters (lag time, maximum thrombin concentration, time to maximum thrombin concentration, and the area under the curve, the endogenous thrombin potential) using ThrombinoscopeTM software (Thrombinoscope BV). Thrombin generation was initiated by addition of a mixture of calcium and fluorogenic substrate in the presence of either human recombinant TF (Dade Innovin®) or FXa (Enzyme Research Laboratories). In experiments with platelet-poor plasma, phospholipid vesicles were added. Dioleoyl-phosphatidylcholine (PC), dioleoyl-phosphatidylserine (PS), and dioleoyl-phosphatidylethanolamine (PE) were from Avanti Polar Lipids. Vesicles (20% PS/60% PC/20% PE) were prepared as described (3, 56).
Prothrombin Activation Kinetics
Initial rates of ProT activation were determined as follows. Twenty μl of prewarmed 25 mm Hepes (pH 7.7 at 37 °C), 175 mm NaCl, 5 mg/ml of bovine serum albumin containing ProT was added to 40 μl of prewarmed buffer containing FVa, CaCl2, and phospholipid vesicles, and after 15 s, reactions were started by addition of 10 μl of FXa. When additional ProT derivatives were present, these were premixed with ProT to reach the desired final concentrations. Final concentrations were 5 mm CaCl2, 5 nm FVa, 1 μm phospholipid vesicles, 2 pm FXa, 5 mg/ml of bovine serum albumin, and ProT or its derivatives as indicated in the figure legends. Aliquots removed after 1 and 2 min of incubation at 37 °C were diluted into ice-cold 50 mm Tris (pH 7.9 at 25 °C), 175 mm NaCl, 20 mm EDTA, and 0.5 mg/ml of ovalbumin. Thrombin was quantified from the rate of d-Phe-l-pipecolyl-l-Arg-p-nitroanilide hydrolysis and rates of ProT activation were calculated (3). Apparent values of Vmax, Km, and Ki were obtained by non-linear least-squares fitting of the competitive inhibition mechanism to the rates of ProT activation as function of inhibitor concentration obtained at three ProT concentrations (50, 150, and 1500 nm). Estimates of error in kinetic and binding parameters represent the 95% confidence interval.
Fluorescence Titrations
Titrations of 20 nm [5F]Hir-(54–65)(SO3−) with ProT and its derivatives were performed with an SLM 8100 spectrofluorometer with excitation at 491 nm and emission at 520 nm (4 nm band pass), using acrylic cuvettes coated with polyethylene glycol 20,000, as described previously (12–14). Experiments were performed in 50 mm Hepes, 110 mm NaCl, 5 mm CaCl2, 1 mg/ml of polyethylene glycol 8000, pH 7.4, at 25 °C. The results expressed as ΔF/Fo = (Fobs − Fo)/Fo) were analyzed by fitting the quadratic binding equation to obtain the dissociation constant (KD) and maximum fluorescence change (ΔFmax/Fo), with the stoichiometric factor fixed at 1. Competitive binding of F1.2 to [6F]FPR-thrombin and native Pre 2 or FPR-Pre 2 were performed by titrations of the decrease in fluorescein fluorescence (495 nm excitation, 520 nm emission, 4 nm band pass) of [6F]FPR-thrombin in the absence and presence of fixed concentrations of native Pre 2 (3.5 and 10 μm) or FPR-Pre 2 (10 μm). Titrations in the absence and presence of competitors were fit simultaneously by the cubic equation for tight competitive binding to obtain the KD for F1.2 binding to [6F]FPR-thrombin and the KD for F1.2 binding to either native Pre 2 or FPR-Pre 2, with stoichiometric factors fixed at 1, as described previously (51).
SDS-Gel Electrophoretic Analysis of ProT Activation
ProT activation reactions and quantitation were performed essentially as described in published procedures (9, 57). Activation reactions containing 5.2 μm ProT, ProTS195A, or FPR-ProT, 50 μm PS/PC/PE vesicles, 60 μm dansylarginine-N-(3-ethyl-1,5-pentanediyl)amide, 50 nm FVa, and 0.7 nm FXa were performed simultaneously at 37 °C, initiated by addition of ProT or ProT derivative. Reactions of ProTR271Q, ProTR320Q, and their FPR derivatives were done the same way except at 0.25 nm FXa. Pre 2 or FPR-Pre 2 cleavage by prothrombinase was performed with 2 μm Pre 2 or FPR-Pre 2 and 0.7 nm FXa. Reactions were sampled (40 μl) at different times and quenched by addition of 14 μl of 50 mm Hepes, 125 mm NaCl, 150 mm EDTA, 1 mg/ml of polyethylene glycol 8000, pH 8.2, containing FPR-CH2Cl to give a final concentration of 250 μm. NuPage LDS sample buffer (Invitrogen) supplemented with 125 mm dithiothreitol and 50 mm EDTA was then added, the samples were boiled for 2 min, centrifuged, and aliquots containing 4 μg of protein were loaded and developed on NuPage Novex 4–12% Bis-Tris gels in NuPage MES SDS Running Buffer as described by Invitrogen. Gels were simultaneously stained for identical duration with the Coomassie Brilliant Blue R-250 analog, Imperial protein stain (Pierce). Images were collected with an EDAS290 digital camera system (Kodak) and densitometry analysis was performed with ImageQuant TL software (GE Biosciences) using the rolling-ball method of integration and normalized to total protein staining in each lane. Fractional molar staining intensities of fProT = 1, fF1.2-A-chain = 0.59, fF1.2 = 0.47, fthrombin B-chain = 0.51, and fPre 2 = 0.43 were determined from the slopes of linear plots of density for purified proteins versus mole of protein loaded, and used to convert densities into relative molar concentrations.
Intravital Microscopy
Intravital video microscopy of the cremaster muscle microcirculation was done as described previously (35). Prior to surgery, mice were anesthetized with intraperitoneal ketamine (125 mg/kg, Abbott Laboratories), xylazine (12.5 mg/kg, Phoenix Pharmaceuticals), and atropine (0.25 mg/kg, American Pharmaceutical Partners). A tracheal tube was inserted and the mouse was maintained at 37 °C on a temperature-controlled rodent blanket. Nembutal (Abbott Laboratories) was administered through a cannulus placed in the jugular vein to maintain anesthesia. Following incision of the scrotum, the testicle and surrounding cremaster muscle were exteriorized onto an intravital microscopy tray. The cremaster preparation was superfused with temperature-controlled (36 °C) and aerated (95% N2, 5% CO2) bicarbonate-buffered saline during the entire experiment. Microvessel data were collected using an Olympus AX microscope with a ×60 0.9 NA water immersion objective. The intravital fluorescence microscopy system has been described previously in detail (58). Digital images were captured with a Cooke Sensicam CCD camera in 640 × 480 format.
Laser-induced Injury
To induce vessel wall injury, a Micropoint Laser System (Andor), focused through the microscope objective and parfocal with the focal plane, was aimed at the vessel wall (37). One or two pulses were typically required to induce injury of the vessel wall of arterioles of 30–50 μm. This laser-injury model spares the endothelium and results in a non-occlusive thrombus. Data were collected from multiple thrombi in a single mouse, with new thrombi formed upstream of earlier thrombi to prevent any contribution from previous thrombi generated in the animal under study. No distinguishing trends in thrombus size or composition were observed in sequential thrombi induced in a single mouse during an experiment. Image analysis was performed using Slidebook (Intelligent Imaging Innovations). Fluorescence data were acquired digitally at speeds up to 50 frames/s and analyzed as described previously (35). Wide-field fluorescence and bright-field images were captured with exposure times of 15 ms. Kinetics of thrombus formation were analyzed from the median fluorescence values determined over time in ∼10–40 thrombi (36, 37, 40, 58). Statistical analysis was performed using the Wilcoxon rank sum test for two independent samples.
RESULTS
ProT Activation in Vitro
Anticoagulant Activity of FPR-ProT
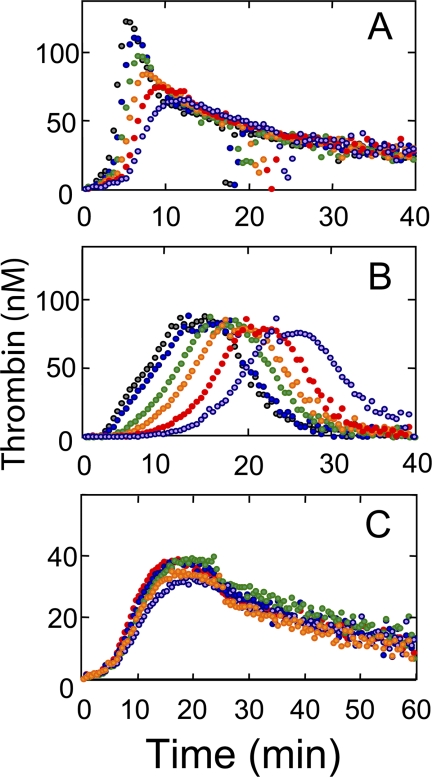
The effect of human FPR-ProT on thrombin generation in normal platelet-rich and platelet-poor mouse and human plasma was studied by CAT (53, 55). Representative thrombin generation curves are shown for TF/Ca2+-initiated coagulation of platelet-rich murine (Fig. 1A) and human plasma (Fig. 1B) as a function of FPR-ProT concentration. Phenomenological analysis of the curves yields 4 parameters: (a) the lag time, representing the time before onset of the thrombin burst, which correlates approximately to the clotting time; (b) the maximum concentration of thrombin reached, which is a function of the maximum rates of thrombin formation and its inhibition; (c) the time required to reach the thrombin maximum; and (d) the endogenous thrombin potential (the area under the curve), which represents the time integral of thrombin formed. Increasing concentrations of FPR-ProT up to the maximum used of 0.5 μm in platelet-rich murine plasma (Fig. 1A) and 1 μm in platelet-rich human plasma (Fig. 1B), equivalent to the plasma ProT concentrations in these experiments, progressively increased the lag time, the time to maximum thrombin, and to a lesser extent the maximum thrombin formed (Fig. 1B), whereas the endogenous thrombin potential was only marginally affected. Similar results were obtained when calcium ionophore was used to activate the platelets (not shown).
FIGURE 1.
Effect of FPR-ProT on thrombin generation in platelet-rich murine and human plasma. A, thrombin generation was determined by CAT at varying concentrations of FPR-ProT as described under “Experimental Procedures” in a total volume of 60 μl containing 20 μl of platelet-rich murine plasma. Concentrations of reactants were: 5 pm TF, 8 mm added CaCl2, 0.42 mm fluorogenic thrombin substrate, 40 μg/ml of corn trypsin inhibitor (to inhibit potential contact activation) and final concentrations of FPR-ProT (μm) at 0 (gray), 0.1 (blue), 0.2 (green), 0.3 (orange), 0.4 (red), and 0.5 (violet). B, thrombin generation in platelet-rich human plasma in a total volume of 125 μl containing 80 μl of plasma. Concentrations of reactants were: 1 pm TF, 16 mm added CaCl2, 0.34 mm fluorogenic substrate, 40 μg/ml of corn trypsin inhibitor, and FPR-ProT (μm) at 0, (gray), 0.2 (blue), 0.4 (green), 0.6 (orange), 0.8 (red), and 1.0 (violet). C, thrombin generation in platelet-rich mouse plasma as in A, but at 10-fold lower TF (0.5 pm) with 0.7 μm of the following ProT derivatives: none (red), F1 (violet), F1.2 (orange), FPR-Pre 1 (blue), or FPR-T (green).
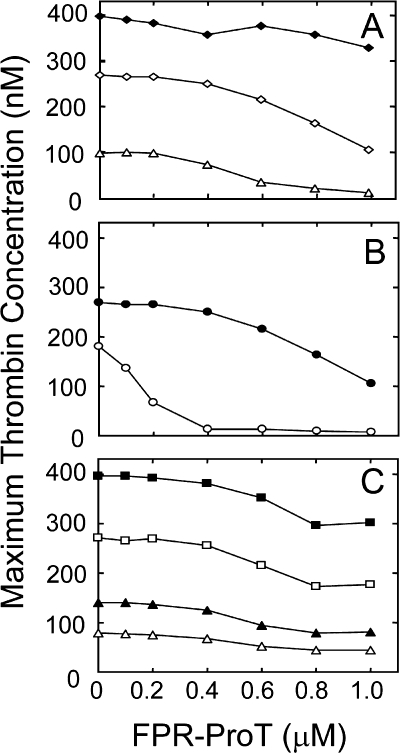
To investigate the roles of ProT domains in the mechanism of inhibition, similar experiments were performed in murine plasma with ProT activation species and domain fragments (FPR-Pre 1, F1, F1.2, or FPR-thrombin) at concentrations of 0.7 μm (equivalent to 140% of plasma ProT). To maximize inhibitory effects, these experiments were carried out at a 10-fold lower TF concentration (0.5 pm; Fig. 1C). With the exception of modest inhibition by F1, the ProT derivatives had no significant effect on thrombin generation. Reaction variables (TF, phospholipid, and plasma ProT concentration) were examined further in TF/Ca2+-initiated activation of human platelet-poor plasma in the presence of PS/PC/PE phospholipid vesicles (Fig. 2). Inhibition of thrombin generation by FPR-ProT in human platelet-poor plasma containing 30 μm phospholipid was dependent on TF concentration over the range of 1.8 to 40 pm, with the lowest concentration producing the largest increase in the lag time from 7 to 15 min at 1 μm FPR-ProT (supplemental Fig. S1). Likewise, the largest effect of FPR-ProT on the peak height of the thrombin generation curve was seen at the lowest TF concentration in platelet-poor human plasma, with a 50% reduction of the maximum thrombin concentration at 0.5 and 0.9 μm FPR-ProT initiated with 1.8 and 7 pm TF, respectively, whereas <10% inhibition was observed at 1 μm FPR-ProT at the highest TF concentration of 40 pm (Fig. 2A). At constant TF (7 pm), the inhibitory effect of FPR-ProT increased with decreasing phospholipid concentration, with a 5-fold decrease in the FPR-ProT concentration required for 50% inhibition at 2 μm phospholipid (200 nm) compared with that at 30 μm (900 nm) (Fig. 2B). Based on the relative changes in lag time (not shown) and peak height of thrombin generation (Fig. 2C), the effect of FPR-ProT at concentrations up to 1 μm was independent of the plasma ProT concentration. In this experiment (Fig. 2C), ProT-deficient plasma was reconstituted with different concentrations of purified ProT, 0.25–1.5 μm in the activation mixtures, corresponding to 25–150% of the plasma ProT present in the experiments. Control experiments using platelet-poor human plasma with FPR-Pre 1, F1, F1.2, and FPR-thrombin showed essentially no inhibition of thrombin generation (supplemental Fig. S2).
FIGURE 2.
Effect of FPR-ProT on thrombin generation in normal human plasma at varying TF, phospholipid, and plasma ProT levels. Thrombin generation in a total reaction volume of 125 μl containing 80 μl of normal human plasma initiated with varying TF concentrations (A) or with 7 pm TF (B and C) and the effect of increasing FPR-ProT concentration on the relative maximum thrombin concentration (%) determined as described under “Experimental Procedures.” Concentrations of reactants were: A, TF (pm) 1.8 (Δ), 7 (♢), or 40 (♦); 16 mm CaCl2, 30 μm phospholipid vesicles, 0.34 mm substrate, 40 μg/ml of corn trypsin inhibitor, and the indicated concentrations of FPR-ProT. B, 7 pm TF and 2 (○) or 30 (●) μm phospholipid vesicles with all other reactants as in A. C, 7 pm TF, ProT-deficient plasma reconstituted with purified human ProT representing 25 (Δ), 50 (▴), 100% (□), and 150% (■) of the concentration of ProT present in normal plasma (1.5 μm). All other reactants are as given in A.
The above results indicated that inhibition of thrombin generation was more pronounced at lower TF and phospholipid concentrations, conditions under which FXa is generated more slowly. Inhibition of thrombin generation by FPR-ProT potently prolonged the lag time and decreased the maximum thrombin generated when coagulation was initiated with low concentrations of FXa (0.7 nm) in the presence of 30 μm PS/PC/PE vesicles (supplemental Fig. S3). Under these conditions, F1 actually had a more pronounced inhibitory effect compared with that in TF/Ca2+-initiated coagulation. Taken together, the data suggest that competition of FPR-ProT for native ProT as a nonproductive substrate of the FXa·Va·membrane complex, and/or competition for membrane-binding sites at low phospholipid concentration, are important for the inhibitory effect of FPR-ProT. Competition for membrane-binding sites must, however, contribute to a minor extent, based on the small inhibitory effect of F1 on thrombin generation and almost no inhibition with F1.2 under the most sensitive experimental conditions. The results demonstrated that all domains of FPR-ProT were necessary for inhibition by FPR-ProT.
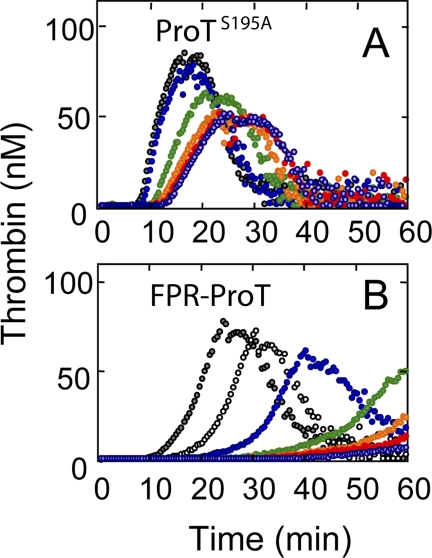
To determine whether competitive inhibition alone was responsible for the inhibition of thrombin generation, the inactive ProT mutant containing the catalytic Ser195 substituted with Ala (ProTS195A) was compared with FPR-ProT as an inhibitor of thrombin generation in platelet-rich human plasma (Fig. 3). At low TF (0.5 pm), ProTS195A did indeed inhibit thrombin generation (Fig. 3A) but the effect was much smaller than that observed with FPR-ProT (Fig. 3B). At 0.1 μm FPR-ProT, the time to maximum thrombin formation was shifted from 25.5 to 31.5 min, whereas 6–10-fold higher concentrations of ProTS195A were required to produce a comparable effect (Fig. 3). Similar results compatible with a ∼10-fold larger inhibition by FPR-ProT compared with ProTS195A were obtained in platelet-poor human plasma at 7 pm TF and 2 μm phospholipid vesicles (not shown). It is apparent that unique properties of FPR-ProT that distinguish it from native ProT and ProTS195A play an important role in its mechanism of inhibition.
FIGURE 3.
Effect of FPR-ProT and ProTS195A on thrombin generation in platelet-rich human plasma. The effect of varying concentrations of ProTS195A (A) and FPR-ProT (B) on TF-initiated thrombin generation in a total volume of 125 μl containing 80 μl of platelet-rich human plasma determined by CAT as described under “Experimental Procedures.” Concentrations of reactants were: 0.5 pm TF, 16 mm added CaCl2, 0.34 mm substrate, 40 μg/ml of corn trypsin inhibitor, and final concentrations: A, ProTS195A (μm) at 0 (gray), 0.2 (blue), 0.4 (green), 0.6 (orange), 0.8 (red), or 1.0 (violet). B, FPR-ProT (μm) at 0 (gray), 0.1 (not filled), 0.2 (blue), 0.4 (green), 0.6 (orange), 0.8 (red), or 1.0 (violet).
Prothrombinase Inhibition by FPR-ProT and ProTS195A
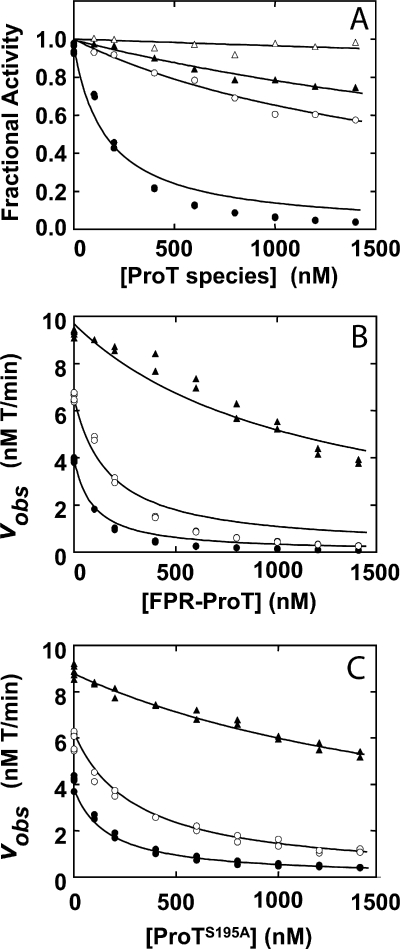
To investigate the mechanism of inhibition of ProT activation by FPR-ProT, initial rate kinetic studies of ProT activation by prothrombinase were performed as a function of substrate and inhibitor concentration. Fig. 4A shows the effect of increasing concentrations of FPR-Pre 1, F1, F1.2, and FPR-ProT on the rate of activation of 0.15 μm ProT by prothrombinase. Compared with the effects seen in plasma, F1 and F1.2 showed somewhat larger inhibition, whereas FPR-Pre 1 did not inhibit ProT activation, in accordance with the findings in plasma that all three domains of FPR-ProT were needed for effective inhibition of prothrombinase. Fitting the model for competitive inhibition to the data yielded apparent inhibition constants (Ki (μm)) of 0.61 ± 0.05 for F1; 1.2 ± 0.1 for F1.2; 9.0 ± 0.3 for FPR-Pre 1 and 0.057 ± 0.008 for FPR-ProT. Fig. 4B shows inhibition of prothrombinase as a function of FPR-ProT concentration (same range as in Fig. 4A) at ProT concentrations of 0.05, 0.15, and 1.5 μm, i.e. below, above, and very high above the apparent Km for ProT. The pattern of inhibition was broadly consistent with competitive binding and a simultaneous fit yielded apparent kinetic constants: Km 79 ± 10 nm for ProT, kcat 81 ± 3 s−1, and Ki of 57 ± 8 nm for FPR-ProT (Fig. 4B). These results, however, showed systematic deviations from a strictly competitive inhibition model. Similar deviations were observed for the kinetics of inhibition by FPR-ProT at 25 μm phospholipid, with apparent constants of Km 160 ± 20 and Ki 280 ± 60 nm, indicating a reduced affinity for inhibition at high phospholipid concentration (not shown). To assess the significance of these deviations, inhibition of ProT activation by ProTS195A was studied similarly over the same range of concentration (Fig. 4C). Analysis of the results showed a much better fit of the competitive inhibition mechanism, with apparent Km and Ki of 71 ± 6 and 98 ± 10 nm, respectively. Although inhibition of single substrate enzymes by nonproductive alternate substrates is predicted to be competitive, the mechanism of inhibition of ProT activation by FPR-ProT is evidently more complex than that of ProTS195A.
FIGURE 4.
Effect of FPR-ProT, ProTS195A, and ProT derivatives on the rate of ProT activation by prothrombinase. Initial rates of ProT activation (vobs) were determined at 37 °C as a function of ProT and FPR-ProT concentrations as described under “Experimental Procedures.” A, changes in initial velocity of 150 nm native ProT activation (Fractional Activity) as a function of total concentrations of FPR-ProT (●), F1 (○), F1.2 (▴), and FPR-Pre 1 (Δ) ([ProT species]). B, initial velocities as a function of total FPR-ProT concentration ([FPR-ProT]) at native ProT concentrations of 50 (●), 150 (○), and 1500 (▴) nm. C, initial velocities as a function of total ProTS195A ([ProTS195A]) concentration as in B. Concentrations of reactants were: 25 mm Hepes, pH 7.5, at 37 °C, 175 mm NaCl, 0.5 mg/ml of bovine serum albumin, 2 pm FXa, 5 nm FVa, and 1 μm phospholipid vesicles. Solid lines represent non-linear least squares fits of the data by the competitive inhibition mechanism with values of apparent Km and Ki for ProT fragments, FPR-ProT, and ProTS195A given under “Results.”
Characterization of Proexosite I on FPR-ProT and the Properties of FPR-ProT as a Prothrombinase Substrate
Previous studies implicated a conformational change in ProT accompanying FPR-CH2Cl labeling of the zymogen catalytic site, reflecting a partial transition of the catalytic domain toward a more proteinase-like state, and resultant enhanced cleavage at Arg271 (57). To investigate the properties of FPR-ProT further, the effect of active site labeling on the properties of proexosite I on zymogens ProT, Pre 1, and Pre 2 was examined in equilibrium binding studies employing fluorescein-labeled hirudin–(54–65) ([5F]Hir-(54–65)(SO3−)) as a previously characterized (pro)exosite I-specific probe (not shown) (12–14, 51). Titrations of [5F]Hir-(54–65)(SO3−) with native ProT gave the typical KD of 2700 ± 600 nm and maximum fluorescence change of −17 ± 1%, compared with full expression of exosite I on FPR-thrombin, to which the peptide bound with a 96-fold higher affinity and a 2.0-fold larger fluorescence decrease (12). ProTS195A bound the peptide similarly to native ProT, with a KD of 1800 ± 300 nm and a fluorescence change of −20 ± 1%. By contrast to native ProT and ProTS195A, FPR-ProT bound [5F]Hir-(54–65)(SO3−) with a KD of 600 ± 100 nm, representing a 4.5-fold higher affinity, and a 2.4-fold larger fluorescence decrease (−40 ± 2%) than observed with ProT. Native Pre 1 and Pre 2 bound [5F]Hir-(54–65)(SO3−) with the same dissociation constants of 520 ± 90 and 530 ± 40 nm, and fluorescence changes of −22 ± 1 and −26 ± 1%, compatible with previous studies (13, 14). FPR-Pre 1 (KD 800 ± 60 nm) and FPR-Pre 2 (KD 1000 ± 70 nm) had affinities slightly lower than those of the native proteins, but showed 2.0- and 1.7-fold larger fluorescence decreases of −44 ± 1 and −43 ± 1%, respectively. The results demonstrated that active-site labeling changes the conformation of the zymogen catalytic domain, enhancing the affinity of ProT for binding a proexosite I ligand and altering the environment of the probe-labeled peptide in complexes with all of the zymogen forms. By contrast, ProTS195A did not display these unique properties.
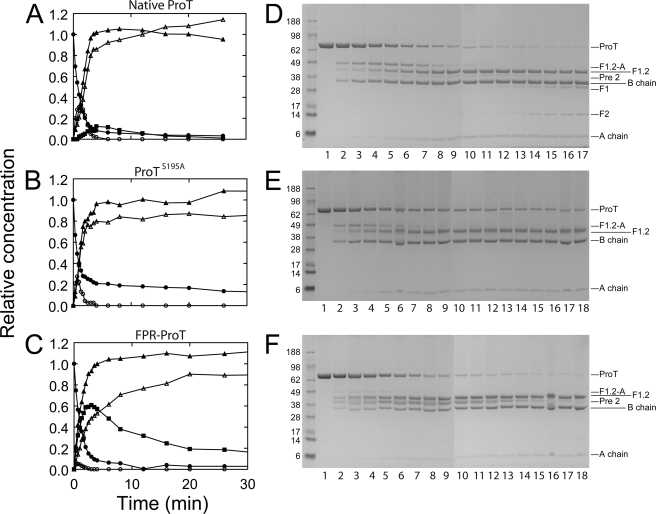
The altered conformation of the catalytic domain in FPR-ProT was associated with functional changes in FPR-ProT as a prothrombinase substrate. Fig. 5 shows results of SDS-gel electrophoretic analysis of full time courses of native ProT, ProTS195A, and FPR-ProT cleavage by prothrombinase. The two alternate reaction pathways of ProT activation are initial cleavage at Arg320, producing meizothrombin (MZT) as the intermediate, or the alternative first cleavage at Arg271 producing the Pre 2·F1.2 noncovalent complex as the intermediate, followed by cleavage of the complementary sites in the intermediates to form the products, thrombin and F1.2. Native ProT cleavage was characterized by the initial, transient formation of the F1.2-A-chain, diagnostic of MZT formation, and subsequent cleavage to the products, the thrombin B-chain and F1.2, with low levels of Pre 2 generated (Fig. 5A). The high concentration of active thrombin produced resulted in the slow appearance of F1 and F2 derived from F1.2 following complete conversion of ProT to thrombin, despite the presence of 60 μm dansylarginine-N-(3-ethyl-1,5-pentanediyl)amide. Consequently, the band migrating at the position of Pre 2 could be Pre 2′ produced by thrombin cleavage of ProT at Arg284. ProTS195A showed a time course of transient MZT formation and product formation similar to that of native ProT (Fig. 5B). The results showed no detectable Pre 2, F1, or F2, but were slightly different in showing the presence of ∼14% residual ProTS195A that was not cleaved. By contrast, for FPR-ProT, FPR-MZT formation was greatly reduced to ∼16% of the maximum seen with native ProT, and FPR-Pre 2 and F1.2 were the major products. FPR-Pre 2 was initially generated ∼2-fold faster than thrombin, accumulated to a maximum of 60%, and was converted slowly to FPR-thrombin (Fig. 5C). The change in the cleavage pathway with active site labeling was consistent with the previous observation that a ProT mutant that could only be cleaved at Arg271 to form Pre 2·F1.2 was cleaved ∼12-fold faster when the active site was FPR-labeled (57). To examine the effect of FPR labeling on the individual cleavages at Arg271 and Arg320, similar experiments were performed with ProTR320Q, ProTR271Q, and their FPR derivatives in which one the cleavages sites was blocked. The results (not shown) indicated that FPR labeling accelerated cleavage at Arg271 in ProTR320Q ∼11-fold and inhibited cleavage at Arg320 in ProTR271Q ∼2-fold.
FIGURE 5.
Time courses of ProT, ProTS195A, and FPR-ProT cleavage by prothrombinase. Aliquots of reaction mixtures containing ProT derivatives (5.2 μm) incubated with 0.7 nm prothrombinase (0.7 nm FXa, 50 nm FVa, and 50 μm PS/PC/PE vesicles) were quenched at times (min) 0, 0.33, 0.67, 1, 1.33, 1.67, 2, 2.5, 3, 3.5, 4, 6, 8, 12, 16, 20, 26, and 32. Relative concentrations of native ProT activation (A) and ProTS195A (B) or FPR-ProT (C) cleavage products. ProT, ProTS195A, or FPR-ProT (●), meizothrombin F1.2-A (○), F1.2 (▴), thrombin B-chain (Δ), and Pre 2 (■). D–F, SDS gels (4–12%), corresponding to the reactions for A (native ProT), B (ProTS195A), and C (FPR-ProT). Lanes 1–18 are samples taken at the above reaction times except for native ProT, where the 32-min sample was deleted because of a gel anomaly. Bands corresponding to ProT, ProTS195A, or FPR-ProT (ProT), F1.2-A, F1.2, Pre 2, thrombin B-chain (B chain), F1, F2, and thrombin A-chain (A chain) are indicated on the right. Positions of molecular weight markers are indicated on the left with the molecular weights in thousands. Reactions were performed and analyzed as described under “Experimental Procedures.”
ProT Localization in Vivo
ProT Binding and Platelet Accumulation during Thrombus Formation
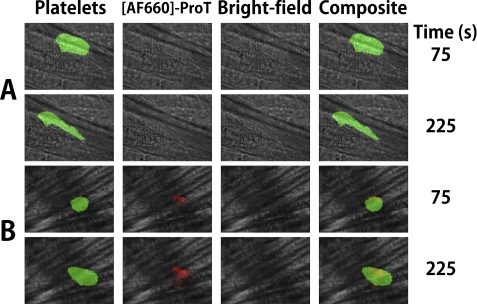
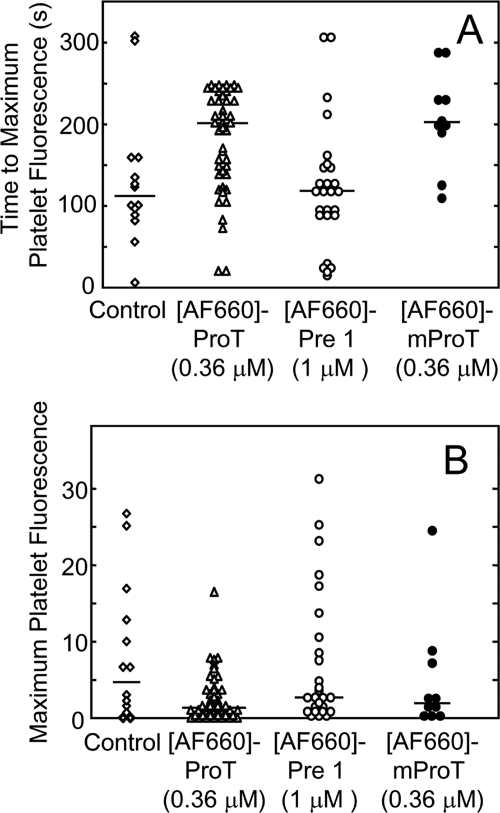
[AF660]FPR-human ProT (0.36 μm final concentration) and anti-CD41 Fab fragments labeled with Alexa Fluor 488 (125 μg/g) to label platelets were infused through the jugular vein of a C57/B6J mouse prior to induction of laser injury of an arteriolar wall in the cremaster muscle. Wide-field fluorescence microscopy was used to quantitate platelet accumulation and [AF660]FPR-human ProT binding at the site of injury. Both in the absence and presence of [AF660]FPR-human ProT, platelet accumulation followed the previously described pattern of rapid accumulation to a maximum, followed by platelet loss, and finally a constant platelet thrombus size (35, 38). However, the time to peak fluorescence intensity of platelets was delayed when [AF660]FPR-human ProT was present (Fig. 6, A and B). Activation of [AF660]FPR-ProT by prothrombinase or ecarin in vitro was not accompanied by a significant fluorescence change (<6%), indicating that the fluorescence signal likely reflected the localized concentration of labeled ProT and/or its labeled cleavage products. Analysis of multiple thrombi showed that [AF660]FPR-human ProT at 0.36 μm inhibited platelet accumulation, increasing the time to maximum accumulation by 100 s (p < 0.02) (Fig. 7A) and decreasing median thrombus size of 75% (p < 0.03) (Fig. 7B). [AF660]FPR-mouse ProT gave results similar to the labeled human zymogen, doubling the time to maximum platelet accumulation to 201 s and decreasing thrombus size 60% (Fig. 7B). In contrast, when [AF660]FPR-human Pre 1 (1 μm), lacking the membrane-binding F1 domain, was present no AF660 fluorescence was detected at the site of injury or platelet thrombus, and there was no effect on platelet accumulation or thrombus size (Fig. 7, A and B). This indicates that the membrane-binding Gla domain of ProT was required for binding of ProT at the site of injury. FPR-human thrombin at concentrations (7 μm) ∼20-fold higher than FPR-ProT showed no statistically significant effect on either the time to platelet accumulation or the maximum fluorescence intensity (not shown).
FIGURE 6.
Bright-field and wide-field fluorescence microscopy of platelet and [AF660]FPR-ProT accumulation during thrombosis. A, wide-field fluorescence intensity of platelets (green) in the absence of [AF660]FPR-human ProT with corresponding bright-field images and composite images at 75 and 225 s after injury. B, wide-field, bright-field, and composite images of platelets (green) in the presence of 0.36 μm [AF660]FPR-human ProT ([AF660]-ProT) (red). Postinjury recording of A and B began at ∼66 and ∼25 s, respectively. Note the smaller size of the thrombus in the presence of labeled ProT. Experiments were performed and analyzed as described under “Experimental Procedures.”
FIGURE 7.
Effect of [AF660]FPR-ProT on the rate and size of thrombus formation. A, time to maximum platelet accumulation following laser injury obtained for controls (2 mice, 13 thrombi) (♢), 0.36 μm [AF660]FPR-human ProT ([AF660]-ProT) (4 mice, 44 thrombi) (Δ), 1 μm [AF660]FPR-human Pre 1 ([AF660]-Pre 1) (3 mice, 26 thrombi) (○), and 0.36 μm [AF660]FPR-mouse ProT ([AF660]-mProT) (2 mice, 10 thrombi) (●). B, maximum thrombus size (same thrombi analyzed in A). Controls (♢), 0.36 μm [AF660]FPR-human ProT (Δ), 1 μm [AF660]FPR-human Pre 1 (○), and 0.36 μm [AF660]FPR-mouse ProT (●) are indicated. The bars denote the medians. Experiments were performed and analyzed as described under “Experimental Procedures.”
Colocalization of [AF660]FPR-Mouse ProT and Platelets
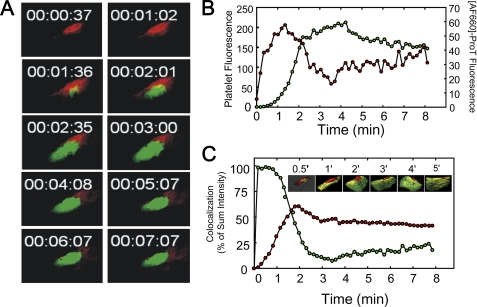
Colocalization of [AF660]FPR-mouse ProT and platelets was examined by confocal microscopy. [AF660]FPR-mouse ProT accumulation was more rapid than platelet accumulation at the site of injury, as indicated in the time-selected images of the maximum intensity projection from three-dimensional time lapse images (Fig. 8A) and the integrated fluorescence intensity of the [AF660]FPR-mouse ProT and platelets (Fig. 8B). Colocalization of platelets and [AF660]FPR-mouse ProT signals was determined from the confocal images. Surface views of time points between 0.5 and 5 min are also shown in Fig. 8C. For some images shown in Fig. 8C, the opacity of the platelet signal was decreased, for presentation purposes only, to reveal the colocalized signal. The maximum amount of [AF660]FPR-mouse ProT colocalized with platelets was 60% at 2 min, which decreased slightly and then remained constant at 50% up to 6 min. After 1 min, 98% of the minimal platelet thrombus present at the site of injury was colocalized with [AF660]FPR-mouse ProT. As the platelet thrombus grew, the percent of the platelet thrombus that colocalized with [AF660]FPR-mouse ProT decreased progressively to 10% at 3 min (Fig. 8C). Therefore, at 3 min, half of the bound [AF660]FPR-mouse ProT was associated with only 10% of the platelet thrombus, and half with the vessel wall. The platelets that were associated with [AF660]FPR-mouse ProT were those proximal to the site of injury.
FIGURE 8.
Colocalization of [AF660]FPR-ProT and platelets. Z-plane images were digitally recorded every 3 μm for 23 slices where the total Z-step distance was 70 μm. A full set of images of the 23 slices was collected every 8 s. A, selected maximum intensity projection images from confocal microscopy of [AF660]FPR-mouse ProT (red) and platelet (green) localization at the indicated times after vessel wall injury. B, platelets (green points) and [AF660]FPR-mouse ProT (red points) total fluorescence intensity for the volume through 70 μm. C, colocalization of platelet fluorescence (green points) and [AF660]FPR-mouse ProT (red points) expressed as percent of the sum of the intensities as a function of time after injury. Inset, selected confocal images of the colocalization (yellow) of platelets (green) and [AF660]FPR-mouse ProT (red). Experiments were performed and analyzed as described under “Experimental Procedures.”
Localization of F1.2 during Thrombosis
Localization of the membrane-binding F1.2, lacking the ProT catalytic domain, was investigated with AF660-labeled F1.2. Primary amines of human F1.2 were labeled in the presence of calcium to protect the NH2 terminus and maintain the functional conformation of the Gla domain. Wide-field fluorescence microscopy was used to examine the time course of labeled F1.2 accumulation after injury. Similar to full-length labeled ProT, [AF660]F1.2 accumulation at the site of injury preceded platelet accumulation, but F1.2 accumulated more slowly and reached a plateau after 3 min (supplemental Fig. S7). The platelet signal followed the typical time course. Unlike [AF660]FPR-ProT, [AF660]F1.2 did not affect the time to peak platelet accumulation at concentrations up to 0.74 μm (supplemental Fig. S8). Colocalization of platelets and [AF660]F1.2 showed that [AF660]F1.2 was found at the vessel wall-platelet interface after injury, similar to full-length [AF660]FPR-ProT, signaling binding to PS-expressing membranes (not shown). F1.2 labeled with AF660 in the absence of calcium, where the NH2 terminus is susceptible to modification, did not bind after laser injury in vivo (not shown). The results indicate that the properly folded Gla domain of labeled ProT are required for localization during thrombus formation and that the catalytic domain of the ProT zymogen is required for inhibition of platelet accumulation. The in vivo results paralleled those of the in vitro studies in showing that FPR-Pre 1 and FPR-thrombin did not inhibit thrombin formation, consistent with their lack of localization in vivo, and that F1.2 localized similarly to labeled FPR-ProT to PS-rich membranes, but did not inhibit thrombus formation. Overall, the results demonstrate that the antithrombotic activity of FPR-ProT in vivo is correlated structurally and functionally with inhibition of thrombin generation in plasma supported by phospholipid vesicles or natural platelet membranes.
Role of FPR-ProT Reaction Products in Prothrombinase Inhibition
FPR-ProT was a ∼10-fold more effective inhibitor of plasma thrombin generation than ProTS195A, presumably due to the different activation pathways, resulting in accumulation of FPR-Pre 2 and FPR-thrombin, and their F1.2 complexes, one or both of which are likely the inhibitory species. Activation of Pre 2·F1.2 to thrombin·F1.2 by prothrombinase follows a two-parallel pathway mechanism affected by the differential affinity of F1.2 for Pre 2 and thrombin (59). F1.2 binds Pre 2S195A with high affinity (KD 200 (59) or 40 nm for native Pre 2; supplemental Fig. S4A), whereas FPR-Pre 2 exhibits much lower affinity (KD 4500 (59) or 3600 nm; supplemental Fig. S4B). Induction of a more proteinase-like conformation in FPR-Pre 2 reduces its affinity for F1.2 and is predicted to inhibit its activation by prothrombinase. To assess the roles of the major products of FPR-ProT activation as inhibitors of native ProT activation, the time courses of native Pre 2 and FPR-Pre 2 in the presence of equimolar F1.2 (2 μm) conversion to thrombin and FPR-thrombin by prothrombinase were quantitated by SDS-gel electrophoresis (supplemental Fig. S5). Analysis of the exponential progress curves gave apparent bimolecular rate constants of 98 μm−1 s−1 for native Pre 2 and 5.5 μm−1 s−1 for FPR-Pre 2. The predicted inhibition of FPR-Pre 2 activation was verified by the 18-fold slower rate of FPR-Pre 2 cleavage by prothrombinase compared with native Pre 2 in the presence of F1.2.
DISCUSSION
The mechanism of ProT substrate recognition provides a basis for understanding the properties of FPR-ProT as a substrate and inhibitor of prothrombinase in vitro and in vivo. ProT recognition by prothrombinase follows a two-step mechanism with initial binding of ProT substrate species to exosites expressed on membrane-bound FXa·FVa, followed by a conformational change in which the bound substrate docks into the FXa active site for cleavage (1, 4, 5, 8, 9). The substrates, ProT, MZT, and Pre 2·F1.2 complex bind to the same exosites with similar apparent affinity (Km), whereas active site docking is substrate-dependent and manifested in kcat (1, 4, 5, 8, 9, 57, 60). Substrate recognition directs the sequential pathway of the two ProT cleavages required to generate thrombin, with initial cleavage at Arg320 to produce MZT, followed by cleavage at Arg271 to generate thrombin and F1.2 (61–63). The MZT pathway is preferred due to the ∼30-fold larger kcat for cleavage of ProT at Arg320 over cleavage at Arg271 (9). Cleavage at Arg271 in MZT, however, proceeds with a ∼30-fold larger kcat than cleavage of the same bond in ProT. This occurs because ProT substrate species bind to prothrombinase in two alternate conformations, dictated by the zymogen to proteinase transition (1, 9, 57, 60). ProT zymogen is bound in a conformation that presents Arg320 to FXa for catalysis, whereas the proteinase, MZT, ratchets to the alternate conformation in which Arg271 is presented for cleavage (57, 60). Evidence for the ratcheting mechanism includes the finding that FPR labeling of the catalytic site of ProTR320Q enhances the rate of cleavage at Arg271 ∼12-fold at 25 °C and ∼11-fold as shown here at 37 °C (57). FPR labeling of ProT results in a 4.5-fold increase in affinity of proexosite I for [5F]Hir-(54–65)(SO3−), consistent with partial exosite activation. The fluorescence of the probe bound to proexosite I on FPR-ProT, FPR-Pre 1, and FPR-Pre 2 was also perturbed, as shown by the increased fluorescence change compared with the unlabeled proteins. These results and the absence of significant changes in proexosite I on ProTS195A confirm the conclusion that the catalytic domain of FPR-ProT is in a more proteinase-like conformation, and demonstrate that FPR-ProT interacts with prothrombinase as a substrate by the exosite binding and active site docking mechanism. As expected from this mechanism, FPR-ProT cleavage was redirected toward enhanced cleavage at Arg271 to form FPR-Pre 2·F1.2 as the principal intermediate.
Initial rate kinetic studies of prothrombinase inhibition by FPR-ProT and ProTS195A showed competitive inhibition by ProTS195A and deviations from this mechanism for FPR-ProT. Similar apparent Ki values were obtained for ProTS195A and FPR-ProT, which were also close to the apparent Km. ProTS195A binds and docks for Arg320 cleavage like native ProT and follows the same pathway. By contrast, FPR-ProT binds like native ProT and ProTS195A, but docks to engage Arg271 for cleavage. The similar apparent inhibition constants are expected, because the observed affinity of substrate binding is determined by exosite binding. These considerations likely account for the deviations from pure competitive inhibition by FPR-ProT and the altered pathway of FPR-ProT activation, but do not exclude a contribution to inhibition from competitive nonproductive binding. Inhibition prothrombinase by ProTS195A and plasma thrombin generation are likely due solely to its competition with native ProT for prothrombinase.
The ratcheting mechanism also partly explains the mechanism of inhibition of native ProT activation by FPR-ProT. Simulation by numerical integration (64, 65) of the Kamath and Krishnaswamy mechanism (59) of Pre 2 activation by prothrombinase was performed, including the effects of F1.2 and active site labeling (supplemental Scheme S1). Following the ratcheting mechanism, the 18-fold ratio of the bimolecular rate constants for FPR-Pre 2 to native Pre 2 cleavage by prothrombinase in the presence of F12 was assumed to reflect a decrease in kcat (1, 4, 5, 8, 9, 57, 60). The previously determined parameters were maintained, with an approximate average KD used for native Pre 2 binding F1.2 of 100 nm. It was also assumed that thrombin·F1.2 and FPR-thrombin·F1.2 bound with similar 4.5 and 12 μm KD values for F1.2, respectively, and with similar affinities for prothrombinase (59). The fraction of total prothrombinase bound in nonproductive (FPR-labeled) complexes was calculated as a function of time for prothrombinase activation of 1 μm Pre 2 or FPR-Pre 2 and equimolar F1.2 (supplemental Fig. S6, A and B). A hypothetical 1:1 mixing experiment at 1 μm of each of the two substrates and 2 μm F1.2 was also performed, which showed that the major accumulating product over a 1500-s time course was FPR-Pre 2·F1.2, which instantaneously occupied ∼20% and maximally ∼55% of prothrombinase before slowly decaying, whereas all other products occupied no more than ∼6% of prothrombinase (supplemental Fig. S6, C and D). This analysis identified FPR-Pre 2·F1.2 as the inhibitory species in the pathway of FPR-ProT cleavage. We conclude that inhibition of native ProT activation by FPR-ProT is initially due to alternate nonproductive substrate inhibition, which is followed by product inhibition of prothrombinase by FPR-Pre 2·F1.2 generated abundantly by the redirected pathway of FPR-ProT cleavage.
Similar, specific inhibition of thrombin generation by human FPR-ProT was demonstrated in both human and mouse platelet-rich and platelet-poor plasmas, consistent with FPR-ProT acting as a reversible inhibitor of prothrombinase. This is supported by results demonstrating the requirement for the intact domain structure of ProT, whereas FPR-Pre 1, FPR-thrombin, F1, and F1.2, lacking either the catalytic or membrane-binding domains, produced little or no inhibition. This indicates that inhibition by FPR-ProT cannot be explained by competition for membrane-binding sites or inhibition by free FPR-thrombin. Analysis of the dependence of inhibition by FPR-ProT on the reaction variables, TF, and phospholipid concentration, as well as native ProT and FPR-ProT concentrations are similarly compatible with FPR-ProT acting directly on endogenously assembled prothrombinase. Similar results obtained by initiating thrombin generation with TF or FXa, and with platelet surfaces or phospholipid vesicles, further support the conclusion that prothrombinase is the target of inhibition.
Results of the in vitro studies suggested that FPR-ProT should be an inhibitor of thrombus formation in vivo. We chose to examine the effect of FPR-ProT on platelet thrombus formation in a laser-induced mouse model of thrombosis. In this model, TF antigen can be detected immediately after laser insult and fibrin accumulates at the site of vascular injury (35, 38, 39, 66). In contrast to other thrombosis models, platelet thrombus formation is thrombin-dependent and collagen- and glycoprotein VI-independent as demonstrated by studies in genetically modified mice. In Par4−/− mice, lacking the primary platelet thrombin receptor, platelet thrombus formation is markedly reduced after laser injury and the platelets in the minimal residual thrombus are activated only after a long delay (40). In contrast, platelet thrombus formation in mice lacking the platelet collagen receptor glycoprotein VI is equivalent to that observed in wild type mice (37).
What does the accumulating AF660 fluorescence signal observed in vivo represent? Cleavage of [AF660]FPR-ProT by prothrombinase in vitro was not accompanied by a significant fluorescence change, suggesting that the intensities observed in vivo reflect the relative concentrations of labeled ProT and its products. Because [AF660]FPR-ProT is only labeled in the catalytic domain and is a prothrombinase substrate, the fluorescence signal may reflect [AF660]FPR-ProT, as well as labeled Pre 2 and thrombin. Several observations support the idea that localization of [AF660]FPR-ProT in vivo reflects primarily its interaction with prothrombinase. Rapid and Gla domain-specific localization of AF660 fluorescence occurred in the presence of [AF660]F1.2, but platelet thrombus growth was not inhibited by F1.2 in vivo nor was thrombin generation inhibited by F1.2 in vitro. This implicates both PS-dependent membrane binding through the F1 domain and the zymogen catalytic domain in the mechanism of inhibition in vivo, and indicates that generation of free F1.2 does not contribute significantly to inhibition. Because the products are active site-blocked, they cannot interact with thrombin substrates, but may interact through exosites I and II. The absence of any inhibition by FPR-thrombin on thrombin generation supported by platelets or on platelet accumulation in vivo indicates that if exosite binding occurred with platelet-associated fibrinogen, fibrin, PAR-4, or glycoprotein Ib, it did not contribute to inhibition. The very similar results of thrombin generation in the presence of platelets or synthetic phospholipid vesicles also support the conclusion that labeled ProT acts on prothrombinase rather than platelet-specific interactions. Furthermore, the similar inhibition of thrombin generation in plasma obtained by initiation with TF or FXa indicates that upstream coagulation factors are not the primary targets of FPR-ProT.
In mice pretreated with [AF660]FPR-ProT, AF660 fluorescence rapidly accumulates on the vessel wall at the site of laser-induced injury and precedes the accumulation of platelets at this site. Subsequently, AF660 fluorescence is observed associated with the developing platelet thrombus proximal to the vessel wall. The localization of AF660 to the vessel wall is Gla domain-dependent, as [AF660]FPR-Pre 1 does not bind. The presence of [AF660]FPR-ProT during laser-induced thrombus formation led to delayed platelet thrombus formation and smaller thrombi, consistent with lower levels of thrombin generation in vivo in mice treated with [AF660]FPR-ProT. These and the above observations support the interpretation that initial [AF660]FPR-ProT localization and cleavage by prothrombinase occurs on PS-expressing, activated endothelial membranes. This is consistent with the observation that fibrin deposition in Par4−/− mice is equivalent to that in wild-type mice after laser injury, suggesting that a membrane surface other than that of activated platelets is responsible for supporting generation of the thrombin required for fibrinogen cleavage in vivo (40). This conclusion is supported directly by the demonstration that endothelial cells are rapidly activated by laser injury, prior to platelet accumulation (41), and, as shown here, prothrombinase is rapidly assembled to generate thrombin before platelet aggregation. The results of the current studies confirm that prothrombinase assembly and consequent thrombin formation occurs initially on activated endothelial cells (41), at least within the context of this in vivo model.
The participation of microparticles in [AF660]FPR-ProT localization and cleavage is also possible, as they accumulate early in thrombus formation (35, 38, 39, 66). P-selectin glycoprotein ligand-1-containing microparticles are expected to associate only with activated platelets. Previous studies of thrombin-dependent platelet activation in vivo showed that platelet population in the thrombus is heterogeneous, with a large number of unactivated platelets that interact only transiently with the thrombus (67). In wild-type mice, P-selectin appears rapidly at the thrombus-vessel wall interface but only at about 2 min postinjury at the lumenal edge of the thrombus (68). These factors may help to explain the localization of labeled ProT during thrombus growth.
There is increasing interest in developing drugs that target allosteric sites of enzymes because of their high specificity among structurally and functionally homologous proteins. FPR-ProT illustrates a highly specific and unique mechanism of action in which altering the exosite interactions and active site docking that control substrate recognition and the prothrombinase reaction pathway is the mechanistic basis of its anticoagulant and antithrombotic activity. The minimal modification of FPR-ProT and long half-life of ProT in plasma of ∼3 days (69) suggest that FPR-ProT has the potential to be used clinically for treatment of thrombosis. The low enthusiasm for parenterally administered, protein-based therapeutics, however, reduces pharmaceutical interest in FPR-ProT. Nevertheless, our studies demonstrate the potential of targeting exosites in the development of new therapeutics for treatment of thrombosis.
Supplementary Material
Acknowledgment
We thank Malabika Laha for outstanding technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 HL038779 and R37 HL071544 (to P. E. B.), K99 HL094533 (to P. P.), PO1 HL87203 and RO1 HL095084 (to B. F.), RO1 HL92125 (to B. C. F.), and PO1 HL74124 (to S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Scheme S1.
- ProT
- prothrombin
- Pre 1
- prethrombin 1
- Pre 2
- prethrombin 2
- MZT
- meizothrombin
- F1
- fragment 1
- F2
- fragment 2
- F1.2
- fragment 1.2
- ProTS195A
- Pre 2S195A, thrombinS195A, MZTS195A, Ser195 to Ala mutants of prothrombin, Pre 2, thrombin, and MZT, respectively
- ProTR271Q
- ProTR320Q, ProT mutants with Arg271 or Arg320 substituted with Gln, respectively
- FPR-CH2Cl
- d-Phe-Pro-Arg-CH2Cl, FPR-ProT, FPR-Pre 1, FPR-Pre 2, ProT, Pre 1, or Pre 2 inactivated with FPR-CH2Cl
- [AF660]FPR-ProT
- prothrombin labeled with Alexa Fluor 660 attached through Nα-[(acetylthio)acetyl]-FPR-CH2Cl
- [6F]FPR-thrombin
- thrombin labeled with 6-(iodoacetamido)fluorescein attached through Nα-[(acetylthio)acetyl]-FPR-CH2Cl
- [AF660]F1.2
- F1.2 labeled with Alexa Fluor 660
- Hir-(54–65)(SO3−)
- Gly-Asp-Phe-Glu-Glu-Ile-Pro-Glu-Glu-Tyr(SO3−)-Leu-Gln
- [5F]Hir-(54–65)(SO3−)
- Hir-(54–65)(SO3−) labeled at the amino terminus with 5-carboxy(fluorescein)
- CAT
- calibrated automated thrombography
- PS
- dioleoyl-phosphatidylserine
- PC
- dioleoyl-phosphatidylcholine
- PE
- dioleoyl-phosphatidylethanolamine
- TF
- tissue factor
- FV
- factor V
- FVa
- thrombin-activated FV
- FXa
- activated factor X
- FXaS195A
- FXa Ser195 to Ala mutant
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Krishnaswamy S. (2005) J. Thromb. Haemost. 3, 54–67 [DOI] [PubMed] [Google Scholar]
- 2. Mann K. G., Nesheim M. E., Church W. R., Haley P., Krishnaswamy S. (1990) Blood 76, 1–16 [PubMed] [Google Scholar]
- 3. Rosing J., Tans G., Govers-Riemslag J. W., Zwaal R. F., Hemker H. C. (1980) J. Biol. Chem. 255, 274–283 [PubMed] [Google Scholar]
- 4. Krishnaswamy S., Betz A. (1997) Biochemistry 36, 12080–12086 [DOI] [PubMed] [Google Scholar]
- 5. Boskovic D. S., Krishnaswamy S. (2000) J. Biol. Chem. 275, 38561–38570 [DOI] [PubMed] [Google Scholar]
- 6. Betz A., Krishnaswamy S. (1998) J. Biol. Chem. 273, 10709–10718 [DOI] [PubMed] [Google Scholar]
- 7. Wilkens M., Krishnaswamy S. (2002) J. Biol. Chem. 277, 9366–9374 [DOI] [PubMed] [Google Scholar]
- 8. Orcutt S. J., Pietropaolo C., Krishnaswamy S. (2002) J. Biol. Chem. 277, 46191–46196 [DOI] [PubMed] [Google Scholar]
- 9. Orcutt S. J., Krishnaswamy S. (2004) J. Biol. Chem. 279, 54927–54936 [DOI] [PubMed] [Google Scholar]
- 10. Beck D. O., Bukys M. A., Singh L. S., Szabo K. A., Kalafatis M. (2004) J. Biol. Chem. 279, 3084–3095 [DOI] [PubMed] [Google Scholar]
- 11. Anderson P. J., Nesset A., Dharmawardana K. R., Bock P. E. (2000) J. Biol. Chem. 275, 16435–16442 [DOI] [PubMed] [Google Scholar]
- 12. Anderson P. J., Nesset A., Dharmawardana K. R., Bock P. E. (2000) J. Biol. Chem. 275, 16428–16434 [DOI] [PubMed] [Google Scholar]
- 13. Anderson P. J., Bock P. E. (2003) J. Biol. Chem. 278, 44489–44495 [DOI] [PubMed] [Google Scholar]
- 14. Anderson P. J., Nesset A., Bock P. E. (2003) J. Biol. Chem. 278, 44482–44488 [DOI] [PubMed] [Google Scholar]
- 15. Arocas V., Lemaire C., Bouton M. C., Bezeaud A., Bon C., Guillin M. C., Jandrot-Perrus M. (1998) Thromb. Haemost. 79, 1157–1161 [PubMed] [Google Scholar]
- 16. Chen L., Yang L., Rezaie A. R. (2003) J. Biol. Chem. 278, 27564–27569 [DOI] [PubMed] [Google Scholar]
- 17. Bock P. E., Panizzi P., Verhamme I. M. (2007) J. Thromb. Haemost. 5, Suppl. 1, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tracy P. B., Nesheim M. E., Mann K. G. (1981) J. Biol. Chem. 256, 743–751 [PubMed] [Google Scholar]
- 19. Rosing J., van Rijn J. L., Bevers E. M., van Dieijen G., Comfurius P., Zwaal R. F. (1985) Blood 65, 319–332 [PubMed] [Google Scholar]
- 20. Miletich J. P., Jackson C. M., Majerus P. W. (1978) J. Biol. Chem. 253, 6908–6916 [PubMed] [Google Scholar]
- 21. Tracy P. B., Eide L. L., Mann K. G. (1985) J. Biol. Chem. 260, 2119–2124 [PubMed] [Google Scholar]
- 22. Dahlbäck B., Stenflo J. (1980) Eur. J. Biochem. 104, 549–557 [DOI] [PubMed] [Google Scholar]
- 23. Maruyama I., Salem H. H., Majerus P. W. (1984) J. Clin. Invest. 74, 224–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rodgers G. M. (1988) FASEB J. 2, 116–123 [DOI] [PubMed] [Google Scholar]
- 25. Rodgers G. M., Shuman M. A. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 7001–7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tracy P. B., Rohrbach M. S., Mann K. G. (1983) J. Biol. Chem. 258, 7264–7267 [PubMed] [Google Scholar]
- 27. Zwaal R. F., Comfurius P., Bevers E. M. (1998) Biochim. Biophys. Acta 1376, 433–453 [DOI] [PubMed] [Google Scholar]
- 28. Nesheim M. E., Taswell J. B., Mann K. G. (1979) J. Biol. Chem. 254, 10952–10962 [PubMed] [Google Scholar]
- 29. Gajdusek C., Carbon S., Ross R., Nawroth P., Stern D. (1986) J. Cell Biol. 103, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bono F., Herault J. P., Avril C., Schaeffer P., Lormeau J. C., Herbert J. M. (1997) J. Cell. Physiol. 172, 36–43 [DOI] [PubMed] [Google Scholar]
- 31. Tijburg P. N., van Heerde W. L., Leenhouts H. M., Hessing M., Bouma B. N., de Groot P. G. (1991) J. Biol. Chem. 266, 4017–4022 [PubMed] [Google Scholar]
- 32. Stern D. M., Nawroth P. P., Kisiel W., Handley D., Drillings M., Bartos J. (1984) J. Clin. Invest. 74, 1910–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodgers G. M., Kane W. H., Pitas R. E. (1988) J. Clin. Invest. 81, 1911–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Furie B., Furie B. C. (2005) J. Clin. Invest. 115, 3355–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Falati S., Gross P., Merrill-Skoloff G., Furie B. C., Furie B. (2002) Nat. Med. 8, 1175–1181 [DOI] [PubMed] [Google Scholar]
- 36. Celi A., Merrill-Skoloff G., Gross P., Falati S., Sim D. S., Flaumenhaft R., Furie B. C., Furie B. (2003) J. Thromb. Haemost. 1, 60–68 [DOI] [PubMed] [Google Scholar]
- 37. Dubois C., Panicot-Dubois L., Merrill-Skoloff G., Furie B., Furie B. C. (2006) Blood 107, 3902–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chou J., Mackman N., Merrill-Skoloff G., Pedersen B., Furie B. C., Furie B. (2004) Blood 104, 3190–3197 [DOI] [PubMed] [Google Scholar]
- 39. Falati S., Liu Q., Gross P., Merrill-Skoloff G., Chou J., Vandendries E., Celi A., Croce K., Furie B. C., Furie B. (2003) J. Exp. Med. 197, 1585–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vandendries E. R., Hamilton J. R., Coughlin S. R., Furie B., Furie B. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 288–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Atkinson B. T., Jasuja R., Chen V. M., Nandivada P., Furie B., Furie B. C. (2010) Blood 116, 4675–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bock P. E. (1992) J. Biol. Chem. 267, 14963–14973 [PubMed] [Google Scholar]
- 43. Bock P. E. (1992) J. Biol. Chem. 267, 14974–14981 [PubMed] [Google Scholar]
- 44. Bock P. E., Craig P. A., Olson S. T., Singh P. (1989) Arch. Biochem. Biophys. 273, 375–388 [DOI] [PubMed] [Google Scholar]
- 45. Dahlbäck B. (1980) J. Clin. Invest. 66, 583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nicolaes G. A., Tans G., Thomassen M. C., Hemker H. C., Pabinger I., Varadi K., Schwarz H. P., Rosing J. (1995) J. Biol. Chem. 270, 21158–21166 [DOI] [PubMed] [Google Scholar]
- 47. Panizzi P., Friedrich R., Fuentes-Prior P., Kroh H. K., Briggs J., Tans G., Bode W., Bock P. E. (2006) J. Biol. Chem. 281, 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krishnaswamy S., Church W. R., Nesheim M. E., Mann K. G. (1987) J. Biol. Chem. 262, 3291–3299 [PubMed] [Google Scholar]
- 49. Fenton J. W., 2nd, Fasco M. J., Stackrow A. B. (1977) J. Biol. Chem. 252, 3587–3598 [PubMed] [Google Scholar]
- 50. Mann K. G., Elion J., Butkowski R. J., Downing M., Nesheim M. E. (1981) Methods Enzymol. 80, 286–302 [DOI] [PubMed] [Google Scholar]
- 51. Bock P. E., Olson S. T., Björk I. (1997) J. Biol. Chem. 272, 19837–19845 [DOI] [PubMed] [Google Scholar]
- 52. van Zutphen L. F. M., Baumans V., Beynen A. C. (eds) (2001) Principles of Laboratory Animal Science, Elsevier, Amsterdam [Google Scholar]
- 53. Tchaikovski S. N., van Vlijmen B. J., Rosing J., Tans G. (2007) J. Thromb. Haemost. 5, 2079–2086 [DOI] [PubMed] [Google Scholar]
- 54. Koenen R. R., Tans G., van Oerle R., Hamulyák K., Rosing J., Hackeng T. M. (2003) Blood 102, 1686–1692 [DOI] [PubMed] [Google Scholar]
- 55. Hemker H. C., Giesen P., Al Dieri R., Regnault V., de Smedt E., Wagenvoord R., Lecompte T., Béguin S. (2003) Pathophysiol. Haemost. Thromb. 33, 4–15 [DOI] [PubMed] [Google Scholar]
- 56. Curvers J., Christella M., Thomassen L. G., de Ronde H., Bertina R. M., Rosendaal F. R., Tans G., Rosing J. (2002) Thromb. Haemost. 87, 483–492 [PubMed] [Google Scholar]
- 57. Bianchini E. P., Orcutt S. J., Panizzi P., Bock P. E., Krishnaswamy S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10099–10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dubois C., Atkinson B., Furie B. C., Furie B. (2007) in Platelets (Michelson A. D. ed) pp. 611–626, Academic Press/Elsevier, Amsterdam [Google Scholar]
- 59. Kamath P., Krishnaswamy S. (2008) J. Biol. Chem. 283, 30164–30173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hacisalihoglu A., Panizzi P., Bock P. E., Camire R. M., Krishnaswamy S. (2007) J. Biol. Chem. 282, 32974–32982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rosing J., Zwaal R. F., Tans G. (1986) J. Biol. Chem. 261, 4224–4228 [PubMed] [Google Scholar]
- 62. Krishnaswamy S., Mann K. G., Nesheim M. E. (1986) J. Biol. Chem. 261, 8977–8984 [PubMed] [Google Scholar]
- 63. Tans G., Janssen-Claessen T., Hemker H. C., Zwaal R. F., Rosing J. (1991) J. Biol. Chem. 266, 21864–21873 [PubMed] [Google Scholar]
- 64. Johnson K. A., Simpson Z. B., Blom T. (2009) Anal. Biochem. 387, 20–29 [DOI] [PubMed] [Google Scholar]
- 65. Johnson K. A., Simpson Z. B., Blom T. (2009) Anal. Biochem. 387, 30–41 [DOI] [PubMed] [Google Scholar]
- 66. Furie B., Furie B. C. (2004) Trends Mol. Med. 10, 171–178 [DOI] [PubMed] [Google Scholar]
- 67. Dubois C., Panicot-Dubois L., Gainor J. F., Furie B. C., Furie B. (2007) J. Clin. Invest. 117, 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gross P. L., Furie B. C., Merrill-Skoloff G., Chou J., Furie B. (2005) J. Leukocyte Biol. 78, 1318–1326 [DOI] [PubMed] [Google Scholar]
- 69. Shapiro S. S., Martinez J. (1969) J. Clin. Invest. 48, 1292–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.