Abstract
Neuron polarization is essential for the formation of one axon and multiple dendrites, establishing the neuronal circuitry. Phosphoinositide 3-kinase (PI3K) signaling promotes axon selection and elongation. Here we report in hippocampal neurons siRNA knockdown of the proline-rich inositol polyphosphate 5-phosphatase (PIPP), which degrades PI3K-generated PtdIns(3,4,5)P3, results in multiple hyperelongated axons consistent with a polarization defect. We identify collapsin response mediator protein 2 (CRMP2), which regulates axon selection by promoting WAVE1 delivery via Kinesin-1 motors to the axon growth cone, as a PIPP-interacting protein by Y2H screening, direct binding studies, and coimmunoprecipitation of an endogenous PIPP, CRMP2, and Kinesin-1 complex from brain lysates. The C-terminal growth cone-targeting domain of PIPP facilitates its interaction with CRMP2. PIPP growth cone localization is CRMP2-dependent. PIPP knockdown in PC12 cells promotes neurite elongation, WAVE1 and Kinesin-1 growth cone localization, whereas knockdown of CRMP2 exhibits the opposite phenotype, with shorter neurites and decreased WAVE1/Kinesin-1 at the growth cone. In contrast, CRMP2 overexpression promotes neurite elongation, a phenotype rescued by full-length PIPP, or expression of the CRMP2-binding PIPP domain. Therefore this study identifies PIPP and CRMP2 exert opposing roles in promoting axon selection and neurite elongation and the complex between these proteins serves to regulate the localization of effectors that promote neurite extension.
Keywords: Axon, Neurobiology, Phosphatidylinositol Signaling, Phosphatidylinositol Phosphatase, PI 3-Kinase, Collapsin Response Mediator Protein 2, Proline-rich Inositol Polyphosphate 5-Phosphatase, Neurite Outgrowth, Neuronal Polarity, Protein-Protein Interaction
Introduction
Neurons are highly polarized cells that contain one long axon, which transmits information, and multiple shorter dendrites, which receive information (1, 2). The formation of axon/dendrite polarity is critical for normal brain function. Neurons form networks connecting via synapses to other neurons, by extending axons in a process of neurite outgrowth and elongation. An actin-rich area at the tip of extending neurites, called the growth cone, enables neurite extension. Many signaling and actin regulatory factors function at the growth cone to promote neurite outgrowth and extension (2).
The phosphoinositide 3-kinase (PI3K)/Akt signaling pathway promotes both neuronal polarity and neurite extension. In response to nerve growth factor (NGF) stimulation PI3K phosphorylates PtdIns(4,5)P23 to form PtdIns(3,4,5)P3, a lipid signal that promotes neurite differentiation (3). In elongating neurons PtdIns(3,4,5)P3 activates Rho GTPases Cdc42 and Rac1 that in turn regulate growth cone actin dynamics facilitating growth cone advancement (4, 5). PtdIns(3,4,5)P3 also binds and activates the serine/threonine kinase Akt, which phosphorylates numerous targets, including Tau, GSK3β, Ezrin, and Pak1 to regulate neuronal polarity, neurite outgrowth, and elongation (6, 7). GSK3β is inactivated by Akt phosphorylation (8) in the nascent growth cone (7, 9, 10). GSK3β effectors include CRMP2 (11, 12) and adenomatous polyposis coli (10, 13), which promote microtubule polymerization in the nascent axon driving neurite elongation (14).
Termination of PI3K/Akt signaling is mediated by PtdIns(3,4,5)P3 degradation by two pathways, either by the 3-phosphatase PTEN, or the inositol polyphosphate 5-phosphatases, forming PtdIns(4,5)P2 or PtdIns(3,4)P2, respectively (15). PTEN inhibits hippocampal neuron differentiation and plays a central role in promoting and maintaining neuronal polarity. The inositol polyphosphate 5-phosphatases contain 10 mammalian members, and many degrade PtdIns(3,4,5)P3 to form PtdIns(3,4)P2 and thereby inhibit Akt signaling (16, 17). Two family members, the proline-rich inositol polyphosphate 5-phosphatase (PIPP) and the SH2 domain 5-phosphatase (SHIP2), inhibit NGF-stimulated PI3K/Akt-dependent neurite elongation (16, 17). RNAi knockdown of PIPP in PC12 cells enhances NGF-stimulated microtubule polymerization, leading to increased PtdIns(3,4,5)P3, phospho-Ser473-Akt, and phospho-Ser9-GSK3β at the growth cone (16). Therefore PIPP, by degrading PtdIns(3,4,5)P3, reduces Akt activation at the growth cone, inhibiting neurite extension.
5-Phosphatases act as inhibitors of PI3K/Akt signaling via their central 5-phosphatase domain that degrades PtdIns(3,4,5)P3, and concomitantly complex via their SH2- or proline-rich domains with other signaling proteins. No such interactions have yet been identified for PIPP, a relatively uncharacterized family member, which contains a central 5-phosphatase catalytic domain flanked by extensive N- and C-terminal proline-rich domains. Here we identify PIPP interacting partners as CRMP2 and tubulin by Y2H screening of brain libraries. CRMP2 is a neuronal adaptor protein that interacts with many binding partners, promoting axon polarity, neurite outgrowth, and elongation (18, 19). PIPP and CRMP2 both independently bind β-tubulin and form a multiprotein complex with the motor protein, Kinesin-1. CRMP2 is essential for the proper localization of PIPP to the growth cone, however, PIPP opposes CRMP2-mediated neurite elongation and localization of its actin regulatory effector, WAVE1, to the growth cone. Collectively, these studies identify PIPP complexes with and opposes the function of CRMP2 to inhibit neurite elongation.
EXPERIMENTAL PROCEDURES
Materials
DNA-modifying and restriction enzymes were obtained from MBI Fermentas (Burlington, Canada), New England Biolabs (Beverly, MA), or Promega (Madison, WI). Oligonucleotides were from Micromon (Monash University, Australia) or Geneworks (Adelaide, Australia). Big Dye version 3.1 sequencing terminators were purchased from Invitrogen. Cell lines were from American Type Culture Collection. Alexa Fluor 488-, Alexa Fluor 594-, Alexa Fluor 568-, or Alexa Fluor 647-conjugated anti-mouse or rabbit antibodies and Texas Red-conjugated or Alexa Fluor 488-conjugated phalloidin were from Molecular Probes (Eugene, OR). Antibodies utilized include anti-CRMP2 (IBL, Hamburg, Germany), anti-HA monoclonal (Covance, Richmond, CA), anti-HA polyclonal (Upstate Biotechnology, Waltham, MA), anti-FLAG M2 (Sigma), anti-GST (Amersham Biosciences), anti-MAP2 (Sigma), anti-Tau-1 (Chemicon, Billerica, MA), anti-β-tubulin (Zymed Laboratories Inc., San Francisco, CA), anti-Myc (Cell Signaling, Beverly, MA), anti-WAVE1 (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-Kinesin heavy chain (KHC)(Chemicon). All other reagents were from Sigma or BDH Chemicals (Kilsyth, Australia) unless otherwise stated.
PIPP-specific Antibodies
A PIPP-specific antipeptide antibody (PIPP-AB1) was generated and characterized as previously described (16). In addition, a PIPP-specific polyclonal antibody (PIPP-AB2) was raised in New Zealand White rabbits to purified full-length recombinant GST-tagged PIPP protein expressed in COS1 cells (supplemental Fig. S1). PIPP-AB2 antibodies were affinity purified by incubating immune sera with GST-PIPP glutathione-Sepharose and eluting antibodies using 0.1 m glycine, pH 2.5. PIPP-specific monoclonal antibodies (PIPP-AB3 and PIPP-AB4) were raised against purified full-length recombinant GST-tagged PIPP by EMBL (Heidelberg, Germany).
Plasmids and Cloning
pGEX-5.1 was purchased from Amersham Biosciences. pCGN (20) was a kind gift from Tony Tiganis (Monash University, Australia). pGBKT7-SKICH/PRD-2 was generated by PCR amplification using primers 5′-GCGAATTCATGCTGTTTGCCTTCAGG-3′ and 5′GCGAATTCTCAGGGCCCCAAACCCCCTTC-3′ and cloned into the EcoRI site of pGBKT7 (Clontech, Mountain View, CA) in-frame with the GAL4 DNA-binding domain bait. pCGN-PIPP has been described previously (16). To generate pCGN-SKICH/PRD-2, the region encoding SKICH/PRD-2 (2179–3012 bp) was PCR amplified using the 5′ primer 5′-GCGGATCCTGATGCTGCAGTTTGCCTTCAGG-3′ and 3′ primer 5′-GCGGATCCTCAGGGCCCCAAACCCCCTTC-3′ and subcloned into the BamHI site of pCGN in-frame with the N-terminal HA tag. pRK5-CRMP1 and other CRMP family constructs are as described (11, 21). pCMV5-GSK3β(S9A) has been described previously (22).
siRNA Design and Construction
Three DNA sequences specific for rat PIB5PA, identified using the Ambion “siRNA target finder,” were used as template for synthesizing siRNA for PIPP knockdown in hippocampal neurons. Double-stranded siRNA was generated from DNA oligonucleotides using the Silencer siRNA construction kit (Ambion, Foster City, CA) according to the manufacturer's instructions. Template sequences were as follows: PIPP1-antisense, 5′-AAGGACCAGCTCAACATGGCCCCTGTCTC-3′, PIPP1-sense, 5′-AAGGCCATGTTGAGCTGGTCCCCTGTCTC-3′, scram1-antisense, 5′-AACGCAGGGAAACTCGCCCATCCTGTCTC-3′, scram1-sense, 5′-AAATGGGCGAGTTTCCCTGCGCCTGTCTC-3′, PIPP2-antisense, 5′-AACTTCCGCATTGAGAGCTATCCTGTCTC-3′, PIPP2-sense, 5′-AAATAGCTCTCAATGCGGAAGCCTGTCTC-3′, scram2-antisense, 5′-AATTCGTCCGACGATTAGATCCCTGTCTC-3′, scram2-sense, 5′-AAGATCTAATCGTCGGACGAACCTGTCTC-3′, PIPP3-antisense, 5′-AACCTACAGACACAAGAAAGCCCTGTCTC-3′, PIPP3-sense, 5′-AAGCTTTCTTGTGTCTGTAGGCCTGTCTC-3′, scram3-antisense, 5′-AAGTAAAGCACACAACGACCACCTGTCTC-3′, scram3-sense, 5′-AATGGTCGTTGTGTGCTTTACCCTGTCTC-3′. For siRNA in PC12 cells, Qiagen siRNA duplexes (AllStars negative control, PIPP target sequence AAGGACCAGCTCAACATGGCC or CRMP2 target sequence AAACTCCTTCCTCGTGTACAT) were labeled with Cy3 using the Silencer® siRNA Labeling Kit (Ambion) according to the manufacturer's instructions.
Analysis of PIPP Knockdown by Quantitative RT-PCR
RNA was extracted from PIPP or control siRNA-transfected PC12 cells using the RNeasy Miniprep kit (Qiagen, AL) and cDNA synthesized using the AffinityScript QPCR cDNA Synthesis kit (Agilent, La Jolla, CA) according to the manufacturer's instructions. QRT-PCR was performed using the Brilliant II SYBR Green QPCR Master Mix kit (Agilent) with PIPP or GAPDH-specific QuantiTect Primer Assays (Qiagen) according to the manufacturer's instructions. Reactions were performed in triplicate in a Corbett 3000 Rotor Gene Cycler using the following conditions: 1 cycle at 95 °C for 10 min, 40 cycles at 95 °C for 30 s and 60 °C for 1 min. Relative expression of PIPP compared with GAPDH was calculated using the ΔΔCt method as described (23).
Hippocampal Neuron Preparation and Transfection
Hippocampal neuron cultures were prepared from pregnant Sprague-Dawley rats (Monash University Animal Ethics number BAM/B/2004/47) at day 18 gestation as described (24), with slight modifications. Briefly, following dissection hippocampi were digested in 0.25% trypsin in Hank's balanced salt solution at 37 °C for 15 min. Cells were manually dissociated by trituration using a fire-polished Pasteur pipette and plated onto 0.01% poly-l-lysine-coated coverslips at a density of 5 × 105 to 1 × 106/mm2. Following adherence of cells to coverslips, plating media (minimal essential medium, 10% fetal calf serum, 0.6% glucose, penicillin/streptomycin) was replaced with Neurobasal media with B27 supplement and 0.5 mm glutamine. Primary hippocampal neurons were transfected with Cy3-labeled siRNA oligonucleotides using Lipofectamine 2000TM. Neurons were fixed in 4% paraformaldehyde, 0.12 m sucrose in PBS for 20 min.
PC12 Culture, Transfection, and Neurite Elongation Analysis
PC12 cells were maintained in DMEM containing 10% fetal calf serum (FCS), 5% horse serum, 2 mm l-glutamine, 100 units/ml of penicillin, and 0.1% streptomycin. Cells were transfected with plasmid constructs either by electroporation or with Lipofectamine LTX. For electroporation, cells were resuspended in 200 μl of the above medium. A 50-μl mixture containing 5 μg of DNA, 0.15 m NaCl was added to the cells and electroporated at 0.2 kV, 0.975 microfarad. The cells were added to 8 ml of the above medium and incubated at 37 °C with 5% CO2 overnight then plated onto 0.01% poly-l-lysine-coated coverslips. For transfection with Lipofectamine LTX, 0.5 μg of DNA (0.2 μg of FLAG-CRMP2) and 0.5 μl of PLUS reagent were diluted in 100 μl of Opti-MEM, incubated for 10 min at room temperature, and 1.25 μl of Lipofectamine LTX added. The mixture was incubated for 25 min at room temperature then added to PC12 cells plated on 13-mm 0.01% poly-l-lysine-coated coverslips in a 24-well dish with 0.5 ml of DMEM, 10% FCS, 5% horse serum and incubated at 37 °C overnight. Transfected cells were differentiated in DMEM containing 1% horse serum and 100 ng/ml of NGF for 3 days. For transient transfections with siRNA duplexes, 160 pmol of labeled siRNA and 10 μl of Lipofectamine were diluted in 500 μl of Opti-MEM, incubated 20 min, then added to PC12 cells plated in a 6-well dish with 2 ml of DMEM, 10% FCS, 5% horse serum. Cells were incubated overnight at 37 °C, plated onto 0.01% poly-l-lysine-coated coverslips, and differentiated in DMEM, 1% horse serum and 50 ng/ml of NGF for 3 days. For transient co-transfections with plasmid constructs and siRNA duplexes, 1 μg of plasmid DNA, 70 pmol of labeled siRNA, and 2 μl of Lipofectamine 2000 were each diluted in 100 μl of Opti-MEM and incubated for 5 min. The DNA/siRNA and Lipofectamine 2000 mixtures were then combined, incubated for 20 min at room temperature, and added to PC12 cells plated in a 12-well dish with 1 ml of DMEM, 10% FCS, 5% horse serum. Cells were incubated overnight at 37 °C, plated onto 0.01% poly-l-lysine-coated coverslips, and differentiated in DMEM, 1% horse serum, and 50 ng/ml of NGF for 3 days.
COS-1 Culture and Transfection
COS-1 cells were maintained in DMEM supplemented with 10% FCS, 2 mm l-glutamine, 100 units/ml of penicillin, and 0.1% streptomycin. For transient transfection, 200 μl of DEAE-dextran/chloroquine (400 μg/ml of DEAE-dextran, 100 μm chloroquine), 5 μg of DNA, and 5 ml of serum-free DMEM was added to a 50% confluent 10-cm dish and incubated 2 h at 37 °C. Cells were treated with 10% dimethyl sulfoxide/PBS for 2 min at room temperature and then incubated in DMEM, 10% fetal calf serum for 2 days at 37 °C.
Yeast Two-hybrid Analysis
The yeast two-hybrid Matchmaker III GAL4-based system (Clontech) was used for all yeast two-hybrid studies. AH109 yeast expressing the pGBKT7-SKICH/PRD-2 “bait” were transformed with the human fetal brain library (Clontech) according to the manufacturer's instructions. Yeast plasmid DNA was extracted from positive clones as described (25). Non-autonomous activation of reporter plasmids was confirmed by co-transformation of positive clone plasmids into p53 bait expressing yeast.
GST Pull-down Analysis and Immunoprecipitations
COS-1 cells were transiently transfected via DEAE-dextran/chloroquine with pEBG-PIPP or PEBG alone and pRK5-FLAG-CRMP1, -2, or -4, grown for 2 days and then harvested in Triton extraction buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 mm NaVO4, Roche Complete mini-protease mixture inhibitors). Triton-soluble lysates, collected following extraction for 3 h and centrifugation at 13,000 × g for 10 min at 4 °C, were applied to glutathione-Sepharose 4B beads for 4 h at 4 °C. Beads were washed 6 times in Tris saline before immunoblotting with FLAG antibodies. Constitutively active Myc-GSK3β(S9A) was cotransfected in some experiments.
Brain Immunoprecipitations
Brains collected from adult mice (Monash University Animal Ethics number BAMB/B/2006/57) were homogenized 4 times for 20 s in 4 volumes of lysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm PMSF, 1 mm NaVO4, 1 mm NaF, Roche Complete mini protease mixture inhibitors). Samples were centrifuged at 100,000 × g for 1 h at 4 °C and Triton-soluble lysates were precleared with protein A-Sepharose beads for 90 min. Triton-soluble lysates (1–1.5 mg of protein) were incubated with protein A-Sepharose and PIPP-AB2 overnight at 4 °C. Beads were washed twice with Tris saline, 1% Triton X-100 and then 4 times with Tris saline before immunoblotting.
Direct Binding Assays
Recombinant GST or GST-tagged CRMP proteins were produced by induction of Escherichia coli transformed with pGEX-5.1, pGEX-CRMP1, pGEX-CRMP2, or pGEX-CRMP4. Cells were grown at 37 °C to an A600 of 0.4 and induced with 0.05 μm isopropyl 1-thio-β-d-galactopyranoside at 26 °C overnight. Cells were collected by centrifugation at 4 °C and lysed in a 5% volume of Triton extraction buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1 mm PMSF, p8849 liquid protease inhibitors (Roche Applied Science)) for 2 h. Triton-soluble lysates collected following centrifugation at 4 °C at 13,000 × g were incubated with glutathione-Sepharose 4B beads (Amersham Biosciences) for 4 h. Beads were washed 6 times in Tris saline. The TnT T7 Coupled Wheat Germ Extract System (Promega) was used as directed by the manufacturer for in vitro transcription/translation of Myc-SKICH/PRD2 from pGBKT7-SKICH/PRD-2 plasmid template. One in vitro transcription/translation reaction was added to each GST sample in a 1-ml volume. Samples were incubated at 4 °C for 4 h, washed 6 times in Tris saline, and subjected to immunoblotting.
Western Blot Analysis
Primary hippocampal neuron cultures or transiently transfected COS-1 cells were washed twice in PBS, lysed in Tris saline (20 mm Tris, pH 7.4, 150 mm NaCl) with 1% Triton X-100, 1 mm PMSF, Roche complete mini protease mixture inhibitor tablet), incubated at 4 °C for 2 h, and ultracentrifuged at 100,000 × g for 1 h. Protein concentration was determined using Bio-Rad DC protein assay kit. 50 μg of protein was separated by SDS-PAGE and subjected to immunoblot analysis using PIPP antibodies. Relative protein levels in each sample were determined by densitometry.
Differentiated, siRNA-transfected PC12 cells were washed once with Tris saline and then lysed in Tris saline with 1% Triton X-100, 1 mm EDTA, 0.2 μg/ml of aprotinin, 0.2 μg/ml of leupeptin, 1 mm PMSF, 1 mm benzamidine, 2 mm β-glycerophosphate, 2 mm sodium pyrophosphate, 1 mm sodium vanadate, 1 mm NaF for 2 h at 4 °C. Lysates were centrifuged at 13,000 × g for 5 min and then the soluble fraction was immunoblotted using polyclonal PIPP-AB2 or monoclonal CRMP2 antibodies. Immunoblots were re-probed with a β-tubulin antibody as a loading control and relative protein expression was determined by densitometry.
Indirect Immunofluorescence
Cells were fixed in 4% paraformaldehyde, 0.12 m sucrose in PBS for 15–20 min, permeabilized in 0.1% Triton X-100/PBS for 2 min, washed three times with PBS, and blocked with 1% BSA/PBS for 15–30 min. Cells were incubated with primary then secondary antibodies as indicated for 1 h each and coverslips were mounted in Slow Fade GOLD. Where indicated, Alexa Fluor-conjugated phalloidin was added during the secondary antibody incubation. Cells were viewed on a Leica TCS-NT confocal microscope with an Ar-Kr triple-line laser at Monash Micro Imaging (Monash University, Australia). Graded fluorescence images were prepared in ImageJ (LUT: Fire). For indirect immunofluorescence of polymerized tubulin, tubulin monomers were extracted and cells were fixed as previously described (26) by incubation in 60 mm PIPES, 25 mm HEPES, 10 mm EGTA, 2 mm MgCl2, pH 6.9, 4% paraformaldehyde, 0.15% glutaraldehyde, 0.2% Triton X-100 for 15 min. Cells were prepared for microscopy, stained with a monoclonal β-tubulin antibody, and imaged as above.
Image Analysis
Image analysis was performed using ImageJ software (27) or AnalysisD. Laser attenuation settings were the same for all images in individual experiments where fluorescence intensity was compared. Fluorescence intensity of phalloidin, WAVE1, KHC, and PIPP immunostaining in the growth cone versus the neurite shaft was determined by outlining the growth cone and neurite shaft of transfected cells and measuring the average pixel intensity. Neurite length was calculated by manually tracing the path of the longest neurite in each cell. Growth cone area was measured by manually tracing around the perimeter of the growth cone of the longest neurite. Levels of polymerized tubulin in neurites of transfected neurites were determined in AnalysisD by masking threshold neurites and recording mean gray value within the masked area. Statistical significance was determined using a paired or unpaired Student's t test where appropriate and in this study p < 0.05 was considered to be statistically significant.
RESULTS
Y2H Screening Identifies βII Tubulin and CRMP1 as PIPP-binding Partners
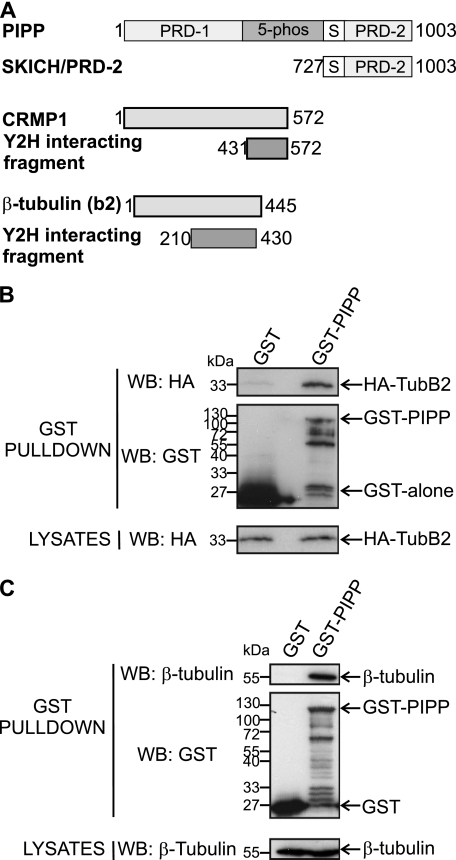
PIPP contains N- and C-terminal proline-rich domains (PRDs), a SKICH domain, and a 5-phosphatase catalytic domain (16, 28) (Fig. 1A). PIPP localizes to the growth cone of NGF-stimulated PC12 cells, and is targeted to this site by its C-terminal SKICH domain (16, 29). We investigated if PIPP complexes with other proteins that regulate neurite outgrowth or differentiation by undertaking a Y2H screen of a human fetal brain library using the C-terminal proline-rich domain (727–1003 amino acids) of PIPP as bait (PIPPSKICH/PRD-2). Two binding partners were identified, TUBB2 (NCBI accession NP001060, amino acids 210–430) and CRMP1 (NCBI accession Q14194, amino acids 431–572) (Fig. 1A). TUBB2, also named βII tubulin, heterodimerizes with α-tubulin. The α- and β-tubulins represent the major components of microtubules (30). To confirm that PIPP and the TubB2 fragment (amino acids 210–430) interact, GST or GST-PIPP were coexpressed in COS-1 cells with HA-TubB2 and lysates were incubated with glutathione-Sepharose. HA-TubB2 was detected in GST-PIPP, but not GST pulldowns (Fig. 1B). To evaluate whether GST-PIPP binds endogenous β-tubulin, cell lysates were incubated with GST or GST-PIPP bound beads. A 55-kDa β-tubulin immunoreactive polypeptide was detected in GST-PIPP, but not GST pulldowns (Fig. 1C).
FIGURE 1.
PIPP complexes with β-tubulin. A, PIPP domains include proline-rich (PRD-1 and PRD-2), catalytic (5-phos), and SKICH domains (S). A DNA-BD/SKICH/PRD-2 fusion protein was used in yeast two-hybrid screening of a fetal brain library identifying CRMP1 (amino acids 431–572) and βII-tubulin (amino acids 210–430). B, COS-1 cells were co-transfected with HA-TubB2 and GST or GST-PIPP and lysates incubated with glutathione-Sepharose, pellets were washed and immunoblotted (WB) with HA (upper panel) or GST (middle panel) antibodies. Lysates were immunoblotted with HA antibodies (lower panel). C, COS-1 cells were transfected with GST or GST-PIPP and lysates were incubated with glutathione-Sepharose, washed, and immunoblotted with β-tubulin (upper panel) or GST (middle panel) antibodies. Lysates were immunoblotted with β-tubulin antibodies (lower panel). WB, Western blot.
PIPP Binds CRMP Family Members
To test for an interaction between PIPP and CRMP family members (1, 2, and 4) and also to confirm that the C-terminal fragment of PIPP (PIPPSKICH/PRD-2) directly binds to CRMP1, an in vitro direct binding assay was performed. CRMPs are multifunction adaptor proteins that complex with many other proteins, so for initial experiments we used a cell-free assay. PIPPSKICH/PRD-2 directly bound GST-CRMP1, GST-CRMP2, and GST-CRMP4, but not GST (Fig. 2A). CRMP-(1–4) proteins exhibit sequence identity and are also highly enriched in the brain (31). To show PIPP and CRMPs form a complex in intact cells, GST-PIPP or GST was co-expressed in COS-1 cells with FLAG-CRMP1, -2, or -4 and complexes were captured on glutathione-Sepharose 4B beads. CRMPs (1, 2, and 4) bound GST-PIPP, but not GST (Fig. 2B). To show an interaction between endogenous CRMPs and endogenous PIPP, we generated polyclonal (PIPPAB2) and monoclonal antibodies (PIPPAB3 and -4) to purified recombinant GST-PIPP (supplemental Fig. S1A). PIPP antibody (PIPP-AB2) detected HA-PIPP and endogenous PIPP, but not other HA-tagged 5-phosphatases expressed in COS-1 cells (supplemental Fig. S1B), and also recognized endogenous PIPP as a ∼107-kDa polypeptide, consistent with its molecular weight, in mouse brain and PC12 cells (supplemental Fig. S1C). Endogenous PIPP was immunoprecipitated from adult mouse brain in complex with endogenous CRMP2 (Fig. 2C), but not CRMP1. However, we did not detect an endogenous PIPP·β-tubulin complex, despite showing endogenous β-tubulin in GST-PIPP pulldowns (see Fig. 1C). β-Tubulin adhered nonspecifically to the Sepharose beads requiring high stringency washing to remove it from the beads and under these conditions we could not detect β-tubulin in PIPP immunoprecipitates. Interestingly, the CRMP2 binding protein, Kinesin-1, was detected in the endogenous brain PIPP·CRMP2 complex as shown by immunoblotting using KHC antibodies (Fig. 2C). Therefore PIPP complexes with CRMP2 and its binding partner the motor protein Kinesin-1.
FIGURE 2.
PIPP binds CRMP1, -2, and -4. A, GST, GST-CRMP1, GST-CRMP2, or GST-CRMP4 were purified from E. coli and incubated with in vitro transcribed/translated Myc-SKICH/PRD-2. GST proteins were captured on glutathione-Sepharose, pellets were washed and immunoblotted with Myc (upper panel) or GST antibodies (lower panel). WB, Western blot. B, COS-1 cells were transiently cotransfected with FLAG-CRMP1, (FLAG-C1), FLAG-CRMP2 (FLAG-C2), or FLAG-CRMP4 (FLAG-C4) and either GST or GST-PIPP. Cell lysates were incubated with glutathione-Sepharose, washed, and immunoblotted with FLAG (upper panel) or GST antibodies (middle panel). Lysates were probed with FLAG antibodies (lower panel). C, mouse brain lysates were immunoprecipitated with non-immune or PIPP antibodies (PIPP-AB2). Lysates and immunoprecipitated proteins were immunoblotted with PIPP (PIPP-AB3), CRMP2, KHC, or β-tubulin antibodies.
PIPP Regulates Axon Polarity and Neurite Length
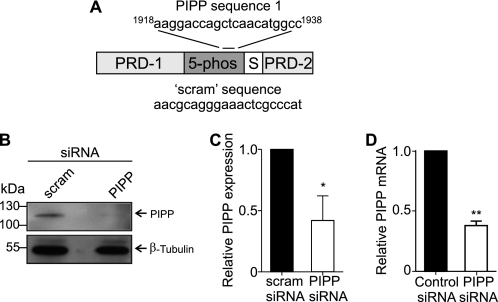
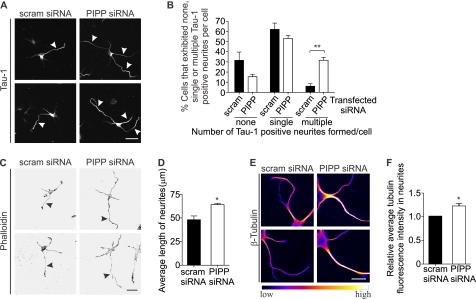
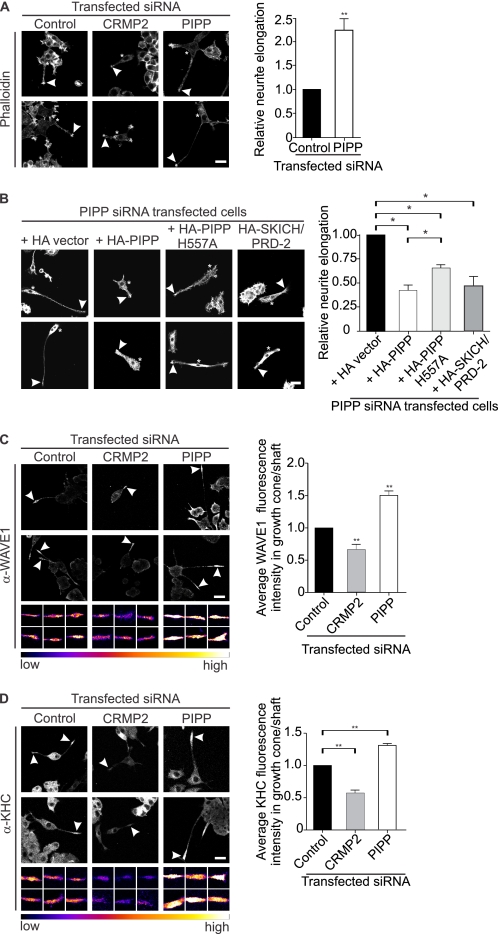
CRMP2 promotes axon/dendrite selection, axon growth cone retraction, and axon elongation (18, 19). We have previously shown that PIPP inhibits neurite extension (16), but have not examined its distribution or function in axons or dendrites. Therefore we investigated the role PIPP plays in neuronal polarization, by its siRNA knockdown in primary hippocampal neurons. Three siRNA oligonucleotides targeted against different regions of the PIPP coding sequence (“PIPP siRNA”) or scrambled controls (“scram siRNA”) were evaluated in the rat-derived PC12 cell line. One sequence (sequence 1, see “Experimental Procedures”) (Fig. 3A) resulted in 63% PIPP protein knockdown by immunoblot analysis (Fig. 3, B and C). Quantitative RT-PCR confirmed a 60% reduction in PIPP mRNA in seq1 siRNA-transfected PC12 cells (Fig. 3D) and this sequence was labeled with Cy3 and transfected into hippocampal neurons. Cy3-siRNA cells exhibited reduced PIPP immunostaining (supplemental Fig. S1D). To assess neuronal polarity, the number of axons were evaluated by analysis of Tau-1, an axon-specific marker. An increase in the percentage of cells that do not form an axon, or that form multiple axons indicates abnormal polarization (32). Cells were scored for having “none,” “single,” or “multiple” Tau-1 positive neurites. PIPP siRNA-transfected cells exhibited multiple axons (PIPP siRNA 31.6 ± 2.7%, versus scrambled siRNA 6.0 ± 2.5%) (Fig. 4, A and B), indicating a polarization defect. The average number of neurites per cell was not affected by PIPP knockdown (PIPP siRNA 4.1 ± 0.2, scrambled siRNA 3.9 ± 0.2), suggesting additional axons are formed at the expense of dendrites. Therefore knockdown of PIPP in hippocampal neurons reveals PIPP is required for proper axon selection. To assess the role of PIPP in regulating neurite elongation, the length of hippocampal neurons transfected with Cy3-labeled siRNAs was determined by manually tracing phalloidin-stained neurites using ImageJ analysis software. At 2 days grown in vitro, PIPP siRNA-transfected cells exhibited a ∼35% increase in neurite length, relative to scrambled siRNA (Fig. 4, C and D), but showed no change in the intensity or distribution of phalloidin-stained F-actin (Fig. 4C). Microtubule polymerization was assessed by extracting tubulin monomers, fixing neurons, and immunostaining polymerized tubulin. PIPP siRNA-transfected neurons exhibited a 1.2-fold increase in neurite-polymerized tubulin (Fig. 4, E and F).
FIGURE 3.
PIPP siRNA knockdown in primary hippocampal neurons. A, PIPP siRNA (PIPP) and control (scram) sequence and PIPP proline-rich domains (PRD-1 and PRD-2), 5-phosphatase (5-phos), and SKICH domain (S) are shown. B, lysates of PC12 cells expressing scrambled or PIPP siRNA were immunoblotted with PIPP (PIPP-AB2) or β-tubulin antibodies. C, PIPP expression was standardized to β-tubulin loading by densitometry and expressed relative to scrambled, which was designated as 1. Bars = mean PIPP expression ± S.E. (n = 4; *, p < 0.05). D, quantitative RT-PCR was performed on cDNA synthesized from mRNA extracted from PC12 cells expressing control or PIPP siRNA using GAPDH as a control. PIPP expression was calculated relative to control, which was designated as 1. Bars = mean PIPP expression ± S.E. (n = 4; **, p < 0.01).
FIGURE 4.
PIPP regulates axon formation and elongation. A, hippocampal neurons were transfected with PIPP or scrambled siRNA and stained with Tau-1 antibodies. Arrowheads indicate Tau1 positive neurites. Bar = 40 μm. B, cells transfected as in A were stained with Tau-1 antibodies and scored for none, single, or multiple Tau-1+ve neurites per cell. C, cells transfected as in A were stained with phalloidin. Arrowheads indicate the longest neurite in each cell. Inverted grayscale images are shown. Bar = 40 μm. D, neurite length of cells treated as in C were measured. Bars = mean length (μm) ± S.E. (20 cells, n = 3, *, p < 0.05). E, cells transfected as in A were stained with β-tubulin antibodies and presented in graded fluorescence. Bar = 20 μm. F, the mean tubulin fluorescence intensity in neurites in cells treated as in E. Bars = mean ± S.E. (20 cells, n = 4, *, p < 0.05). Bars = mean ± S.E. (20 cells, n = 3, **, p < 0.01).
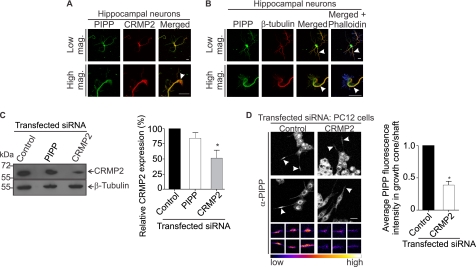
Next PIPP was co-localized with its binding partners CRMP2 or tubulin in hippocampal neurons at 2 and 3 days in vitro (Fig. 5, A and B). PIPP was observed in both MAP2 (dendrite marker) and Tau-1 (axon) positive neurites, in the cell body, along neurites, and in neurite tips (growth cones) at all stages of differentiation, indicating PIPP does not accumulate selectively in the axon or dendrites (supplemental Fig. S2). In neurons CRMP2 is enriched in the axon shaft and growth cone (33). PIPP and CRMP2 colocalized in the cell body, the axon shaft, and showed partial co-localization in the growth cone (see high mag in Fig. 5A). The neuronal growth cone contains an actin-rich peripheral domain with lamellipodial veils and spike-like filopodia and a microtubule-enriched central domain (2, 34). PIPP colocalized with β-tubulin in the central growth cone and also in the axon shaft (Fig. 5B). In control studies PIPP antiserum was pre-absorbed with the antigen to which it was raised, and this serum was not immunoreactive (supplemental Fig. S1E).
FIGURE 5.
CRMP2 and PIPP co-localize in the growth cone and axon shaft and PIPP growth cone localization is CRMP2 dependent. A, hippocampal neurons were stained with PIPP (PIPP-AB1) and CRMP2 antibodies and imaged by confocal microscopy. Arrowhead shows colocalization of PIPP and CRMP2 in the growth cone. Higher power images are shown below. Bar = 20 μm. B, hippocampal neurons were costained with PIPP (PIPP-AB1) and β-tubulin antibodies and with phalloidin (blue) staining. Colocalization of β-tubulin and PIPP is indicated by arrowheads. Higher magnification images are shown below. Bar = 20 μm. C, PC12 cells expressing control, PIPP, or CRMP2 siRNA were immunoblotted with CRMP2 antibodies, then re-probed with β-tubulin antibodies. Relative CRMP2 expression was determined by densitometry and expressed as a % of the scrambled, designated 100%. Bars = mean ± S.E. (n = 5, *, p < 0.05). D, cells transfected with control or CRMP2 siRNA were differentiated for 3 days and costained with PIPP antibodies (PIPP-AB4) and imaged by confocal microscopy at the same laser attenuation. Arrowheads indicate growth cones of Cy3 siRNA-transfected cells. Bar = 20 μm. The smaller panels below show representative examples of additional growth cones imaged at the same laser attenuation and at higher magnification for each indicated transfected siRNA. Growth cones are presented in graded fluorescence. Bars = mean fluorescence intensity of PIPP in the growth cone relative to the neurite shaft ± S.E., where the control is designated 1 (≥10 cells, n = 3, *, p < 0.05).
CRMP2-dependent neurite extension is in part mediated by Kinesin-1-dependent transport of tubulin, Sra-1/WAVE1, and TrkB to the growth cone (35–37). CRMP2 is essential for growth cone localization of these effectors, as it links Sra-1/WAVE1 to Kinesin-1. PIPP also localizes to the growth cone by unknown mechanisms (16). As PIPP complexes with CRMP2, we investigated if CRMP2 is required for access of PIPP to the growth cone. We undertook CRMP2 knockdown in PC12 cells resulting in a ∼50% decrease in CRMP2 protein expression by immunoblot analysis (Fig. 5C). In control knockdown cells PIPP localization was prominent at the growth cone of differentiated PC12 cells, however, in CRMP2 knockdown cells the intensity of PIPP antibody staining at the growth cone was significantly decreased (Fig. 5D, arrowheads). The fluorescence intensity of PIPP immunostaining was determined by the average pixel intensity at the growth cone, relative to the neurite shaft at the same laser attenuation revealing a >50% decrease in PIPP growth cone staining in CRMP2 knockdown cells (Fig. 5D, see graded fluorescence images of growth cones). Therefore PIPP requires its association with CRMP2 to access the growth cone.
The CRMP2·PIPP Complex Is Reduced by GSK3β Phosphorylation of CRMP2
We next investigated whether PIPP and CRMP2 form a constitutive or regulated complex. PIPP is relatively uncharacterized and the regulators of its activity, localization, and/or function(s) are unknown. In contrast, CRMP2 is dynamically regulated by phosphorylation by Cdk5, DYRK2, GSK3β, Rho kinase (ROCK), or Fes and this may affect its interaction with its many binding partners (11, 12, 21, 38–41). CRMP2 phosphorylation is complex and requires an initial Cdk5 priming phosphorylation at Ser522, followed by GSK3β phosphorylation at Ser518, Thr514, and Thr509, which reduces CRMP2-tubulin binding (11, 40, 42) (Fig. 6A). To determine whether CRMP2 phosphorylation is required for PIPP association, a CRMP2(S522A) mutant, which cannot be phosphorylated by Cdk5 or subsequently by GSK3β (11), was evaluated for PIPP binding. CRMP2(S522A) bound GST-PIPP marginally less than wild type CRMP2, however, analysis of 4 independent experiments revealed this reduction was not statistically significant (Fig. 6, B and C). Therefore PIPP·CRMP2 association does not require CRMP2 phosphorylation by Cdk5 or GSK3β. However, this does not exclude the possibility that CRMP2 phosphorylation by GSK3β may regulate the relative level of PIPP binding to CRMP2. AGC family kinases phosphorylate Ser9 of GSK3β, which in turn inhibits GSK3β activity toward its primed substrates, including CRMP2 (12, 43). GSK3β-Ser9 mutation to alanine prevents its Akt-mediated inactivation, rendering it constitutively active (44). GST or GST-PIPP were coexpressed with FLAG-CRMP2 and increasing levels of Myc-GSK3β(S9A) resulting in a decrease (∼60%) in GST-PIPP-bound FLAG-CRMP2 (Fig. 6, D and E). In control studies no difference in the PIPP·CRMP2 complex was noted when GST-PIPP was coexpressed with CRMP2(S522A), which cannot be phosphorylated by Cdk5 and subsequently by GSK3β and Myc-GSK3β(S9A) indicating the decrease in the complex was due to CRMP2 rather than PIPP phosphorylation (Fig. 6F). In further control studies coexpression of Myc-GSK3β(S9A) with FLAG-CRMP2 induced the formation of a CRMP2 doublet, evidence of CRMP2 phosphorylation (supplemental Fig. S3). These results suggest a complex feedback loop may regulate PIPP and CRMP2 association. PIPP inhibits PI3K/Akt-dependent neurite extension, conditions under which GSK3β is active, and this in turn may reduce the association of PIPP with CRMP2.
FIGURE 6.
The PIPP·CRMP2 complex is regulated by GSK3β. A, GSK3β sequentially phosphorylates CRMP2 at Ser518, Thr514, and Thr509 following priming phosphorylation of Ser522 by Cdk5. B, COS-1 cells expressing GST or GST-PIPP and either FLAG-CRMP2 wild type (WT) or FLAG-CRMP2(S522A) were captured on glutathione-Sepharose, washed, and immunoblotted with FLAG (upper panel) or GST antibodies (middle panel). Lysates were immunoblotted (WB) with FLAG antibodies (lower panel). C, relative FLAG-CRMP2 WT or S522A mutant binding to GST-PIPP was quantitated using densitometry, standardized to GST-PIPP levels in pulldowns. Bars = mean ± S.E., where the FLAG-CRMP2 WT is designated 1 (n = 4). D, COS-1 cells were transfected with FLAG-CRMP2 and GST or GST-PIPP and increasing amounts of Myc-GSK3β(S9A) (0–5 μg) and lysates were incubated with glutathione-Sepharose, pellets were washed and immunoblotted with FLAG (upper panel) or GST antibodies (middle panel). Lysates were immunoblotted with FLAG and Myc antibodies (lower panels). E, relative FLAG-CRMP2 detected in PIPP pulldowns in the presence or absence of Myc-GSK3β(S9A) (5 μg) was quantitated using densitometry, standardized to GST-PIPP levels in pulldowns. Bars = mean ± S.E., where the no GSK3β(S9A) control is designated 1 (n = 6, **, p < 0.001). F, PC12 cells were transfected with the indicated constructs and GST pulldown was performed followed by immunoblot with FLAG or GST antibodies. WB, Western blot.
Next we determined if CRMP2 regulates the ability of PIPP to degrade PtdIns(3,4,5)P3 (16). To this end in vitro [32P]PtdIns(3,4,5)P3 5-phosphatase assays using purified GST-PIPP were performed in the presence or absence of GST-CRMP2 or GST revealing no change in 5-phosphatase activity (supplemental Fig. S4A). The same results were obtained using FLAG-CRMP2 isolated from COS-1 cells (supplemental Fig. S4B). Thus CRMP2 binds the C terminus of PIPP and this does not affect the central 5-phosphatase catalytic domain.
PIPP and CRMP2 Exhibit Opposing Functions in Regulating Neurite Elongation
To evaluate the function of the PIPP·CRMP2 complex, we examined the effects of loss of function of either protein, or overexpression of CRMP2 and PIPP on PC12 cell neurite elongation. CRMP2 promotes axon extension in part by its microtubule stabilizing activity and also by Kinesin-1-dependent WAVE1/Sra-1 transport to the growth cone (11, 33, 45). In contrast, PIPP inhibits neurite extension by regulating PI3K/Akt signaling at the growth cone, and perhaps by additional mechanisms (16). To directly evaluate in the same cell type if PIPP and CRMP2 compete to regulate neurite extension, we used PC12 cells, a well established cell line whereby following 3-day NGF stimulation, differentiated PC12 cells bearing long neurites (∼20–30% of cells) and undifferentiated cells can be detected by phalloidin staining (16). We undertook PIPP or CRMP2 siRNA knockdown in PC12 cells and assessed NGF-stimulated neurite elongation by staining with phalloidin, which binds F-actin. Only cells with a neurite length greater than the width of the cell body were scored, as neurite length (μm) divided by cell body width (μm). CRMP2 knockdown resulted in decreased neurite extension (Fig. 7A, arrowheads) consistent with the known role of CRMP2 in promoting neurite elongation. In contrast, PIPP knockdown resulted in hyperelongated neurites (Fig. 7A), a phenotype rescued by expression of non-targeted HA-PIPP (Fig. 7B). Interestingly, a construct containing the CRMP2 binding domain in PIPP, HA-PIPPSKICH/PRD-2, also significantly reduced the length of neurites in PIPP siRNA PC12 cells (Fig. 7B), perhaps by acting as a dominant-negative construct and binding and sequestering endogenous CRMP2. A 5-phosphatase catalytically inactive construct, HA-PIPPH557A, also reduced the hyperelongated neurite phenotype, but to a lesser extent (Fig. 7B). Therefore PIPP exhibits opposing function to CRMP2. PIPP-CRMP2 binding, in addition to the PtdIns(3,4,5)P3 5-phosphatase activity of PIPP, may contribute to the inhibition of PIPP neurite elongation.
FIGURE 7.
PIPP and CRMP2 exhibit opposing functions in regulating neurite elongation. A, PC12 cells expressing control, PIPP, or CRMP2 Cy3-labeled siRNA were differentiated for 3 days and stained with phalloidin to outline differentiated neurites. Asterisks indicate transfected cells. Growth cones are indicated by arrowheads. Bar = 20 μm. Neurite elongation of cells (defined as neurite length divided by cell body width) was measured. Bars = the mean neurite length ± S.E., relative to control siRNA, designated 1 (≥15 cells, n = 3, **, p < 0.01). B, PC12 cells were co-transfected with PIPP Cy3-labeled siRNA and plasmids encoding HA-vector, HA-PIPP, HA-PIPPH557A, or HA-SKICH/PRD-2 and differentiated for 3 days. Cells were fixed and stained with HA antibodies (not shown) and phalloidin. Asterisks indicate transfected cells. Arrowheads indicate growth cones. Bar = 20 μm. Neurite elongation of cells was measured as in A. Bars = the mean neurite length ± S.E., relative to HA-vector, designated 1 (≥10 cells, n = 3, *, p < 0.05). C and D, PC12 cells expressing control, PIPP, or CRMP2 Cy3-labeled siRNA were differentiated for 3 days and stained with WAVE1 antibodies (C) or Kinesin-1 heavy chain antibodies (D) and imaged by confocal microscopy at the same laser attenuation. Arrowheads show WAVE1 or Kinesin-1 staining at growth cones of transfected cells. Bar = 20 μm. The smaller panels below show examples of additional growth cones imaged at the same laser attenuation and at higher magnification for each indicated transfected siRNA. Growth cones are presented in graded fluorescence. Fluorescence intensity of WAVE1 or Kinesin-1 in the growth cone relative to the neurite shaft of transfected cells. Bars = mean ± S.E., control is designated 1, (10 cells, n = 3, **, p < 0.05).
A significant mechanism by which CRMP2 promotes neurite elongation is via Sra-1/WAVE1. At the growth cone WAVE1 activates the Arp2/3 complex, which nucleates actin monomers, allowing their incorporation into microfilaments (46). RNAi knockdown of CRMP2 results in loss of WAVE1 at the growth cone, and short neurites (36). We next examined the localization of WAVE1 to the growth cone under conditions of PIPP or CRMP2 siRNA knockdown (Fig. 7C). CRMP2 knockdown resulted in shorter neurites with decreased WAVE1 at the growth cone, as assessed by the intensity of WAVE1 staining, as described (36)(Fig. 7C). In contrast, PIPP knockdown increased neurite length associated with enhanced WAVE1 staining (1.5-fold) in the growth cone. CRMP2 functions as a cargo receptor, which links the partner proteins responsible for axon extension such as WAVE1 to motor proteins such as Kinesin-1 (36, 37, 47). Kinesins transport organelles, membrane vesicles, and protein complexes along microtubules to maintain neuronal polarity and promote elongation (36, 48–50). Kinesin-1 is a heterotetramer composed of two KHC and two KLC subunits providing a physical scaffold for cargo delivery to the growth cone (51, 52). However, it remains unclear how the association of Kinesin-1 to CRMP2 or other cargo is selectively transported to a growing axon. Because PIPP complexes with both CRMP2 and Kinesin-1, we evaluated the effect of PIPP or CRMP2 knockdown on Kinesin-1 localization, using antibodies to KHC (Fig. 7D). CRMP2 knockdown resulted in shorter neurites that exhibited growth cones with less intense staining for Kinesin-1. In contrast, PIPP knockdown led to longer neurites with increased intensity of Kinesin-1 at the growth cone (Fig. 7D). In control studies, the area of the growth cone, and the ratio of actin staining in the growth cone relative to the neurite shaft was equal in control, CRMP2, and PIPP knockdown cells (supplemental Fig. S5, A and B) revealing changes in WAVE1 or Kinesin-1 localization in PIPP or CRMP2 knockdown cells is not a consequence of altered neurite length or growth cone size. Therefore PIPP promotes neurite elongation and growth cone localization of the CRMP2 effector WAVE1 and its motor protein Kinesin-1. Knockdown of CRMP2 results in the diametrically opposite phenotype under the same conditions in PC12 cells.
PIPP Inhibits CRMP2-mediated Neurite Elongation
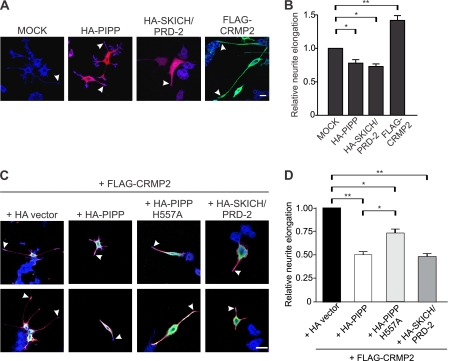
We next examined whether PIPP expression could inhibit CRMP2-induced neurite outgrowth in PC12 cells. After 3 days of NGF stimulation, cells were fixed and stained with HA antibodies to identify transfected cells, and phalloidin to outline neurites (Fig. 8A). PIPP expression significantly reduced neurite elongation relative to vector controls (Fig. 8, A and B) (16). Interestingly, the PIPP·CRMP2 binding domain, PIPPSKICH/PRD-2, which lacks 5-phosphatase activity, also inhibited neurite elongation to the same extent as full-length PIPP, perhaps by sequestering endogenous CRMP2. In contrast, expression of CRMP2 increased PC12 neurite elongation (Fig. 8, A and B). Upon HA-PIPP co-expression with CRMP2, only short neurites were formed indicating PIPP can effectively inhibit CRMP2-induced neurite elongation (Fig. 8, C and D). PIPPSKICH/PRD-2, which, as reported here binds CRMP2, also inhibited CRMP2-dependent neurite elongation to the same extent as wild type PIPP (Fig. 8, C and D). In addition, catalytically inactive PIPPH557A significantly reduced neurite length compared with vector control, but not to the same degree as HA-PIPP or HA-PIPPSKICH/PRD-2. Therefore PIPP inhibits neurite elongation by two mechanisms first via negative regulation of PI3K signaling and second by binding to and inhibiting CRMP2.
FIGURE 8.
PIPP inhibits CRMP2-mediated NGF-stimulated PC12 cell neurite elongation. A, PC12 cells expressing HA-vector (mock), HA-PIPP, HA-SKICH/PRD-2, or FLAG-CRMP2 were treated with NGF for 3 days. Cells were fixed and stained with FLAG (green) and HA (red) antibodies and phalloidin (blue). Bar = 20 μm. Arrowheads indicate growth cones. B, neurite elongation of cells as in A was measured. Bars = the mean neurite length ± S.E., relative to mock, designated 1 (20 cells, n = 3, *, p < 0.01; **, p < 0.001). C, PC12 cells were co-transfected with plasmids encoding FLAG-CRMP2 and HA-vector, HA-PIPP, HA-PIPPH557A, or HA-SKICH/PRD-2 and differentiated for 3 days. Cells were fixed and stained with HA (green) and FLAG (red) antibodies and phalloidin (blue). Bar = 20 μm. Arrowheads indicate growth cones. D, neurite elongation of cells as in C was measured. Bars = the mean neurite length ± S.E., relative to HA-vector, designated 1 (>10 cells, n = 4, *, p < 0.05; **, p < 0.01).
To exclude the possibility that PIPP regulates CRMP2 function via regulation of its expression and/or phosphorylation, we examined the total levels and distribution (growth cone versus neurite shaft) of phospho-Thr509/Thr514-CRMP2, revealing they were unchanged in PIPP knockdown PC12 cells (supplemental Fig. S6). Additionally, total CRMP2 protein levels were not affected by PIPP knockdown (Fig. 5C).
DISCUSSION
The PI3K signal PtdIns(3,4,5)P3 activates Akt, which inactivates GSK3β at the growth cone, to facilitate proper axon selection and elongation (3). Expression of constitutively active PI3K (p110α) or Akt (myr-Akt) results in multiple axon formation (53). We have shown here that a negative regulator of PI3K/Akt signaling, PIPP, a PtdIns(3,4,5)P3 5-phosphatase, inhibits axon selection, neurite outgrowth, and elongation. PIPP knockdown in hippocampal neurons results in multiple Tau-1 positive axons. Similarly, knockdown of another negative regulator of PI3K/Akt, PTEN, also leads to multiple axons in hippocampal neurons (7, 12). PIPP, like PTEN, degrades PtdIns(3,4,5)P3 and thereby inhibits Akt activation and GSK3β inactivation at the growth cone (3, 4, 6, 16). Therefore, the polarization defect observed in PIPP knockdown axons is consistent with the role of PIPP in inhibiting PI3K/Akt signaling.
We have also identified here a second molecular mechanism by which PIPP regulates neurite differentiation. PIPP complexes with and inhibits CRMP2 activity in promoting WAVE1 localization to the growth cone. CRMP2 is an adaptor protein that regulates the cytoskeleton, microtubules, and vesicular trafficking by binding numerous effectors, such as WAVE1, actin, amyloid precursor protein, Slp1/TrkB receptor neurofilament L and M, tubulin, and Kinesin-1 (36–38, 47, 54, 55). We demonstrate here that PIPP is a novel inhibitory CRMP2 interacting protein as shown by Y2H, direct purified protein-protein binding studies, GST pulldown in mammalian cells, coimmunoprecipitation of endogenous proteins, and co-localization of CRMP2 and PIPP in the axon of primary hippocampal neurons. As CRMP2 binds PIPP in purified component assays, in the absence of Kinesin-1, tubulin, or any other CRMP2 binding partner, the PIPP·CRMP2 complex is a direct protein-protein interaction. We have shown the PIPP C-terminal proline-rich domain (SKICH/PRD) facilitates its interaction with CRMP2. This C-terminal domain is adjacent to the PtdIns(3,4,5)P3 5-phosphatase domain of PIPP. The SKICH/PRD domain has been shown previously to facilitate phosphatase access to both the plasma membrane and also the neurite growth cone (16, 29). Notably, we demonstrate here the PIPP growth cone localization is CRMP2 dependent, as revealed by decreased PIPP staining in the growth cones of CRMP2 knockdown cells. This suggests CRMP2 connects PIPP to Kinesin-1, thus acting as a cargo adaptor as reported for tubulin heterodimers, Sra-1·WAVE1, and TrkB·Slp1 complexes (35–37).
CRMP2 and PIPP Exhibit Opposing Roles in Axon Selection and Elongation
CRMP2 promotes neurite extension by multiple mechanisms, including binding and stabilizing tubulin at the plus ends of microtubules (56, 57), microtubule-based anterograde transport of tubulin heterodimers, the Sra-1·WAVE1 complex, and the Slp1·TrkB receptor complex to the distal part of the neurite via directly binding KLC (35–37) and regulation of anterograde transport. CRMP2 also inhibits retrograde transport (47) and links endocytic regulatory proteins to dynein (35–37). In PC12 cells, used here as a model of early neurite elongation, we have shown CRMP2 and PIPP knockdown induce strikingly opposite phenotypes in terms of neurite extension, and localization of CRMP2 cargo WAVE1, and binding partner Kinesin-1 to the growth cone. Furthermore, we have demonstrated by co-expression studies in PC12 cells that PIPP opposes CRMP2 function in promoting neurite elongation. In PC12 cells, expression of full-length PIPP, or the domain of PIPP that bound CRMP2, SKICH/PRD-2 inhibited CRMP2-dependent neurite outgrowth. Although we have not delineated the molecular mechanism(s) by which PIPP inhibits CRMP2 function, which is beyond the scope of these studies, PIPP may displace CRMP2/Kinesin-1 cargo or transport effectors or may affect the interaction of CRMP2 with tubulin. Consistent with this contention, in PIPP-depleted PC12 cells, the CRMP2 cargo WAVE1 showed increased growth cone localization. In contrast, WAVE1 exhibited decreased growth cone localization in CRMP2 knockdown cells.
Some CRMP2 functions are regulated by sequential phosphorylation by Cdk5 and GSK3β although this area of investigation is complex (11, 12, 38). Hyperphosphorylated CRMP2 is associated with neurofibrillary tangles in Alzheimer-diseased brains where it colocalizes with WAVE1 (58–61). The association between PIPP and CRMP2 does not require CRMP2 phosphorylation, rather this complex significantly reduces when GSK3β is active, conditions under which PI3K/Akt signaling is not activated. Therefore, we propose a model whereby when PI3K/Akt signaling is activated by NGF stimulation, GSK3β will be inhibited by Akt phosphorylation, and PIPP will maximally associate with CRMP2 to inhibit CRMP2-dependent neurite elongation.
In summary, our studies identify a significant complex between a negative regulator of PI3K/Akt signaling, PIPP, with a positive regulator of neuronal differentiation, CRMP2. The CRMP2·PIPP complex is likely to dynamically regulate both neurite outgrowth and axon polarity in the normal brain.
Supplementary Material
This work was supported by National Health and Medical Research Council, Australia, Grant 436638 and Australian Research Council, Australia, Grant DP110103655.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PtdIns(3,4,5)P2
- phosphatidylinositol 3,4,5-trisphosphate
- CRMP
- collapsin response mediator protein
- KHC
- Kinesin heavy chain
- KLC
- Kinesin light chain
- GSK3β
- glycogen synthase kinase 3β
- N-WASP
- neural Wiskott-Aldrich syndrome protein
- PIPP
- proline-rich inositol polyphosphate 5-phosphatase
- PRD
- proline-rich domain
- WAVE1
- WASP family verprolin-homologous protein 1
- Y2H
- yeast two-hybrid
- QRT
- quantitative reverse transcription.
REFERENCES
- 1. Craig A. M., Banker G. (1994) Annu. Rev. Neurosci. 17, 267–310 [DOI] [PubMed] [Google Scholar]
- 2. Dent E. W., Gertler F. B. (2003) Neuron 40, 209–227 [DOI] [PubMed] [Google Scholar]
- 3. Ménager C., Arimura N., Fukata Y., Kaibuchi K. (2004) J. Neurochem. 89, 109–118 [DOI] [PubMed] [Google Scholar]
- 4. Schwamborn J. C., Püschel A. W. (2004) Nat. Neurosci. 7, 923–929 [DOI] [PubMed] [Google Scholar]
- 5. Weiner O. D., Neilsen P. O., Prestwich G. D., Kirschner M. W., Cantley L. C., Bourne H. R. (2002) Nat. Cell Biol. 4, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi S. H., Jan L. Y., Jan Y. N. (2003) Cell 112, 63–75 [DOI] [PubMed] [Google Scholar]
- 7. Jiang H., Guo W., Liang X., Rao Y. (2005) Cell 120, 123–135 [DOI] [PubMed] [Google Scholar]
- 8. Sutherland C., Leighton I. A., Cohen P. (1993) Biochem. J. 296, 15–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cross D. A., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. (1995) Nature 378, 785–789 [DOI] [PubMed] [Google Scholar]
- 10. Zhou F. Q., Zhou J., Dedhar S., Wu Y. H., Snider W. D. (2004) Neuron 42, 897–912 [DOI] [PubMed] [Google Scholar]
- 11. Cole A. R., Knebel A., Morrice N. A., Robertson L. A., Irving A. J., Connolly C. N., Sutherland C. (2004) J. Biol. Chem. 279, 50176–50180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshimura T., Kawano Y., Arimura N., Kawabata S., Kikuchi A., Kaibuchi K. (2005) Cell 120, 137–149 [DOI] [PubMed] [Google Scholar]
- 13. Shi S. H., Cheng T., Jan L. Y., Jan Y. N. (2004) Curr. Biol. 14, 2025–2032 [DOI] [PubMed] [Google Scholar]
- 14. Zhou F. Q., Snider W. D. (2005) Science 308, 211–214 [DOI] [PubMed] [Google Scholar]
- 15. Astle M. V., Horan K. A., Ooms L. M., Mitchell C. A. (2007) Biochem. Soc. Symp. 74, 161–181 [DOI] [PubMed] [Google Scholar]
- 16. Ooms L. M., Fedele C. G., Astle M. V., Ivetac I., Cheung V., Pearson R. B., Layton M. J., Forrai A., Nandurkar H. H., Mitchell C. A. (2006) Mol. Biol. Cell. 17, 607–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aoki K., Nakamura T., Inoue T., Meyer T., Matsuda M. (2007) J. Cell Biol. 177, 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soutar M. P., Thornhill P., Cole A. R., Sutherland C. (2009) Curr. Alzheimer Res. 6, 269–278 [DOI] [PubMed] [Google Scholar]
- 19. Hensley K., Venkova K., Christov A., Gunning W., Park J. (2011) Mol. Neurobiol. 43, 180–191 [DOI] [PubMed] [Google Scholar]
- 20. Tanaka M., Herr W. (1990) Cell 60, 375–386 [DOI] [PubMed] [Google Scholar]
- 21. Cole A. R., Causeret F., Yadirgi G., Hastie C. J., McLauchlan H., McManus E. J., Hernández F., Eickholt B. J., Nikolic M., Sutherland C. (2006) J. Biol. Chem. 281, 16591–16598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu H. K., Perrier S., Lipina C., Finlay D., McLauchlan H., Hastie C. J., Hundal H. S., Sutherland C. (2006) BMC Mol. Biol. 7, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dussault A. A., Pouliot M. (2006) Biol. Proced. Online 8, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goslin K., Banker G. (1991) Rat Hippocampal Neurons in Low Density Culture, MIT Press, Cambridge, MA [Google Scholar]
- 25. Dyson J. M., O'Malley C. J., Becanovic J., Munday A. D., Berndt M. C., Coghill I. D., Nandurkar H. H., Ooms L. M., Mitchell C. A. (2001) J. Cell Biol. 155, 1065–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He Y., Yu W., Baas P. W. (2002) J. Neurosci. 22, 5982–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abramoff M., Magelhaes P., Ram S. (2004) Biophotonics Int. 11, 36–42 [Google Scholar]
- 28. Mochizuki Y., Takenawa T. (1999) J. Biol. Chem. 274, 36790–36795 [DOI] [PubMed] [Google Scholar]
- 29. Gurung R., Tan A., Ooms L. M., McGrath M. J., Huysmans R. D., Munday A. D., Prescott M., Whisstock J. C., Mitchell C. A. (2003) J. Biol. Chem. 278, 11376–11385 [DOI] [PubMed] [Google Scholar]
- 30. Cleveland D. W. (1983) Cell 34, 330–332 [DOI] [PubMed] [Google Scholar]
- 31. Wang L. H., Strittmatter S. M. (1996) J. Neurosci. 16, 6197–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dotti C. G., Sullivan C. A., Banker G. A. (1988) J. Neurosci. 8, 1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Inagaki N., Chihara K., Arimura N., Ménager C., Kawano Y., Matsuo N., Nishimura T., Amano M., Kaibuchi K. (2001) Nat. Neurosci. 4, 781–782 [DOI] [PubMed] [Google Scholar]
- 34. Gordon-Weeks P. R. (1991) Bioessays 13, 235–239 [DOI] [PubMed] [Google Scholar]
- 35. Arimura N., Kimura T., Nakamuta S., Taya S., Funahashi Y., Hattori A., Shimada A., Ménager C., Kawabata S., Fujii K., Iwamatsu A., Segal R. A., Fukuda M., Kaibuchi K. (2009) Dev. Cell 16, 675–686 [DOI] [PubMed] [Google Scholar]
- 36. Kawano Y., Yoshimura T., Tsuboi D., Kawabata S., Kaneko-Kawano T., Shirataki H., Takenawa T., Kaibuchi K. (2005) Mol. Cell. Biol. 25, 9920–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kimura T., Watanabe H., Iwamatsu A., Kaibuchi K. (2005) J. Neurochem. 93, 1371–1382 [DOI] [PubMed] [Google Scholar]
- 38. Arimura N., Ménager C., Kawano Y., Yoshimura T., Kawabata S., Hattori A., Fukata Y., Amano M., Goshima Y., Inagaki M., Morone N., Usukura J., Kaibuchi K. (2005) Mol. Cell. Biol. 25, 9973–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arimura N., Inagaki N., Chihara K., Ménager C., Nakamura N., Amano M., Iwamatsu A., Goshima Y., Kaibuchi K. (2000) J. Biol. Chem. 275, 23973–23980 [DOI] [PubMed] [Google Scholar]
- 40. Uchida Y., Ohshima T., Sasaki Y., Suzuki H., Yanai S., Yamashita N., Nakamura F., Takei K., Ihara Y., Mikoshiba K., Kolattukudy P., Honnorat J., Goshima Y. (2005) Genes Cells 10, 165–179 [DOI] [PubMed] [Google Scholar]
- 41. Mitsui N., Inatome R., Takahashi S., Goshima Y., Yamamura H., Yanagi S. (2002) EMBO. J. 21, 3274–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rembutsu M., Soutar M. P., Van Aalten L., Gourlay R., Hastie C. J., McLauchlan H., Morrice N. A., Cole A. R., Sutherland C. (2008) Biochemistry 47, 2153–2161 [DOI] [PubMed] [Google Scholar]
- 43. Grimes C. A., Jope R. S. (2001) Prog. Neurobiol. 65, 391–426 [DOI] [PubMed] [Google Scholar]
- 44. Stambolic V., Woodgett J. R. (1994) Biochem. J. 303, 701–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nishimura T., Fukata Y., Kato K., Yamaguchi T., Matsuura Y., Kamiguchi H., Kaibuchi K. (2003) Nat. Cell Biol. 5, 819–826 [DOI] [PubMed] [Google Scholar]
- 46. Pollitt A. Y., Insall R. H. (2009) J. Cell Sci. 122, 2575–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arimura N., Hattori A., Kimura T., Nakamuta S., Funahashi Y., Hirotsune S., Furuta K., Urano T., Toyoshima Y. Y., Kaibuchi K. (2009) J. Neurochem. 111, 380–390 [DOI] [PubMed] [Google Scholar]
- 48. Horiguchi K., Hanada T., Fukui Y., Chishti A. H. (2006) J. Cell Biol. 174, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jacobson C., Schnapp B., Banker G. A. (2006) Neuron 49, 797–804 [DOI] [PubMed] [Google Scholar]
- 50. Schwamborn J. C., Khazaei M. R., Püschel A. W. (2007) J. Biol. Chem. 282, 35259–35268 [DOI] [PubMed] [Google Scholar]
- 51. Bloom G. S., Wagner M. C., Pfister K. K., Brady S. T. (1988) Biochemistry 27, 3409–3416 [DOI] [PubMed] [Google Scholar]
- 52. Vale R. D., Reese T. S., Sheetz M. P. (1985) Cell 42, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yoshimura T., Arimura N., Kawano Y., Kawabata S., Wang S., Kaibuchi K. (2006) Biochem. Biophys. Res. Commun. 340, 62–68 [DOI] [PubMed] [Google Scholar]
- 54. Pawlik M., Otero D. A., Park M., Fischer W. H., Levy E., Saitoh T. (2007) Biochem. Biophys. Res. Commun. 355, 907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hensley K., Christov A., Kamat S., Zhang X. C., Jackson K. W., Snow S., Post J. (2010) J. Neurosci. 30, 2979–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fukata Y., Itoh T. J., Kimura T., Ménager C., Nishimura T., Shiromizu T., Watanabe H., Inagaki N., Iwamatsu A., Hotani H., Kaibuchi K. (2002) Nat. Cell Biol. 4, 583–591 [DOI] [PubMed] [Google Scholar]
- 57. Chae Y. C., Lee S., Heo K., Ha S. H., Jung Y., Kim J. H., Ihara Y., Suh P. G., Ryu S. H. (2009) Cell. Signal. 21, 1818–1826 [DOI] [PubMed] [Google Scholar]
- 58. Takata K., Kitamura Y., Nakata Y., Matsuoka Y., Tomimoto H., Taniguchi T., Shimohama S. (2009) Am. J. Pathol. 175, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoshida H., Watanabe A., Ihara Y. (1998) J. Biol. Chem. 273, 9761–9768 [DOI] [PubMed] [Google Scholar]
- 60. Gu Y., Hamajima N., Ihara Y. (2000) Biochemistry 39, 4267–4275 [DOI] [PubMed] [Google Scholar]
- 61. Cole A. R., Noble W., van Aalten L., Plattner F., Meimaridou R., Hogan D., Taylor M., LaFrancois J., Gunn-Moore F., Verkhratsky A., Oddo S., LaFerla F., Giese K. P., Dineley K. T., Duff K., Richardson J. C., Yan S. D., Hanger D. P., Allan S. M., Sutherland C. (2007) J. Neurochem. 103, 1132–1144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.