Abstract
The C-terminal domain (Ct-FDH) of 10-formyltetrahydrofolate dehydrogenase (FDH, ALDH1L1) is an NADP+-dependent oxidoreductase and a structural and functional homolog of aldehyde dehydrogenases. Here we report the crystal structures of several Ct-FDH mutants in which two essential catalytic residues adjacent to the nicotinamide ring of bound NADP+, Cys-707 and Glu-673, were replaced separately or simultaneously. The replacement of the glutamate with an alanine causes irreversible binding of the coenzyme without any noticeable conformational changes in the vicinity of the nicotinamide ring. Additional replacement of cysteine 707 with an alanine (E673A/C707A double mutant) did not affect this irreversible binding indicating that the lack of the glutamate is solely responsible for the enhanced interaction between the enzyme and the coenzyme. The substitution of the cysteine with an alanine did not affect binding of NADP+ but resulted in the enzyme lacking the ability to differentiate between the oxidized and reduced coenzyme: unlike the wild-type Ct-FDH/NADPH complex, in the C707A mutant the position of NADPH is identical to the position of NADP+ with the nicotinamide ring well ordered within the catalytic center. Thus, whereas the glutamate restricts the affinity for the coenzyme, the cysteine is the sensor of the coenzyme redox state. These conclusions were confirmed by coenzyme binding experiments. Our study further suggests that the binding of the coenzyme is additionally controlled by a long-range communication between the catalytic center and the coenzyme-binding domain and points toward an α-helix involved in the adenine moiety binding as a participant of this communication.
Keywords: Crystal Structure, Enzyme Catalysis, Enzyme Mechanisms, Enzymes, Folate Metabolism, NAD, NADH, Protein Structure, Aldehyde Dehydrogenase
Introduction
Enzymes belonging to the family of aldehyde dehydrogenases (ALDH)3 are involved in conversions of a broad variety of aliphatic and aromatic aldehydes to their respective carboxylic acids (1–3). These enzymes are either dimers or tetramers of identical subunits (1). Numerous crystal structures showed that subunits of different ALDHs have very similar fold with each composed of catalytic, nucleotide binding, and oligomerization domains (4–10).
Two members of the family, cytosolic ALDH1 and mitochondrial ALDH2, are essential components of the pathway metabolizing ethanol (2, 11). ALDH2 is also involved in mediating the vasodilator activity of nitroglycerine (12). Furthermore, activation of this enzyme correlates with reduced ischemic heart damage in rodent models (13). The polymorphism in the ALDH2 gene found in about 40% of the Asian population causes decreased tolerance to alcohol in heterozygous and homozygous individuals (14). This ALDH2 genetic variant, ALDH2*2, has a lysine instead of a glutamate at position 487 within the oligomerization domain of the tetrameric enzyme. This substitution induces conformational changes at the subunit interface, which are reflected by structural alterations within the catalytic center and the coenzyme binding site (5, 15, 16). The final outcome is a strong decrease in the affinity for NAD+ binding and a severely reduced catalytic activity (17). Recent studies have demonstrated that the activity of the catalytically deficient mutant is restored in the presence of a small molecule identified by a high-throughput screening (13). One of the effects of this activator was a significant reduction in the apparent Km for NAD+ (18).
It is generally accepted that the ALDH reaction occurs in two principal steps, acylation and deacylation (Fig. 1) (5, 7, 19–21). In the first step, the invariant active site cysteine 302 (the numbering is according to human ALDH2) conducts a nucleophilic attack on the carbonyl carbon of the aldehyde molecule to form a thiohemiacetal intermediate. The hydride ion is then transferred from an aldehyde to the C4 atom of the nicotinamide ring of NAD(P)+, which causes a collapse of the thiohemiacetal to a thioester intermediate. In the second step, an activated water molecule hydrolyzes the thioester, releasing the product. It has been suggested that deprotonation of the catalytic cysteine prior to the nucleophilic attack on the substrate, a required step in this mechanism, involves acceptance of a proton by either glutamate 268 or 399 (5, 7, 20, 22–24). The resulting thiolate ion is likely stabilized by the positively charged nicotinamide ring of the coenzyme and/or adjacent main chain amide groups. It has also been suggested that, in tetrameric class 1 and 2 ALDHs, the proton abstracted by Glu-268 is shuttled to bulk water (10, 25). In addition, Glu-268 is also the most likely residue activating a water molecule in the deacylation step of the reaction (5, 7, 10, 23, 26).
FIGURE 1.
Proposed mechanism of the aldehyde dehydrogenase catalysis. In the active site of an apoALDH, a glutamate (Glu-268 in class 1/2 ALDHs or Glu-673 in Ct-FDH) is hydrogen bonded to a cysteine (Cys-302 in class 1/2 ALDHs or Cys-707 in Ct-FDH). Binding of NAD(P)+ (step 1) results in the rotation of the glutamate side chain away from the cysteine, which simultaneously loses a proton; in some ALDHs the glutamate might assist in the proton withdrawal. Negatively charged sulfur of the cysteine forms a transient covalent bond (shown as a gray oval) with the C4 atom of the nicotinamide ring of the coenzyme. The aldehyde dehydrogenase catalysis includes two phases, acylation (steps 2–4) and deacylation (steps 5 and 6). In this mechanism, Cys-707 functions as a catalytic nucleophile, whereas Glu-673 is postulated to activate a water molecule in the deacylation step. The structures studied in this paper are shown in boxes (they include apoproteins and their complexes with NADP+ or NADPH).
A distinct feature of ALDHs is the apparent requirement for coenzyme flexibility in the process of catalysis (10, 27–29). For hydride transfer to occur, in the first step of this reaction the C4 atom of the nicotinamide ring has to be in close proximity to the catalytic cysteine. After the hydride transfer, the reduced nicotinamide ring must exit the catalytic site to allow for the accessibility of a water molecule, which hydrolyzes the acyl-sulfur bond releasing the product. In agreement with this mechanism, numerous crystal structures of ALDH revealed two common conformations of the coenzyme, depending on its oxidation state: the extended (“hydride transfer”) conformation of NAD(P)+, with the nicotinamide ring positioned close to Cys-302; and the contracted (“hydrolysis”) conformation of NAD(P)H, with the nicotinamide ring found outside of the catalytic site or disordered (4, 5, 7, 10, 25, 27, 30). The mechanism by which ALDHs sense the oxidation state of the bound coenzyme and control its conformation is not fully understood.
In humans there are 19 known aldehyde dehydrogenases (2). One member of the ALDH family, 10-formyltetrahydrofolate dehydrogenase (FDH, ALDH1L1), is the product of a natural fusion of three unrelated genes (31). A part of the ALDH1L1 gene encoding for the 500-amino acid residue long C-terminal domain (Ct-FDH) was derived from an ancestor ALDH gene. Expressed separately, this domain displays enzymatic and structural features of canonical class 1 or 2 ALDHs including the presence of two catalytically essential residues, glutamate 673 and cysteine 707 (corresponding to Glu-268 and Cys-302 of ALDH) (10, 32). Previously solved crystal structures of apo-Ct-FDH and its complexes with reduced or oxidized coenzyme indicated interactions between the two catalytic residues and the nicotinamide ring. Specifically, a reversible rotation of the glutamate side chain upon NADP+ binding and a covalent bond formation between the cysteine and the C4 atom of nicotinamide were observed (10). In the current work, we solved crystal structures of the E673A, E673A/C707A, E673Q, and C707A mutants of Ct-FDH and their complexes with NADP+ or NADPH to investigate the role of these catalytic residues in conformational flexibility of the bound coenzyme and in the retaining oxidized and discharging reduced coenzyme.
EXPERIMENTAL PROCEDURES
Generation of Mutants
Site-directed mutagenesis was carried out using a QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing of the mutant constructs. The mutant proteins were expressed in Escherichia coli and purified as previously described (10).
Crystallization and Data Collection
Crystals were grown by the vapor diffusion technique in hanging drops over wells containing 1.4–1.9 m ammonium sulfate and 0.1 m MES, pH 6.4–6.8, 0.1 m HEPES, pH 7.0, or 0.1 m Tris-HCl, pH 7.5–8.1, as described previously (10). Crystals of binary complexes of the E673Q and C707A mutants with NADP+ or NADPH were produced by soaking protein crystals overnight in a solution containing 0.1 m Tris-HCl, pH 7.5, 5 mm DTT, 5 mm of the corresponding coenzyme, and the concentration of ammonium sulfate equal to that in the mother liquor. To maintain equivalent conditions, the crystals of apoproteins were soaked in the same solution containing no coenzymes before mounting. Prior to data collection, all crystals were passed through the same solution containing 27.5% glycerol and flash-frozen in situ at 100 K using an X-Stream Cryostream (Rigaku MSC). In the case of the binary complexes, the cryoprotecting solution also contained 5 mm of the corresponding coenzyme.
The data sets were collected on an RAXIS IV++ image plate detector mounted on a RU-H3R rotating anode x-ray generator operating at 50 kV and 100 mA (Medical University of South Carolina) and at a wavelength of 1.0 Å on MAR300 CCD detector (MAR-USA) at the SER-CAT beamline ID22 at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). All the crystals belong to space group C2 with cell dimensions a = 258.6, b = 194.8, c = 97.4, and β = 108.8. The data were processed using HKL2000 (33). The statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics
| E673A | E673A/C707A | E673Q apo | E673Q NADP+ | C707A apo | C707A NADP+ | C707A NADPH | |
|---|---|---|---|---|---|---|---|
| Synchrotron (Y/N) | Y | Y | Y | Y | N | N | N |
| Resolution (Å) | 1.9 | 2.0 | 2.38 | 2.25 | 2.5 | 2.1 | 2.3 |
| Completeness, % | 98.6(89.2)a | 96.4(78.8) | 96.9(81.2) | 99.9(99.9) | 98.2(96.7) | 95.7(93.4) | 99.8(99.8) |
| Mean I/σI | 9.8(1.9) | 10.4(2.0) | 6.7(1.9) | 9.7(5.5) | 7.4(2.9) | 8.1(2.6) | 6.9(2.1) |
| Redundancy | 4.2(2.2) | 3.9(1.9) | 4.7(2.1) | 5.3(4.9) | 4.8(4.7) | 4.9(4.8) | 3.9(3.8) |
| Rmerge (%) | 11.8(49.8) | 8.8(36.3) | 14.3(47.5) | 13.0(54.7) | 15.2(43.8) | 11.6(60.5) | 12.4(60.2) |
| Rcryst (%) | 19.0 | 17.6 | 18.4 | 16.6 | 22.5 | 17.3 | 16.7 |
| Rfree (%) | 20.4 | 19.0 | 20.7 | 18.5 | 25.7 | 19.1 | 18.5 |
| Root mean square deviation bond lengths (Å) | 0.010 | 0.008 | 0.011 | 0.009 | 0.011 | 0.009 | 0.009 |
| Root mean square deviation bond angles (°) | 1.25 | 1.13 | 1.29 | 1.19 | 1.33 | 1.15 | 1.21 |
| Ramachandran plot | |||||||
| Most favored, % | 90.6 | 90.9 | 89.1 | 90.7 | 89.9 | 90.7 | 90.6 |
| Additionally allowed, % | 8.9 | 8.7 | 9.7 | 8.9 | 9.5 | 8.9 | 9.0 |
| Generously allowed, % | 0.2 | 0.2 | 0.7 | 0.2 | 0.4 | 0.2 | 0.2 |
| Disallowed, % | 0.2 | 0.2 | 0.5 | 0.2 | 0.2 | 0.2 | 0.2 |
a The numbers in parentheses refer to the highest resolution shell of the data.
Model Building and Refinement
All the structures were obtained by molecular replacement with Molrep (34) using the structure of apo-Ct-FDH (PDB code 2O2P) (10) as a search model. The coenzyme molecules were identified by examining the |Fo| − |Fc| and 2|Fo| − |Fc| electron density maps. The models were refined by alternating rounds of restrained refinement in REFMAC5 (35) and manual building using O (36) and Coot (37). Water molecules were introduced using Coot and inspected manually. During refinement, 5% of the data were reserved to calculate the free R-factor and this assignment was maintained for all data sets. In all the structures, unidentified electron density of varying intensity was observed in the substrate entrance tunnel. In some subunits it could be filled with a MES molecule at 0.5 occupancy or with a combination of a glycerol molecule and a water molecule, as in the structures of wild-type Ct-FDH (10). The refinement statistics are shown in Table 1. The stereochemistry of the models was verified by PROCHECK (38) (Table 1). The figures were prepared using PyMol (www.pymol.org). Coordinates and structure factors have been deposited in the Protein Data Bank with codes 3RHJ, 3RHL, 3RHM, 3RHO, 3RHP, 3RHQ, and 3RHR for the E673A mutant in complex with co-purified NADP+, E673A/C707A double mutant in complex with co-purified NADP+, E673Q mutant, E673Q mutant in complex with NADP+, C707A mutant, C707A mutant in complex with NADP+, and C707A mutant in complex with NADPH, respectively.
Determination of the Oxidation State of the Coenzyme
To determine the oxidation state of the coenzyme that was co-purified with E673A and E673A/C707A mutants, a protein sample was diluted 1:80 with deionized water and concentrated using an Amicon Ultra centrifugal filter device (Millipore). The protein was then denatured by addition of NaOH to increase the pH to 11.5. After a 1-h incubation, the lower molecular weight components were separated by filtration as described above. Concentrations of NADP+ and NADPH were then estimated from the absorbance spectra of the filtrates recorded in the 240–380 nm range using extinction coefficients 1.8 × 104 m cm−1 (NADP+ at 260 nm), 1.5 × 104 m cm−1 (NADPH at 260 nm), and 6.22 × 103 m cm−1 (NADPH at 340 nm). The presence of NADPH in the filtrate was additionally verified using a fluorescence spectrum excited at 330 nm and recorded from 400 to 550 nm. The protein concentration in the high molecular weight fraction was estimated by the method of Bradford. The approximate ratio of the coenzyme molar concentration to the protein subunit molar concentration was calculated from these data.
Equilibrium Dialysis
The E673A and E673A/C707A mutants of Ct-FDH were subjected to 5 consecutive 24-h rounds of dialysis at 4 °C against either 20 mm Tris-HCl buffer, pH 7.8, containing 50 mm NaCl, 0.5 mm DTT, and 0.02% NaN3 or 20 mm MES buffer, pH 5.6, containing 0.5 m NaCl and 0.5 mm DTT, at a protein sample to buffer ratio of 1:500. The sample was then concentrated and crystallized as described above or used for the coenzyme binding study.
Coenzyme Binding Study
Wild-type Ct-FDH or mutant proteins were incubated with increasing concentrations of NADP+ (up to 5 × 10−5 m) or NADPH (up to 6 × 10−5 m) for 1 h at room temperature in 20 mm BisTris propane buffer, pH 7.5, containing 100 mm NaCl and 0.5 mm DTT. Emission fluorescence spectra of proteins were recorded on a Hitachi F-2500 fluorescence spectrophotometer by scanning from 300 to 400 nm with excitation at 295 nm. Changes in NADPH fluorescence at 450 nm were recorded upon excitation at 330 nm. Kd values were determined by non-linear fitting of experimental data points to equations described under supplemental materials.
RESULTS
All crystals produced in this study share the same C2 space group with unit cell dimensions and crystal packing identical to those of the crystals of WT Ct-FDH (10). Root mean square deviations in positions of all Cα atoms between mutant structures and WT Ct-FDH (PDB code 2O2P) were within 0.32 Å, with the exception of the structure of the E673Q mutant in the absence of NADP+, which showed a root mean square deviation of 0.62 Å. In each structure, the four subunits maintain the same fold, and there were no large scale structural rearrangements due to introduction of the mutations or binding of NADP+/NADPH.
Crystal Structure of the E673A Mutant of Ct-FDH Reveals a Co-purified NADP+ Molecule in the Active Site
The structure of the E673A mutant of Ct-FDH, crystallized without addition of the coenzyme, was determined at 1.9-Å resolution. The electron density in the active site of each subunit suggested the presence of a co-purified NADP+ molecule bound in the extended conformation (Fig. 2A). In contrast, WT Ct-FDH was crystallized as the apoenzyme in the absence of NADP+/NADPH (10). After denaturing the protein, the bound coenzyme was extracted and identified as NADP+ based on the absorption spectrum (Fig. 2B) and the absence of fluorescence at 460 nm upon excitation at 330 nm, typical of NADPH. NADP+ molecules were introduced into the density and assigned occupancies of 0.75 based on minimizing the peaks in the |Fo| − |Fc| electron density map (Fig. 2C). This occupancy value is in agreement with the spectrophotometrically determined NADP+ to protein subunit molar ratio of 0.8 (see “Experimental Procedures”). Soaking the crystals in the presence of NADP+ increased the occupancy of the cofactor to 1.0 (data not shown).
FIGURE 2.
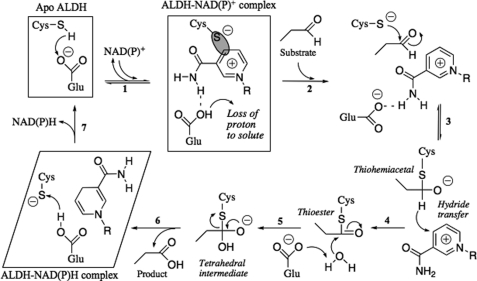
NADP+ molecule in the E673A mutant structure of Ct-FDH. A, stereoview of the active site region. The |Fo| − |Fc| electron density map of co-purified NADP+ (shown in green) was obtained by refinement in the absence of the coenzyme and contoured at 3 σ. The NADP+ molecule (shown in stick mode, gray) from the holo-structure of WT Ct-FDH (PDB code 2O2Q) is displayed for reference. B, the presence of NADP+ in the E673A mutant demonstrated by absorption spectroscopy: solid curve, the absorption spectrum of the purified E673A mutant (the shoulder at 320 nm is a signature of a covalent bond between Cys-707 and the NC4 atom of NADP+); dotted curve, the absorption spectrum of the low molecular weight component separated after denaturation of the protein. C, stereoview of a superposition of the active sites of the E673A mutant (colored) and the WT Ct-FDH/NADP+ complex (gray). The 2|Fo| − |Fc| (1 σ) electron density map of Cys-707, Ala-673, and the coenzyme is shown in blue.
The structure of the active site of the mutant is identical to that of WT Ct-FDH in complex with NADP+ (Fig. 2C). The coenzyme is bound through a non-classical Rossmann fold with the pyrophosphate part open to the solvent and the nicotinamide moiety buried in a pocket adjacent to the active site, with no noticeable deviations between positions of amino acid residues involved in contacts with NADP+. Likewise, conformations of NADP+ in the two structures were identical. As we previously observed in the WT Ct-FDH structure (10), two conformations of the sulfur atom of the active site cysteine 707 were observed in electron density of the E673A mutant. However, the C4 atom of the nicotinamide ring apparently forms a covalent bond with one of these conformers only in subunits B and D (supplemental Figs. S1 and S2), whereas in the wild-type protein such covalent bonds were formed in all four subunits. The covalent adduct with NADP+ was confirmed by the presence of a small but reliably detected absorbance peak at 320 nm in the spectrum of the E673A mutant (Fig. 2B). We showed previously that this peak is associated with formation of the Ct-FDH/NADP+ covalent adduct (10).
NADP+ Is Trapped in the Active Site of the E673A Mutant
We attempted to remove NADP+ from the active site of the E673A mutant using extensive dialysis of the protein sample prior to crystallization trials. The resulting 2.7-Å structure showed strong electron density for NADP+ in all four subunits (supplemental Fig. S3A). The presence of bound NADP+ was confirmed using the aforementioned spectrophotometric assay (data not shown). In another approach, multiple rounds of soaking the E673A crystals in mother liquor in the absence of the cofactor did not influence the electron density with respect to NADP+. In contrast, a single “back-soaking” completely removes the coenzyme from WT Ct-FDH (10). These results indicate that replacing the catalytic glutamate of Ct-FDH with an alanine traps NADP+ in the active site without inducing noticeable structural rearrangements of the protein molecule.
To strip the E673A mutant of NADP+ we had to destabilize the complex by decreasing the pH to 5.6 and increasing the sodium chloride concentration to 0.5 m. Prolonged dialysis at this condition resulted in removal of most of the coenzyme as demonstrated by the corresponding crystal structure solved at 2.0 Å (supplemental Fig. S3B). Of note, a further decrease in pH resulted in irreversible protein precipitation.
We have previously demonstrated that the E673A mutant of Ct-FDH has essentially no ALDH activity (10). In agreement with this finding, soaking the crystals of the mutant in the presence of propanal, a substrate for the Ct-FDH ALDH reaction, had no noticeable effect on the conformation of either the active site residues or the coenzyme (data not shown). In contrast, the same treatment of WT Ct-FDH in complex with NADP+ altered the positions of both the Glu-673 side chain and the nicotinamide part of the coenzyme (due to reduction of the coenzyme upon induction of the enzymatic reaction by propanal) (10).
The E673A/C707A Double Mutant
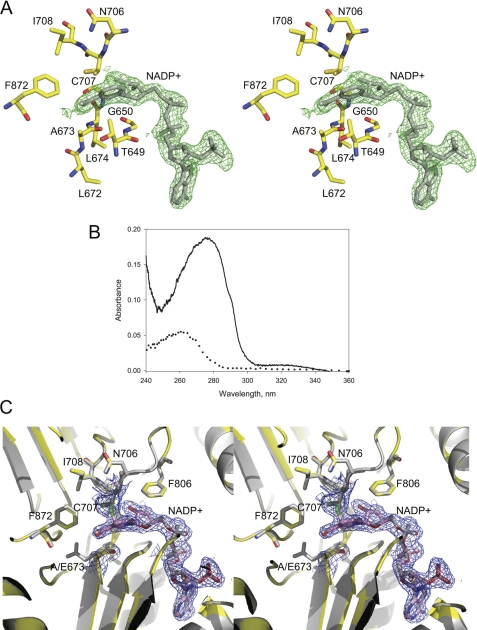
The structure of the E673A/C707A double mutant, determined at a 2.0-Å resolution, is essentially identical to the structure of the E673A mutant. Examination of the |Fo| − |Fc| and 2|Fo| − |Fc| electron density maps in the active center revealed the presence of co-purified cofactor molecules in all four subunits (Fig. 3) at occupancy of 0.8. The bound coenzyme was identified as NADP+ by absorption spectroscopy after extraction from the protein (data not shown). Despite the absence of the contact with residue 707, the coenzyme in this structure maintains exactly the same conformation as in the E673A mutant and WT Ct-FDH, as demonstrated by superimposition of Cα atoms of these structures (Fig. 3). Attempts to remove NADP+ from the active site by dialysis of protein preparations or crystal back-soaking were not successful: the structure of the mutant still revealed prominent electron density for coenzyme molecules. These results confirm that the increased affinity of the E673A mutant toward NADP+ was not caused by a stronger covalent bond between the nicotinamide ring of the coenzyme and Cys-707 (as a possible result of lacking a glutamate residue at position 673).
FIGURE 3.
Stereoview of a superposition of the active sites of the E673A/C707A mutant (colored) and the E673A mutant (gray). The 2|Fo| − |Fc| electron density map of the active site Ala-707 and Ala-673 and co-purified NADP+ for the double mutant is shown in blue and contoured at 1 σ.
The Structures of the E673Q Mutant of Ct-FDH
The active site of the 2.38-Å apo-structure of the E673Q mutant of Ct-FDH is substantially different from that of the WT enzyme in the absence of NADP+ (Fig. 4A). Only one conformation of the sulfur atom of Cys-707, corresponding to SNat1 in WT Ct-FDH (10), was observed in the electron density map. Similar to Glu-673, Gln-673 points toward the active site cysteine, but an apparent rotation around the Cγ atom places the side chain of Gln-673 farther from Cys-707 (the distances between the sulfur atom of Cys-707 and oxygen/nitrogen of the carbamide group of Gln-673 or oxygens of the carboxyl group of Glu-673 are 4.0 and 4.9 Å versus 3.3 and 3.7 Å, respectively). This results in the loss of the direct contact between Cys-707 and the side chain of residue 673, observed in the wild-type protein structure.
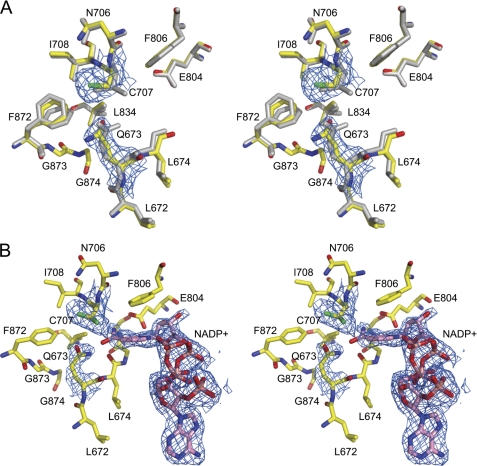
FIGURE 4.
Active sites of the E673Q mutant of Ct-FDH. A, a superposition of the active sites of the mutant in the absence of NADP+ (colored) and WT Ct-FDH (gray). The 2|Fo| − |Fc| electron density map of active site cysteine 707 and glutamine 673 of the mutant protein contoured at 1 σ is shown in blue. B, stereoview of the active site of the E673Q mutant in the presence of NADP+. Two conformers of NADP+ are shown. The 2|Fo| − |Fc| electron density map of the catalytic cysteine 707, glutamine 673, and NADP+ of the mutant protein contoured at 1 σ is shown in blue.
The active site of the complex between the E673Q mutant and NADP+ significantly deviates from the active sites of both the native protein complexed with NADP+ and the E673Q mutant in the absence of NADP+. In this 2.25-Å structure, two conformations of Cys-707 are clearly seen. Of note, upon binding of the coenzyme, the side chain of residue 673 is not displaced by the nicotinamide ring (as seen in WT Ct-FDH) but maintains the position close to Cys-707 typical of the apo-structure of Ct-FDH (Fig. 4B). The position of the adenine moiety of NADP+ is identical to that observed in the wild-type protein but electron density clearly indicates two conformations of the pyrophosphate of the nicotinamide part, each with 0.5 occupancy. Although the overall strong electron density for NADP+ is somewhat distorted in the vicinity of Cys-707, the positions of the nicotinamide ring and the ribose of one of the NADP+ conformers are well defined. However, these positions are significantly different from what is observed in the extended and contracted conformations of the coenzyme in ALDH (Fig. 4B), apparently due to the inability to displace the side chain of Gln-673 from the active site. The nicotinamide ring and the ribose of the second conformer of NADP+ are not clearly defined and thus were not modeled. As in WT Ct-FDH, the coenzyme can be removed from the active site by a single round of soaking in the mother liquor.
Conformational Changes of Helix G
Helix G (residues 652–667) of the nucleotide-binding domain, which forms one-half of a cleft for binding of the adenine part of the coenzyme in ALDH, displayed a high degree of disorder in all four subunits of the apo-structure of the E673Q mutant (Fig. 5A): no electron density was seen for the side chains and some of the main chain atoms of amino acids of this helix. Therefore, residues 652–667 were built as alanines. The average temperature factor of the residues in helix G is 87.0 versus 42.8 Å2 for the entire structure. This is in sharp contrast with the structures of WT Ct-FDH and other mutants analyzed in this study, which show high-quality electron density (Fig. 5A) and no increase in temperature factors in this region. Interestingly, binding of NADP+ to the E673Q mutant stabilizes helix G, as reflected by strong electron density in this region, which allows a reliable assignment of positions of all amino acid side chains (Fig. 5A). Of note, a superposition of the apo- and holo-structures of the E673Q mutant revealed no significant conformational differences except for the altered position of the side chain of Arg-644, a residue located in a loop adjacent to helix G (Fig. 5B). In WT Ct-FDH and in the complex of E673Q with NADP+, Arg-644 of chains A and C forms hydrogen bonds with Glu-831 of chain B and D, respectively (Fig. 5B). Arg-644 of chains B and D participates in symmetrical interactions with Glu-831 of chains A and C. In the coenzyme-free E673Q structure, however, Arg-644 contacts the main chain oxygen atom of Lys-876 of the neighboring subunit (Fig. 5B).
FIGURE 5.
Disorder in the nucleotide-binding domain (helix G) of the Ct-FDH E673Q mutant. A, schematic representation of dimer of subunits A and B of WT Ct-FDH (central panel, helices G are shown in yellow; NADP+ is shown in cyan) and stick representations of helices G in the apo- (left panel) and holo-E673Q (right panel). The 2|Fo| − |Fc| electron density maps (1 σ) are shown in blue. B, replacement of the active site glutamate with glutamine leads to disordering of helix G and an altered position of Arg-644. Schematic/stick representations of helix G and surrounding residues of the E673Q mutant in the absence (left) and presence (right) of NADP+ are shown. The red sphere represents a water molecule (see “Discussion” for details).
The Structures of the C707A Mutant
Three structures of the C707A mutant were determined, including the coenzyme-free form at 2.5-Å resolution, the complex with NADP+ at 2.1 Å, and the complex with NADPH at 2.3 Å. The binary complexes were obtained by soaking in the presence of NADP+ or NADPH. Active sites of these structures are virtually identical to that of WT Ct-FDH. Inspection of the electron density in the active center of the coenzyme-free structure showed no presence of NADP+ or NADPH. Despite the lack of the contact with the sulfur atom in the absence of a cysteine residue at position 707, the side chain of Glu-673 of the mutant occupies exactly the same position as observed in the apo-structure of WT Ct-FDH (Fig. 6A).
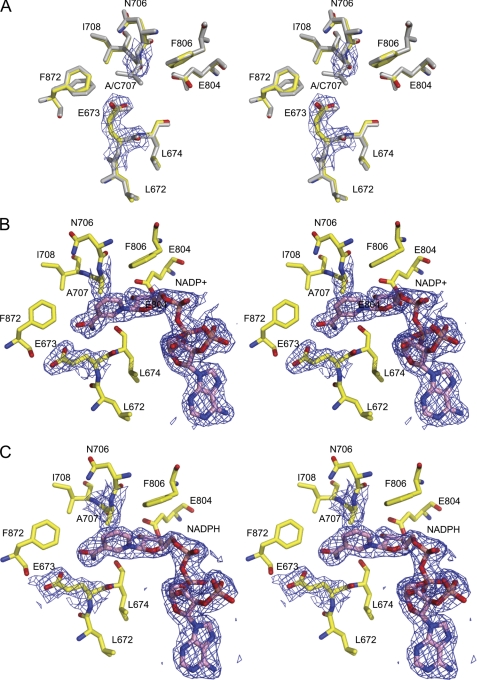
FIGURE 6.
Stereoview of the active site of the apo-Ct-FDH C707A mutant (A) and its complexes with NADP+ (B) or NADPH (C). The 2|Fo| − |Fc| electron density maps of the active site alanine 707, glutamate 673, and NADP+/NADPH contoured at 1 σ are shown in blue. Panel A also shows the superimposed active site of WT Ct-FDH (gray).
In the NADP+-bound structure of the C707A mutant, the coenzyme was found in the extended conformation, with the nicotinamide ring positioned between Ala-707 and Glu-673 (Fig. 6B). Because of the absence of the restraint imposed by the covalent bond, the ring is positioned slightly farther from residue 707. This is very similar to the coenzyme position in the structures of ALDHs lacking the covalent adduct between the catalytic cysteine and nicotinamide (5, 27). As in the WT enzyme, the nicotinamide ring of NADP+ displaces the side chain of glutamate 673 away from residue 707 by an apparent rotation around the Cα-Cβ bond. The back-soaking of crystals of the C707A mutant in mother liquor in the absence of the coenzyme resulted in a structure that showed no traces of electron density corresponding to NADP+ (data not shown).
The structure of the C707A mutant in complex with NADPH shows a high degree of similarity to the NADP+-bound structure (Fig. 6C). An unusual feature of this structure, however, is a well ordered nicotinamide moiety of NADPH in the same (extended) conformation as observed in the complex with NADP+. This is in striking contrast with WT Ct-FDH where this part of NADPH is completely disordered (10). Of note, based on the overall analysis of the conformation of NADPH bound to WT Ct-FDH, we previously suggested that this coenzyme is bound in the extended conformation (10), and the structure of the C707A mutant now gives direct evidence for this conclusion. Thus, the extended conformation of NADPH with a disordered nicotinamide ring observed in the complex with WT Ct-FDH is a functional analog of the contracted conformation of the reduced coenzyme in canonical ALDHs.
Determination of Equilibrium Dissociation Constants
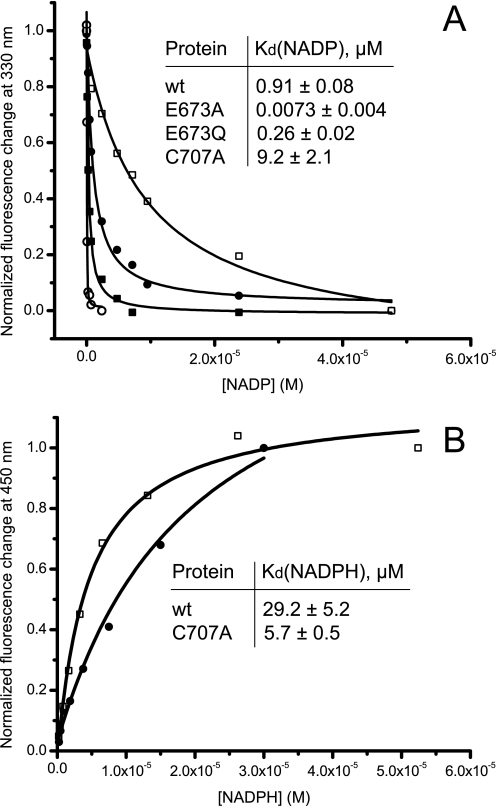
Kd values for NADP+ were determined based on quenching of tryptophan fluorescence upon binding of the coenzyme (39). The candidate tryptophan residues in the structure of Ct-FDH with bound NADP+ (PDB code 2O2Q) are Trp-573 located about 4 Å from the pyrophosphate group of the coenzyme and Trp-582 located about 8 Å from the nicotinamide ring. In our experiments, protein fluorescence was decreased by up to 40% for wild-type and mutant Ct-FDH upon NADP+ addition (data not shown). In agreement with the fact that the coenzyme was bound irreversibly to the E673A mutant at physiological pH, we observed a strong decrease in Kd for NADP+ binding indicating an about 100-fold increase in the affinity (Fig. 7A). Mutation of the glutamate to a glutamine also resulted in a decrease in Kd in about 3-fold (Fig. 7A). In contrast, the replacement of Cys-707 with an alanine has increased Kd about 10-fold (Fig. 7A).
FIGURE 7.
Equilibrium binding of NADP+ (A) or NADPH (B) to wild-type Ct-FDH and its mutants. Insets show Kd values calculated from these experiments. All measurements were done in duplicates. Non-linear curve fitting was performed using Origin 8 (OriginLab). Closed circles, wild-type Ct-FDH; open squares, C707A; open circles, E673A; closed squares, E673Q.
The above method cannot be used to measure Kd values for NADPH binding due to the fluorescence resonance energy transfer from tryptophan residues to NADPH (data not shown). Instead, changes in NADPH fluorescence at 450 nm were monitored after excitation at 330 nm (binding to Ct-FDH leads to dequenching of NADPH fluorescence) (39). These experiments demonstrated that replacement of Cys-707 with an alanine decreases Kd for NADPH about 5-fold (Fig. 7B).
DISCUSSION
Catalytic Glutamate Is Critical for Mobility and Correct Positioning of Coenzyme
Several studies have suggested that the invariant catalytic glutamate of ALDHs (Glu-268 in class 1 and 2 enzymes) participates in both acylation and deacylation stages of the ALDH catalysis (5, 7, 23, 25). Specifically, it has been proposed that in the first step it deprotonates the catalytic cysteine prior to nucleophilic attack on the substrate, whereas in the second step it activates a water molecule to facilitate the hydrolysis of the acyl-enzyme intermediate. In agreement with this mechanism, we have previously shown that the conserved glutamate of Ct-FDH (Glu-673) is an essential catalytic residue (10). In the present study we demonstrate that, in addition to its role in catalysis, Glu-673 of Ct-FDH defines the positioning of the nicotinamide moiety of NADP+ in the active center as well as prevents excessively tight binding of the coenzyme.
Replacement of glutamate with an alanine traps NADP+ in the extended conformation within the binding pocket, resulting in a coenzyme-protein complex that is essentially irreversible at physiological conditions. Indeed, the apparent Kd for the mutant was in a low nanomolar range, a 100-fold decrease compared with the wild-type protein. The tighter binding of the coenzyme could potentially result from conformational changes or misfolding induced by the mutation. However, the two crystal structures harboring the E673A mutation (E673A mutant and E673A/C707A double mutant) do not show any detectable conformational deviations from WT Ct-FDH and display the conformations of NADP+ indistinguishable from that in the wild-type protein. Another possible explanation for the enhanced coenzyme binding in the E673A mutant could be the inability to reverse the transient covalent bond between Cys-707 and the C4 atom of the nicotinamide ring demonstrated in WT Ct-FDH (10). The structure of the E673A/C707A double mutant, which cannot form a covalent bond with the coenzyme, ruled out this possibility: this mutant also irreversibly binds NADP+, indicating that the transient covalent bond does not play a role in this enhanced binding of the coenzyme. Therefore, we propose that enabling of coenzyme flexibility is an intrinsic role of Glu-673.
In the crystal structure of the E673Q mutant in complex with NADP+, the glutamine is not displaced by the nicotinamide ring of the coenzyme and maintains contacts with the catalytic cysteine. This arrangement prevents binding of NADP+ in the extended conformation (because Gln-673 blocks the entrance to the catalytic center) but allows binding of the coenzyme in a new conformation that is similar to the canonical contracted conformation in that the nicotinamide ring lies outside of the active site (Fig. 4B). A significant increase in affinity of NADP+ binding (about 3-fold) was observed for the complex between the coenzyme and E673Q mutant. We speculate that this might be a result of enhanced interactions between the coenzyme and helix G (see below for a more detailed discussion). This complex, however, should be catalytically inactive because attaining the position of the nicotinamide ring close to the sulfur atom of the catalytic cysteine is an essential requirement for the hydride transfer in the ALDH reaction (5, 27, 28). These results suggest that the conserved glutamate controls the binding of the coenzyme within the catalytic center.
Of note, replacement of the corresponding conserved glutamate in nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans did not enhance NAD+ binding but trapped the covalent thioacyl-enzyme intermediate (40). Thus, it is not clear at present whether the newly discovered function of the glutamate is a common phenomenon in ALDH enzymes or it is a unique feature of Ct-FDH. However, because Ct-FDH is more closely related to class 1 and 2 ALDH, it would be plausible to suggest that replacement of the glutamate in some members of these two classes produces a similar effect on the coenzyme binding.
Active Site Cysteine Distinguishes between Oxidized and Reduced Forms of Coenzyme
Although numerous studies have demonstrated that oxidized and reduced forms of the coenzyme are bound to ALDHs in distinct conformations (5, 7, 10, 27–29), the mechanism of recognition of NAD(P)+ versus NAD(P)H remains unclear. In our study, in the structures of the binary complexes of the C707A mutant of Ct-FDH with NADP+ or NADPH, the coenzymes were bound in the same (extended) conformation with the nicotinamide moieties clearly present in the active site, which is typically observed only for NAD(P)+ (Fig. 6, B and C). Of note, the binding constants for NADP+ and NADPH were significantly different between the mutant and wild-type protein: the affinity for NADPH was increased and the affinity for NADP+ was simultaneously decreased in the mutant (Fig. 7). Although the enhanced NADPH binding could be explained by a better accommodation of the nicotinamide ring within the active center (as compared with the disordered ring in the wild-type protein), the drop in NADP+ affinity is apparently a result of the absence in the mutant of the transient covalent bond between the cysteine and C4 atom of the nicotinamide ring observed in the wild type protein (10).
Overall, the C707A mutant demonstrates very similar affinity for NADP+ and NADPH, which is in agreement with the fact that both coenzymes occupy the same position in this protein, and is in sharp contrast with the wild-type enzyme properties. Thus, replacement of the catalytic cysteine with an alanine results in loss of the ability to discriminate between the oxidized and the reduced coenzymes, indicating that this residue is important to prevent the nicotinamide of NADPH from entering the active site. Apparently, the thiol of the cysteine experiences a steric clash with the reduced nicotinamide ring of NADPH forcing the ring out of the active site. This is likely to constitute the mechanism of the coenzyme elimination from the catalytic center after the reaction. Of note, the C302S mutant of ALDH2 has been shown to correctly position NAD+ (extended conformation) and NADH (contracted conformation) (27) that indicates the importance of a nucleophile in the catalytic center to differentiate between oxidized and reduced forms of the coenzyme.
Activation of the Catalytic Cysteine
It was postulated that the catalytic cysteine is deprotonated in the initial phase of the ALDH catalysis (23). In the case of class 1 and 2 ALDHs, the likely candidate to abstract a proton from Cys-302 is Glu-268 (5, 10, 20, 22). The glutamate then shuttles the proton to bulk water before participating in the deacylation step of the reaction (10, 25). Crystal structures of wild type Ct-FDH in complex with NADP+ gave further support of the cysteine deprotonation: the covalent bond between its sulfur atom and the nicotinamide ring, predicted based on quantum mechanical/molecular mechanical simulations in ALDH2 (41, 42) and directly observed in our structure, is possible only upon the loss of a proton from the sulfhydryl group (10). Based on the postulated mechanism, we proposed that the glutamate acts as the proton acceptor. If this is true, the substitution of the glutamate with an alanine should prevent deprotonation of the cysteine. However, the present study indicates a covalent bond between the sulfur atom of the cysteine and the C4 atom of the nicotinamide ring in two out of four subunits of the E673A mutant. Thus, in Ct-FDH the catalytic cysteine can be activated in the absence of glutamate 673, presumably through stabilization of a thiolate by the positive charge of either the nicotinamide ring of NADP+ or the main chain amides of surrounding residues, as suggested earlier (5). Regardless, Glu-673 undoubtedly facilitates formation of the thiolate. We propose that these two activation mechanisms are involved simultaneously to accelerate the deprotonation of the catalytic cysteine.
Communication between the Active Center and the Nucleotide-binding Domain
Numerous studies of ALDHs showed that the adenine moiety of the coenzyme is largely responsible for binding, whereas the nicotinamide ring and the adjacent ribose make just a few contacts with the protein (5, 7, 8, 10, 27). Our data suggest that Glu-673 plays a critical role in the accommodation of the coenzyme within the Ct-FDH molecule. However, rather than providing additional binding contacts, the glutamate plays the role of a restrictor by preventing tighter binding of the coenzyme. Indeed, the replacement of this residue with either alanine or glutamine strongly increases binding affinity. It is unlikely that the mechanism for this phenomenon is a direct repulsion because the interactions of Glu-673 and the carboxyamide group of the nicotinamide ring are limited to only two potential hydrogen bonds (10). Instead our studies point toward a different mechanism that involves communication between the catalytic and nucleotide-binding domains. In support of this view, introducing a glutamine residue in place of catalytic Glu-673 of Ct-FDH induces a prominent conformational disorder in helix G, which forms one-half of the site for binding of the adenine part of the coenzyme (Fig. 5). Of note, a disorder in the adenine-binding site was described earlier in the “asian variant” (E487K) mutant (15, 16) or the R475Q mutant (43) of human ALDH2. These two residues participate in inter-subunit interactions to stabilize the dimer interface. In the case of Ct-FDH, a minor Glu/Gln alteration in residue 673 located in the catalytic domain propagates to the nucleotide-binding domain of the same subunit indicating communication between these domains (Fig. 5). Therefore, we hypothesize that the exceptionally strong binding of NADP+ in Ct-FDH with an alanine instead of a glutamate at position 673 may be a result of the distant enhancement of interactions with the adenine moiety of NADP+. Although we do not observe significant alterations between the native and the mutant structures, such an enhancement may result from minor movements of the secondary structure elements forming the nucleotide-binding domain.
Interestingly, the E673Q mutation also alters the inter-subunit contacts within the dimer that forms the Ct-FDH tetramer. In particular, Arg-644 hydrogen bonded to Glu-831 of the neighboring subunit in WT Ct-FDH displaces a water molecule to contact the main chain oxygen atom of Lys-876 instead (Fig. 5B). Of note, Arg-644 is located about 20 Å from the active center. Curiously, in a recent paper, Gonzáles-Segura and coauthors (25) suggested that this “crystallographic water” molecule bound at the C terminus of helix G could be a potassium atom. Located in the inter-subunit interface, this atom seems to be critical for maintaining the tetrameric state of Pseudomonas aeruginosa betaine ALDH (44). The authors proposed that binding of a stabilizing potassium or sodium atom at this position could be a general feature of ALDH. Whether the “signals” from the active site that we have demonstrated here may influence the stability of the tetramer of Ct-FDH will await experimental verification.
Supplementary Material
Acknowledgments
We thank Dr. Christopher Davies for helpful discussions. The x-ray crystallography facility used for this work is supported by the Medical University of South Carolina's Research Resource Facilities program. Data were also collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory.
This work was supported, in whole or in part, by National Institutes of Health Grant DK54388 (to S. A. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and materials.
- ALDH
- aldehyde dehydrogenase
- FDH
- 10-formyltetrahydrofolate dehydrogenase
- Ct-FDH
- C-terminal domain of FDH
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Perozich J., Nicholas H., Wang B. C., Lindahl R., Hempel J. (1999) Protein Sci. 8, 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marchitti S. A., Brocker C., Stagos D., Vasiliou V. (2008) Expert Opin. Drug. Metab. Toxicol. 4, 697–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang M. F., Han C. L., Yin S. J. (2009) Chem. Biol. Interact. 178, 36–39 [DOI] [PubMed] [Google Scholar]
- 4. Liu Z. J., Sun Y. J., Rose J., Chung Y. J., Hsiao C. D., Chang W. R., Kuo I., Perozich J., Lindahl R., Hempel J., Wang B. C. (1997) Nat. Struct. Biol. 4, 317–326 [DOI] [PubMed] [Google Scholar]
- 5. Steinmetz C. G., Xie P., Weiner H., Hurley T. D. (1997) Structure 5, 701–711 [DOI] [PubMed] [Google Scholar]
- 6. Lamb A. L., Newcomer M. E. (1999) Biochemistry 38, 6003–6011 [DOI] [PubMed] [Google Scholar]
- 7. Moore S. A., Baker H. M., Blythe T. J., Kitson K. E., Kitson T. M., Baker E. N. (1998) Structure 6, 1541–1551 [DOI] [PubMed] [Google Scholar]
- 8. Johansson K., El-Ahmad M., Ramaswamy S., Hjelmqvist L., Jörnvall H., Eklund H. (1998) Protein Sci. 7, 2106–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cobessi D., Tête-Favier F., Marchal S., Azza S., Branlant G., Aubry A. (1999) J. Mol. Biol. 290, 161–173 [DOI] [PubMed] [Google Scholar]
- 10. Tsybovsky Y., Donato H., Krupenko N. I., Davies C., Krupenko S. A. (2007) Biochemistry 46, 2917–2929 [DOI] [PubMed] [Google Scholar]
- 11. Zakhari S. (2006) Alcohol Res. Health 29, 245–254 [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Z., Foster M. W., Zhang J., Mao L., Rockman H. A., Kawamoto T., Kitagawa K., Nakayama K. I., Hess D. T., Stamler J. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12159–12164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen C. H., Budas G. R., Churchill E. N., Disatnik M. H., Hurley T. D., Mochly-Rosen D. (2008) Science 321, 1493–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoshida A., Huang I. Y., Ikawa M. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larson H. N., Weiner H., Hurley T. D. (2005) J. Biol. Chem. 280, 30550–30556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larson H. N., Zhou J., Chen Z., Stamler J. S., Weiner H., Hurley T. D. (2007) J. Biol. Chem. 282, 12940–12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farrés J., Wang X., Takahashi K., Cunningham S. J., Wang T. T., Weiner H. (1994) J. Biol. Chem. 269, 13854–13860 [PubMed] [Google Scholar]
- 18. Perez-Miller S., Younus H., Vanam R., Chen C. H., Mochly-Rosen D., Hurley T. D. (2010) Nat. Struct. Mol. Biol. 17, 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feldman R. I., Weiner H. (1972) J. Biol. Chem. 247, 267–272 [PubMed] [Google Scholar]
- 20. Sheikh S., Ni L., Hurley T. D., Weiner H. (1997) J. Biol. Chem. 272, 18817–18822 [DOI] [PubMed] [Google Scholar]
- 21. Marchal S., Cobessi D., Rahuel-Clermont S., Tête-Favier F., Aubry A., Branlant G. (2001) Chem. Biol. Interact. 130–132, 15–28 [DOI] [PubMed] [Google Scholar]
- 22. Mann C. J., Weiner H. (1999) Protein Sci. 8, 1922–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X., Weiner H. (1995) Biochemistry 34, 237–243 [DOI] [PubMed] [Google Scholar]
- 24. Wymore T., Hempel J., Cho S. S., Mackerell A. D., Jr., Nicholas H. B., Jr., Deerfield D. W., 2nd (2004) Proteins 57, 758–771 [DOI] [PubMed] [Google Scholar]
- 25. González-Segura L., Rudiño-Piñera E., Muñoz-Clares R. A., Horjales E. (2009) J. Mol. Biol. 385, 542–557 [DOI] [PubMed] [Google Scholar]
- 26. Marchal S., Rahuel-Clermont S., Branlant G. (2000) Biochemistry 39, 3327–3335 [DOI] [PubMed] [Google Scholar]
- 27. Perez-Miller S. J., Hurley T. D. (2003) Biochemistry 42, 7100–7109 [DOI] [PubMed] [Google Scholar]
- 28. Hammen P. K., Allali-Hassani A., Hallenga K., Hurley T. D., Weiner H. (2002) Biochemistry 41, 7156–7168 [DOI] [PubMed] [Google Scholar]
- 29. Talfournier F., Pailot A., Stinès-Chaumeil C., Branlant G. (2009) Chem. Biol. Interact. 178, 79–83 [DOI] [PubMed] [Google Scholar]
- 30. Ahvazi B., Coulombe R., Delarge M., Vedadi M., Zhang L., Meighen E., Vrielink A. (2000) Biochem. J. 349, 853–861 [PMC free article] [PubMed] [Google Scholar]
- 31. Krupenko S. A. (2009) Chem. Biol. Interact. 178, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krupenko S. A., Wagner C., Cook R. J. (1997) J. Biol. Chem. 272, 10266–10272 [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 34. Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 35. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 36. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 37. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 38. Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 39. Takahashi K., Weiner H., Hu J. H. (1980) Arch. Biochem. Biophys. 205, 571–578 [DOI] [PubMed] [Google Scholar]
- 40. D'Ambrosio K., Pailot A., Talfournier F., Didierjean C., Benedetti E., Aubry A., Branlant G., Corbier C. (2006) Biochemistry 45, 2978–2986 [DOI] [PubMed] [Google Scholar]
- 41. Hempel J., Nicholas H. B., Jr., Brown S. T., Wymore T. (2007) in Enzymology and Molecular Biology of Carbonyl Metabolism (Weiner H., Maser E., Lindahl R., Plapp B. eds) pp. 9–13, Purdue University Press, West Lafayette, IN [Google Scholar]
- 42. Wymore T., Deerfield D. W., 2nd, Hempel J. (2007) Biochemistry 46, 9495–9506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hurley T. D., Perez-Miller S., Breen H. (2001) Chem. Biol. Interact. 130–132, 3–14 [DOI] [PubMed] [Google Scholar]
- 44. Valenzuela-Soto E. M., Velasco-García R., Mújica-Jiménez C., Gaviria-González L. L., Muñoz-Clares R. A. (2003) Chem. Biol. Interact. 143–144, 139–148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.