Abstract
Reduced axonal mitochondrial transport has been observed in major neurodegenerative diseases, including fALS patients and SOD1G93A mice. However, it is unclear whether this defect plays a critical role in axonal degeneration or simply reflects sequelae of general transport alteration. Using genetic mouse models combined with time-lapse imaging of live neurons, we previously discovered that axon-targeted syntaphilin (SNPH) acts as a docking receptor specific for axonal mitochondria. Deletion of the snph gene in mice results in a substantially higher proportion of axonal mitochondria in the mobile state without any effect on the transport of other axonal organelles. Here we address whether increased (rescued) axonal mitochondrial mobility changes the disease course by crossing fALS-linked transgenic SOD1G93A and snph−/− knock-out mice. We found that a 2-fold increase in axonal mitochondrial mobility in SOD1G93A/snph−/− mice did not affect the onset of ALS-like symptoms. Both SOD1G93A and SOD1G93A/snph−/− mice exhibit similar weight loss, deterioration in motor function and motor neuron loss, significant gliosis, and a lifespan of 152–154 days. Thus, for the first time, our study provides genetic and pathological evidence that the impairment of mitochondrial transport seen in SOD1G93A mice plays a minimal role in the rapid-onset of fALS-linked pathology.
Keywords: Amyotropic Lateral Sclerosis (Lou Gehrig's disease), Axon, Mitochondrial Transport, Neurodegeneration, Trafficking, Axonal Mitochondrial Transport, Axonal Degeneration, Mitochondrial Mobility
Introduction
Mitochondria are essential organelles for neuronal survival and function. Neurons require specialized mechanisms to regulate mitochondrial transport along axons and retain them in growth cones, nodes of Ranvier, and synaptic terminals, where energy production and calcium homeostasis are critical (1–5). Defects in mitochondrial transport could cause local energy depletion and toxic changes in Ca2+ buffering, triggering synaptic dysfunction and loss. Altered transport and distribution of axonal mitochondria has been implicated in the neurodegenerative diseases such as Alzheimer, Huntington, and Amyotrophic Lateral Sclerosis (ALS).3 However, it has not been established whether altered mitochondrial mobility plays a critical role in axonal degeneration.
Characterization of mitochondrial transport in mature neurons from aged and disease-stage neurodegenerative disease models is an important step in understanding the cellular mechanisms of neurodegeneration. ALS is a late-onset neurodegenerative disease that causes motor neuron loss (6). Mutations in Cu/Zn superoxide dismutase (SOD) genes have been found to cause familial ALS (7). Transgenic mice expressing mutant human SOD1 are clinically and pathologically similar to human ALS patients (8), becoming paralyzed in one or more limbs because of a loss of motor neurons in the spinal cord. Several laboratories have reported altered transport of axonal mitochondria in ALS patients and SOD1G93A mice, as well as neurons and motor neuron NSC34 cell lines expressing SOD1G93A (9–11). The presence of dysfunctional mitochondria at distal axons could be a consequence of impaired recycling or degradation of abnormal mitochondria because of reduced mobility and altered axonal transport. To slow axonal degeneration, it is proposed that increased mitochondrial transport might aid the efficient delivery of healthy mitochondria to axons and/or the removal of damaged mitochondria from distal synapses for degradation or repair. Therefore, it is important to assess whether increased (rescued) axonal mitochondrial mobility in mutant SOD1 disease models has a beneficial impact on the pathogenesis of motor neuron degeneration.
We previously discovered that axon-targeted syntaphilin (SNPH) acts as a docking receptor specific for axonal mitochondria (12). Deleting snph in mice results in a substantially higher proportion of axonal mitochondria in the mobile state without any effect on the transport of other axonal organelles. In the present study, we first examine snph−/− mice and found that increased axonal mitochondrial mobility by deleting snph has no impact on lifespan or motor function, and snph−/− mice display no observable neurodegeneration phenotype. Second, by crossing SOD1G93A and snph−/− mice we address whether increasing (rescued) mitochondrial mobility has any impact on the pathogenesis of the fALS-linked SOD1G93A mouse model by comparing clinical and histological observations of SOD1G93A mice to the crossed SOD1G93A/snph−/− mice. To our surprise, although the crossed SOD1G93A/snph−/− mice exhibit a 2-fold increase in axonal mitochondrial mobility at disease stages, there is no observable improvement in the deterioration of motor function or in disease progression and lifespan. Furthermore, immunostaining of spinal cord sections from both SOD1G93A and SOD1G93A/snph−/− mice show similar ALS-like disease histopathology that includes motor neuron loss and gliosis. Thus, our study provides genetic, cellular, and pathological evidence that challenges the prevailing hypothesis that defective transport of axonal mitochondria contributes to rapid-onset motor neuron degeneration in this mouse model.
EXPERIMENTAL PROCEDURES
Animals and Neuronal Cultures
B6.Cg-Tg (SOD1G93A) 1Gur/J mice were originally described by Gurney et al. (8) and purchased from The Jackson Laboratory. Transgenic mice carrying this SOD1G93A express high copy number of a G93A mutant form of human SOD1. Hemozygotes are viable and fertile and exhibit a phenotype similar to ALS in humans becoming paralyzed in one or more limbs due to loss of motor neurons from the spinal cord, and have a life span of 157.1 ± 9.3 days.4 Transgenic mice on a mixed B6SJL genetic background were backcrossed to C57BL/6J for at least 10 generations in The Jackson Laboratory to generate this congenic strain (Stock No. 004435). This strain was crossed with C57BL/6 background snph−/− mice to generate SOD1G93A/snph+/− heterozygous mice. These mice were then crossed with snph+/− mice to generate WT, SOD1G93A, snph−/− and SOD1G93A/snph−/− littermates. Dorsal root ganglion (DRG) neuron cultures from P120-P125 mice were dissected in Hank's buffered salt solution (HBSS), followed by digestion in 2.5 units/ml dispase II and 200 units/ml collagenase (Worthington Biochemical). Tissues were triturated in Neurobasal medium and plated on coverslips coated with 100 μg/ml poly-d-lysine and 5 μg/ml laminin (Invitrogen) supplemented with B27 (Invitrogen) and 100× Glutamax (Invitrogen). The final density of cells is 2,000 per coverslip.
Mitochondrial Mobility Studies
Time-lapse imaging was performed using a Zeiss LSM 510 META confocal microscope (Zeiss Microimaging) with a P-Apochromat 40×/1.3 oil objective. Mitochondria in DIV2 DRG neurons were labeled with 100 nm MitoTracker red CMX-Ros (Invitrogen) in Neurobasal medium for 20 s. Proximal axons were captured for time-lapse imaging in a coverglass chamber (Nalge Nunc International) with Tyrode's solution at 37 °C. Kymograghs and mobility analysis were carried out as previously described (12).
Immunocytochemistry
Mice were anesthetized by intraperitoneal injection of 100 mg/kg of ketamine (Fort Dodge) and 10 mg/kg of xylazine (Akorn Inc) and euthanized by transcardial perfusion with PBS for 5 min followed by perfusion-fixation with 4% paraformaldehyde in PBS for 5 min. The spinal cord was removed and post-fixed in 4% paraformaldehyde in PBS. The lumbar region of the spinal cord and motor axons were embedded in paraffin and sectioned at 8-μm thickness. To quantify the motor neuron number, every 13th section was sampled for a total of 30 sections for each animal. Motor neurons were stained with SMI32 (Sternberger Monoclonal/Covance) and Nissl. Only large neurons with a clear nucleus and nucleolus were counted. Astrocytes and microglia were stained by GFAP (Sigma) and Iba-1 (Wako Chemicals), respectively. Three animals were used for each genotype.
For astrocyte and microglia studies, 9 spinal cord transverse sections were randomly selected from each genotype. For microglia quantification, only cells in the lateral ventral horns that were in focus and completely within the field of view were counted. NIH Image J was used to quantify the mean densities of Iba-1 and GFAP. For astrocytes, the region of interest for cell counts (lateral ventral horn) was defined using landmarks and reference points from the mouse spinal cord (15). Motor axon transverse sections from the lumbar spinal level were co-stained with antibodies against SNPH (12) and SMI32. DIV2 DRG neurons from WT and snph−/− were fixed with 4% paraformaldehyde in PBS for 30 min, followed by co-staining with antibodies against SNPH and cytochrome c (BD Pharmingen).
Determination of Disease Onset and End Point
Disease onset and animal lifespan were defined by weight and motor function. Upon reaching 13 weeks of age, animals were weighed weekly. Onset of disease was defined as the week following peak weight based on the individual weight curve for the mouse (13). Motor performance was evaluated by weekly Rotarod testing (San Diego Instruments). Mice were placed on a rotating rod that was set to accelerate from 0 to 40 rpm over the course of 180 s. The amount of time the mouse was able to remain on the rotating rod before falling off was used to indicate motor function. Disease end point (or life span) was defined as either the loss of the righting reflex (inability to right the torso within 10 s) or a greater than 20% weight loss (14).
Western Blot
Spinal cords from P150 WT, SOD1G93A, snph−/−, and SOD1G93A /snph−/− mice were homogenized. 20 μg of homogenates were loaded and detected with monoclonal antibodies against hSOD1, cytochrome c, p115, and a polyclonal antibody against SNPH. Proteins were detected by ECL systems. Results were scanned and analyzed by Gel pro (Roper Industries).
Statistical Analysis
Statistical analyses of means for more than two groups were performed using one-way analysis of variance (ANOVA) with the categories of genotype and age as independent factors followed by the Tukey-Kramer post-hoc test for pair-wise comparisons. For analyses of means involving only two groups with a sample size n < 30, the F-test was used to determine if the variances between the two groups was significantly different. For samples with a significant difference in variance, the Welch's t test was applied. Student's t test was applied for the samples with an insignificant difference in variance or where n ≥ 30. The null hypothesis was rejected at the 0.05 levels. Results for the mobility studies are expressed as the mean percentage of observed mitochondria in the mobile state ± S.E. Disease onset and lifespan are expressed as mean postnatal days ± S.D. All statistical computations were carried out using Prism (Graphpad Software).
RESULTS
The SOD1G93A/snph−/− Mice Exhibit Increased Axonal Mitochondrial Mobility
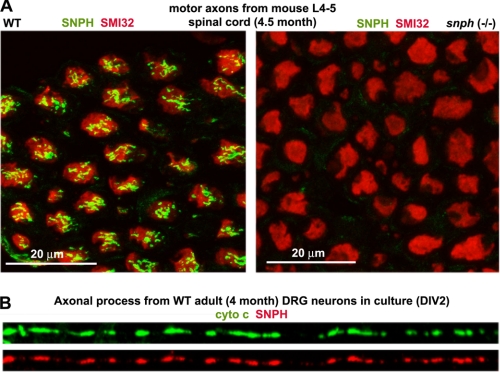
Our previous study demonstrated that SNPH, an axonal mitochondria-associated protein, serves as a docking receptor that specifically immobilizes axonal mitochondria. Deleting murine snph results in a dramatic increase in the percentage of axonal mitochondria in the mobile state (76 ± 20%) in cultured hippocampal neurons relative to wild-type controls (36 ± 15%) (12). To determine whether SNPH is also expressed in motor neuron axons, we immunostained transverse sections of motor axons sending out from the spinal cords of adult wild-type (WT) and snph−/− mice at postnatal day 150 (P150), and confirmed that SNPH is localized within motor axon bundles from WT but not snph−/− mouse spinal cords (Fig. 1A). SNPH targets to axonal mitochondria in cultured embryonic hippocampal neurons (12) and mature dorsal root ganglion (DRG) neurons isolated from adult mice (Fig. 1B). Thus, our snph−/− mice provide an ideal genetic tool to assess the role of increased mobility of axonal mitochondria in the pathogenesis of SOD1G93A mice by generating crossed SOD1G93A/snph−/− mice.
FIGURE 1.
SNPH is expressed in axons from spinal cord and targeted to axonal mitochondria in adult DRG neurons. A, transverse sections of ventral roots (axons) from lumbar spinal cords of adult WT and snph−/− mice (P150) were co-stained with antibodies against SNPH (green) and neurofilaments (SMI32) (red). SNPH is localized within motor axon bundles from WT but not snph−/− mouse spinal cords. Scale bars: 20 μm. B, representative image showing co-localization of SNPH (red) and mitochondrial marker cytochrome c (green) in the axons of cultured DRG neurons from 4-month-old WT mice.
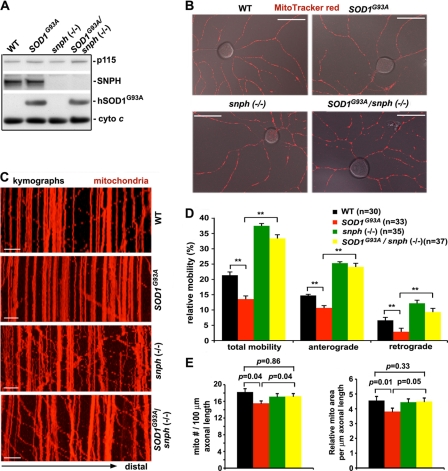
We first examined hSOD1 levels in spinal cord homogenates from adult (P150) mice of four genotypes: WT, SOD1G93A, snph−/−, and the crossed SOD1G93A/snph−/− (Fig. 2A). No SNPH was detected in SOD1G93A/snph−/− mice. Crossbreeding did not significantly alter hSOD1G93A expression: normalized intensity of hSOD1 in SOD1G93A/snph−/− mice was 90.25 ± 15.98% relative to that of SOD1G93A mice (p = 0.28, n = 9 from 3 littermates).
FIGURE 2.
Cultured DRG neurons from adult snph−/− and SOD1G93A/snph−/− mice exhibit enhanced axonal mitochondrial mobility. A, representative immunoblot showing SNPH and SOD1 expression in mice of four different genotypes. Equal amounts (20 mg) of spinal cord homogenate from P150 mice were sequentially blotted on the same membranes, which were stripped between each antibody application. Golgi marker p115 and mitochondrial marker cytochrome c serve as loading controls. The blots are representative of nine repeats from three pairs of littermates. B, representative DIV2 DRG neurons from adult mice labeled with MitoTracker red CMX-Ros. Scale bars: 50 μm. C, representative kymographs showing the relative mobility of axonal mitochondria in cultured DRG neurons (DIV2) from four genotypes of mice at ages between P120 and P125. Vertical lines represent stationary mitochondria and oblique lines or curves indicate mobile ones. The arrow in the bottom points toward distal axonal processes. Time-lapse sequences of 1,024 × 1,024 pixels (8 bits) were collected at ∼10-s intervals for 50 images during 8.3 min. Scale bars: 10 μm. D, quantitative analysis of the percentage of total mobile axonal mitochondria and anterogradely versus retrogradely transported mitochondria. The number of DRG neurons examined for each genotype (from three littermates) is indicated in parentheses. Total axonal length measured was 4008.62 μm (WT), 4327.84 μm (snph−/−), 4715.33 μm (SOD1G93A), and 3744.38 μm (SOD1G93A/snph−/−). Error bars, S.E. **, p < 0.001. E, quantitative analysis of axonal mitochondrial density in cultured adult DRG neurons (DIV2). Relative mitochondrial density was measured in the distal axonal processes (at least >100 μm away from the soma) and is expressed as the number of mitochondria per 100 μm axonal length (left) and normalized relative mitochondrial area per μm axonal length (right). Total axonal length measured was 2016.90 μm (WT), 3049.27 μm (SOD1G93A), 2096.23 μm (snph−/−), and 2137.13 μm (SOD1G93A/snph−/−).
To confirm that snph deletion resulted in increased axonal mitochondrial mobility in the crossed SOD1G93A/snph−/− mice, we selected live DRG neurons from adult disease stage mice instead of embryonic motor neurons for time-lapse imaging analysis due to four critical factors. First, motor neuron degeneration and symptoms of ALS are progressive and age-dependent, thus it would be more clinically relevant to assess mitochondrial transport in neurons isolated from adult mice exhibiting disease symptoms rather than from embryos. Second, it is technically impossible to culture motor neurons from adult mice. Third, both motor neurons and DRG neurons show similar altered axonal transport in SOD1G93A mice (16), thus DRG neurons from aged disease-stage mice have been widely used for study of defective axonal transport in the ALS-linked mouse models. Fourth, SNPH targets to axonal mitochondria in DRG neurons from adult mice (Fig. 1B), deleting snph substantially increases axonal mitochondrial mobility in cultured DRG neurons from adult mice relative to that from WT littermates (Fig. 2, C and D), providing an ideal live neuronal model to assess mitochondrial mobility.
We examined axonal mitochondrial mobility in live DRG neurons from four genotypes of age-matched adult mice (P120–125). Mitochondria were labeled at DIV2 with MitoTracker red CMX-Ros (100 nm for 20 s) (Fig. 2B), followed by time-lapse imaging. Kymographs were used to show the relative mobility of axonal mitochondria (Fig. 2C). Consistent with the axonal transport defects observed in embryonic motor neurons and adult DRG neurons (10, 11, 16), axonal mitochondria in SOD1G93A DRG neurons showed reduced mobility (13.54 ± 0.38%, mean ± S.E.), anterograde transport (10.66 ± 0.77%), and retrograde transport (2.88 ± 0.46%) compared with age-matched WT DRG neurons (total mobility: 21.35 ± 1.08%, p < 0.001; anterograde: 14.74 ± 1.05%, p < 0.001; retrograde: 6.61 ± 0.78%, p < 0.001) (Fig. 2D). In contrast, axonal mitochondria in both snph−/− and SOD1G93A/snph−/− DRG neurons exhibited a statistically significant increase in total mobility (37.49 ± 1.74%, p < 0.001; 33.37 ± 1.22%, p < 0.001, respectively), anterograde transport (25.30 ± 1.16%, p < 0.001; 24.06 ± 1.20%, p < 0.001, respectively), and retrograde transport (12.18 ± 1.02%, p < 0.001; 9.31 ± 1.22%, p < 0.001, respectively) relative to WT DRG neurons (Fig. 2D, also see supplemental Movies S1–S4 available online). In addition, the axonal mitochondrial density in SOD1G93A DRG neurons was also significantly reduced (Fig. 2E), which is consistent with a reduced axonal mitochondrial density observed in SOD1G93A embryonic motor neurons (10). Interestingly, enhanced axonal mitochondrial mobility in SOD1G93A/snph−/− mice rescued the deficit in mitochondrial density. Thus, the crossed SOD1G93A /snph−/− mice provide a valuable model to assess the impact of increased (rescued) axonal mitochondrial mobility and density on motor neuron degeneration in ALS-linked mutant mice.
Both SOD1G93A and Crossed SOD1G93A/snph−/− Mice Display Similar ALS-like Disease Phenotypes
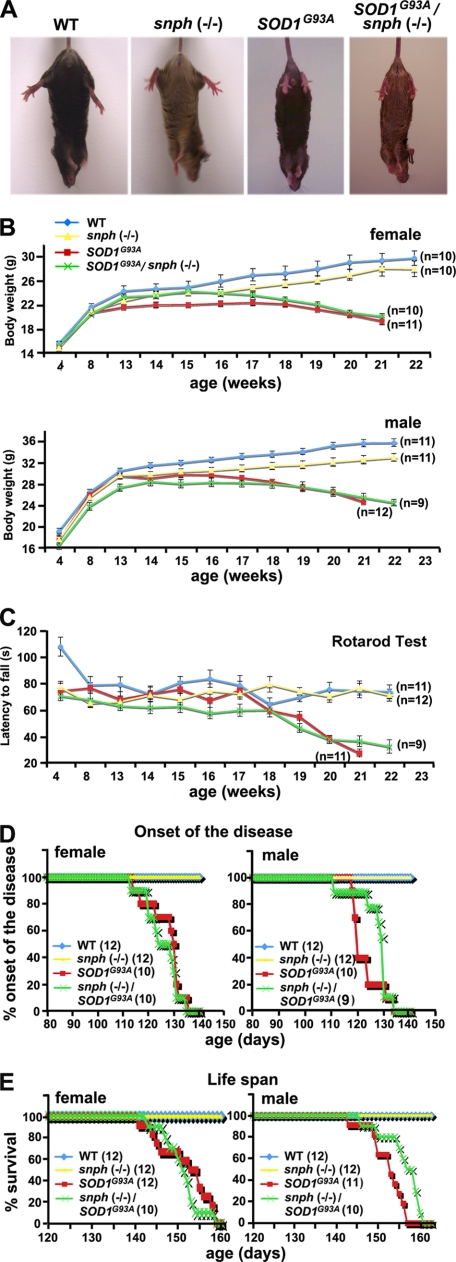
First, we evaluated the SOD1G93A and SOD1G93A /snph−/− phenotypes against WT and snph−/− controls using body weight and motor coordination parameters. We found that both SOD1G93A and SOD1G93A /snph−/− mice exhibited similar clinical evidence of motor neuron disease. By postnatal day 150 (P150), both genotypes had developed weakness accompanied by hind limb muscle wasting. When suspended by the tail, both SOD1G93A and SOD1G93A /snph−/− mice failed to extend their hind limbs (Fig. 3A). We weighed the animals and tested motor coordination monthly during the first 3 months of age and then weekly after that. Both WT and snph−/− mice continue to grow with age, and although male mice were ∼20% heavier than their female counterparts in both snph−/− and WT genotypes, snph−/− mice were slightly lighter than their WT littermates. In contrast, both male and female SOD1G93A and SOD1G93A/snph−/− mice began to lose weight at 17–18 weeks of age (Fig. 3B) and exhibited deterioration in motor function beginning at ∼18 weeks of age (Fig. 3C). Rotarod tests did not detect any significant difference in motor coordination between SOD1G93A and SOD1G93A/snph−/− mice (p = 0.21, Student's t test). As a control, snph−/− mice kept alive throughout the duration of the experiment and did not display any signs of motor deterioration, further confirming that increased axonal mitochondrial mobility in snph−/− mice has no observable impact on motor neuron function.
FIGURE 3.
Both SOD1G93A and crossed SOD1G93A/snph−/− mice display similar ALS-like disease phenotypes. A, at P150, both SOD1G93A, and SOD1G93A/snph−/− mice fail to extend their hind limbs when suspended by their tail. The behavioral alteration is consistent across these two genotypes of mice. B, both SOD1G93A and SOD1G93A/snph−/− mice begin losing body weight at 17 weeks of age. Body weight of female (upper) and male (lower) mice was measured weekly. There was no significant difference in body mass between SOD1G93A and SOD1G93A/snph−/− mice at 21 weeks (p = 0.33 for female, p = 0.39 for male). C, both SOD1G93A and SOD1G93A/snph−/− mice exhibit similar deterioration in motor function beginning at 18 weeks as the disease progresses. Motor coordination was assessed weekly by the rotarod test. The apparatus was set to accelerate from 0 to 40 rpm over 180 s and the latency to fall was recorded. There was no significant difference in between SOD1G93A and SOD1G93A/snph−/− mice at age 21 weeks (p = 0.12). As a control, WT and snph−/− mice showed no signs of motor deterioration during the time frame of the experiment. D and E, enhanced axonal mitochondrial transport has no observable effect on the disease onset (D) and life span (E) of the crossed SOD1G93A/snph−/− mice. The littermates of both SOD1G93A and SOD1G93A/snph−/− mice had similar disease onset, defined as the day following peak weight, and life span, defined as the loss of the righting reflex (loss of ability to right the torso after 10 s) or a >20% weight loss. The number of mice measured for each genotype is indicated in parentheses.
Second, we compared the average onset of disease and lifespan. We did not find any significant difference between SOD1G93A and SOD1G93A/snph−/− mice. The average disease onset in female SOD1G93A and SOD1G93A/snph−/− mice was 127 ± 7 days (n = 10) and 126 ± 7 days (n = 10), respectively, versus 123 ± 5 days (n = 10) and 127 ± 7 days (n = 9) for male SOD1G93A and SOD1G93A/snph−/− mice, respectively (Fig. 3D). The average lifespan of C57BL6 mice carrying a high copy number of the human mutant SOD1G93A allele was 152 days, females had an average lifespan of 152 ± 6 days (n = 12) and males lived 152 ± 4 days (n = 11). Deleting snph had no significant impact on lifespan; female SOD1G93A/snph−/− mice lived 151 ± 4 days (n = 10) and males lived 156 ± 5 days (n = 10) (Fig. 3E). Overall, enhancing axonal mitochondria mobility by crossing snph−/− and SOD1G93A mice did not affect disease onset, progression or lifespan.
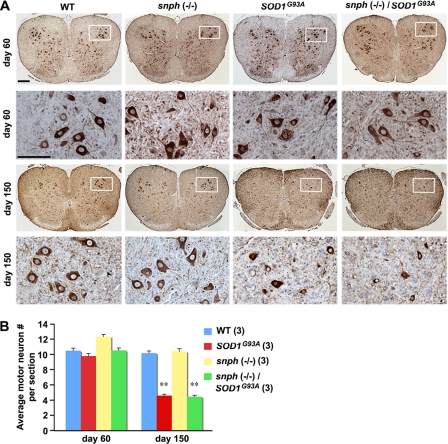
We next examined lumbar spinal cord motor neurons from P60 and P150 mice by labeling them with anti-SMI32 antibody (brown) and counterstaining with Nissl (purple). At the presymptomatic age (P60) (Fig. 4A, upper two panels), there was no significant difference in the spinal cord motor neuron count between WT and SOD1G93A mice (p = 0.06) or between WT and SOD1G93A/snph−/− mice (p = 0.91). When animals reached the end stage of the disease at about 150 days (Fig. 4A, lower two panels), there was a significant reduction in motor neuron number in both SOD1G93A (54.90%, p < 0.001) and SOD1G93A/snph−/− mice (55.88%, p < 0.001) compared with aged-matched WT and snph−/− mice (Fig. 4B).
FIGURE 4.
Enhanced axonal mitochondrial transport does not prevent motor neuron loss in SOD1G93A/snph−/− mice. A, lumbar spinal cord motor neurons from P60 and P150 mice were labeled with anti-SMI32 antibody (brown) and counterstained with Nissl (purple). The second and fourth rows are high magnification images corresponding to the boxed region in each spinal cord section. Scale bars: 200 μm in low magnification and 100 μm in high magnification. B, average number of motor neurons per spinal cord section at P60 and P150. Note that at the presymptomatic age of 60 days, the spinal cord motor neuron count in SOD1G93A and SOD1G93A/snph−/− mice did not display any significant difference from the WT littermate (p = 0.12 for SOD1G93A, p = 0.91 for SOD1G93A/snph−/−. In contrast, near the disease end stage (P150), a significant reduction in motor neuron count in both SOD1G93A (54%, p < 0.001) and SOD1G93A/snph−/− mice (55%, p < 0.001) was observed when compared with age-matched WT and snph−/− littermates. Three animals and a total of 90 spinal cord sections were examined for each genotype. Error bars: S.E. **, denote p < 0.001.
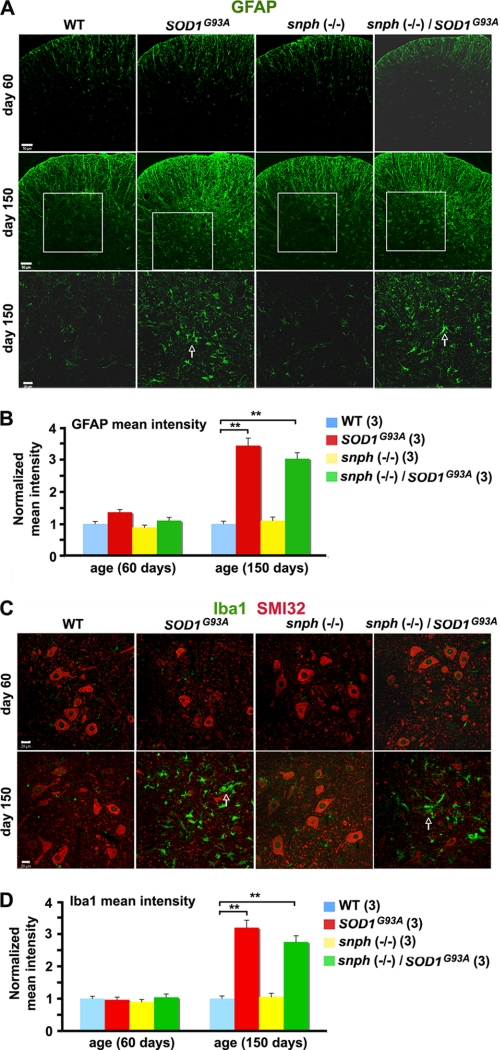
As spinal cord and brain gliosis is a prominent feature in ALS (17, 18), next we immunostained GFAP (glial fibrillary acidic protein) and Iba-1 (ionized calcium binding adaptor molecule 1) to examine astrocytes and microglia, respectively. At 60 days of age, there was no significant difference in the normalized mean intensity of GFAP staining when SOD1G93A/snph−/− and snph−/− mice were compared with WT mice (Fig. 5A). However, by P150 the normalized mean intensity of GFAP increased by 243.00 ± 23.45% in SOD1G93A and by 201.51 ± 19.30% in SOD1G93A/snph−/− mice compared with their WT littermates (p < 0.001). Interestingly, no significant difference in the mean intensity of GFAP was found between SOD1G93A and SOD1G93A/snph−/− mice (p = 0.19) (Fig. 5B). Consistently, we did not observe any difference in the normalized mean intensity of Iba-1 staining between any of the four genotypes at the presymptomatic age of 60 days (Fig. 5C). However, the normalized mean intensity of Iba-1 in the lateral ventral horn of P150 mice was significantly increased by 218.79 ± 18.80% in SOD1G93A and by 175.06 ± 13,67% in SOD1G93A/snph−/− mice compared with their WT littermates (p < 0.001). Again, there was no significant difference in the mean intensity of Iba-1 between SOD1G93A and SOD1G93A/snph−/− mice at their late disease stage (p = 0.08) or between the WT and snph−/− controls of the same age (p = 0.50) (Fig. 5D).
FIGURE 5.
Similar increased density pattern of astrocytes and microglia in the spinal cord of SOD1G93A and SOD1G93A/snph−/− mice. A, increased GFAP staining in the lumbar spinal cord of SOD1G93A and SOD1G93A/snph−/− P150 mice indicates increased astrocyte density. The transverse sections of the lumbar region of the spinal cord derived from P60 or P150 mice were stained with an anti-GFAP antibody (green). The boxed regions in the second row are magnified and displayed in the third row of the panel. Arrows point to GFAP-labeled astrocytes. Scale bars: 50 μm at low magnification (rows 1 and 2) and 20 μm at high magnification (row 3). B, quantification of normalized GFAP mean intensity. C, increased microglia density (labeled with Iba1) and reduced motor neuron density (labeled with SMI32) in the spinal cord of SOD1G93A and the crossed SOD1G93A/snph−/− mice at P150. Transverse spinal cord sections were co-stained with antibodies against SMI32 (red) and Iba1 (green). Arrows indicate Iba1-labeled microglia. Scale bars: 20 μm. D, quantification of the normalized mean intensity of Iba-1. 9 slice sections from three mice for each genotype were examined. Error bars: S.E. ** denotes p < 0.001.
DISCUSSION
ALS is one of the most common motor neurodegenerative disorders. Whereas 90% of ALS cases are sporadic, 10% are familial, of which 20% are associated with mutations in SOD1 (7). Although the mechanisms underlying the selective degeneration of motor neurons in mutant SOD1 fALS remain elusive, the toxic mutant SOD1 gain of function is involved in the pathogenesis. Several hypotheses have been proposed, including the sequestration of essential cellular components in SOD1 aggregates (19), secretion of toxic factors by astrocytes expressing mutant SOD1 (20, 21), disruption of oxidative stress and calcium homeostasis, mitochondria-dependent apoptotic pathway (22), caspase-mediated apoptosis (23), and the ubiquitin-proteasome system (see reviews by 24, 25). Mutant human SOD1 accumulates in mitochondria of the brain, spinal cord, and motor neurons (26–28). Both morphological changes and dysfunction of axonal mitochondria have been observed in ALS patients and mutant SOD1 mouse models (15, 29–40).
There is also growing evidence that the perturbation and impairment of axonal transport (9, 14, 16, 41–47) is associated with the pathogenesis of ALS patients and mutant SOD1 mice. Mutant SOD1 has been shown to interact with both the anterograde kinesin-2 motor protein complex via kinesin-associated protein 3 (KAP3) (48), and the retrograde dynein-dynactin motor protein complex (49). Mitochondrial morphology and membrane dynamics, required for maintaining proper function, depend on proper mitochondrial mobility in neurons (50). Several studies have suggested that the onset of ALS symptoms is caused by alterations in mitochondrial transport induced by mutant SOD1 expression (10, 11). In the SOD1G93A mouse model, disease onset and motor neuron death is immediately preceded by the accumulation of dysfunctional mitochondria in the distal region of the axon near the neuromuscular junction. However, whether altered mitochondrial transport plays a critical role in axonal degeneration or is merely a side effect of general defective axonal transport has not been addressed directly.
By genetically crossing snph−/− and SOD1G93A mice, we provide the first genetic, cellular and pathological evidence that alteration of axonal mitochondrial mobility in SOD1G93A mice plays a minor role in fALS-linked pathology. The deletion of snph, the docking receptor for axonal mitochondria, substantially increases the number of axonal mitochondria in the mobile state (12). We found that the lifespan and motor function of snph−/− mice dose not differ significantly from the C57BL/6 background WT mice used in our study despite a significant increase in mobile axonal mitochondria. This suggests that increasing axonal mitochondrial mobility by deleting snph does not adversely affect lifespan or motor function. Thus, crossing the SOD1G93A transgenic mice with snph knock-out mice allowed us to directly investigate the role of mitochondrial mobility in axonal degeneration by comparing clinical and histological observations of SOD1G93A mice to SOD1G93A/snph−/− mice. The crossed SOD1G93A/snph−/− mice demonstrate significantly increased axonal mitochondrial mobility relative to SOD1G93A. Additionally, both anterograde and retrograde transport was significantly increased in the SOD1G93A/snph−/− mice, thus providing a unique model to directly assess the impact of increased axonal mitochondrial transport on motor neuron degeneration in the ALS-linked mutant mice.
Surprisingly, despite more than a 2-fold increase in axonal mitochondrial mobility in the crossed SOD1G93A/snph−/− mice, we did not observe any clinical or histological difference in the disease course of this crossed mouse line compared with SOD1G93A mice. Both SOD1G93A and SOD1G93A/snph−/− mice exhibited a similar weight loss pattern accompanied by deterioration of motor function beginning at ∼17 weeks. Furthermore, both strains of mice exhibited 55% motor neuron loss and significant gliosis at the disease end stage. Our findings suggest that the role of mutant SOD1 in the pathogenesis of this ALS model is unlikely mediated through altering mitochondrial transport. The high mitochondrial mobility in both SOD1G93A/snph−/− and snph−/− mice dose not correlate with lifespan or motor function, which further suggests that altered mitochondrial mobility probably plays a minor role in the pathogenesis of this mouse model.
We previously demonstrated that snph−/− embryonic hippocampal neurons have a slightly lower mitochondrial density than WT mice (12). In the present study, we did not observe a statistical difference in mitochondrial density in the distal axonal processes between adult snph−/− and WT mice. This may be attributable to developmental regulation of axonal mitochondrial transport. It was reported that mitochondria are more mobile in young neurons relative to mature neurons (51). Deleting snph in mice results in a more robust increase in axonal mitochondrial mobility of embryonic hippocampal neurons (76 ± 20%) relative to that of adult DRG neurons (37.49 ± 1.74%). Such a large increase in mobile mitochondria in embryonic neurons may impact their density in axons.
In contrast, there was a lower mitochondrial density in axons of SOD1G93A DRG neurons relative to WT neurons. Deleting snph (SOD1G93A/snph−/−) rescued the deficit. Given the fact that the SOD1G93A/snph−/− phenotype was clinically similar to the uncrossed SOD1G93A phenotype, the reduced axonal mitochondrial density observed in SOD1G93A mice does not critically contribute to the pathogenesis of fALS. It is likely that other factors including, but not limited to, protein aggregation and mitochondrial dysfunction caused by mutant SOD1 play a more significant role in the disease process. It is important to note that while many studies related to mitochondrial transport in ALS mice models have been carried out using the SOD1G93A mutant, the disease process in this strain progresses rapidly with an average lifespan of 152 days. With such a short lifespan, we cannot exclude the possibility that the crossed SOD1G93A/snph−/− mice do not live long enough for the genetic rescue to occur.
Our study suggests that the impairment of mitochondrial transport seen in SOD1G93A mice may be a secondary effect of a more general alteration of axonal transport caused by mutant aggregates or motor protein defects. In SOD1G93A mice, a specific mutation in the dynein heavy chain produces a complete recovery of the axonal retrograde transport defect and rescues the phenotype (14). A Cra1 mutation in Dnchc1 improves the phenotype of SOD1G93A mice and prolongs their survival period (52). Recent findings that the SOD1-dependent pathogenic mechanism impairs axonal transport in both familial and sporadic ALS indicate a general transport defect associated with ALS-linked neurodegeneration (46). Alterations of other transport cargos may play a more important role than defective mitochondrial transport. A shift in retrograde transport from survival-promoting to death-promoting signaling may be responsible for the rapid onset of symptoms in the SOD1G93A mouse model (16). Misfolded mutant SOD1 associates with the outer membrane of mitochondria in spinal cord and motor neurons (26, 28) and damages mitochondrial membrane conductance and metabolites by directly binding to the voltage-dependent anion channel (VDAC) (40). Thus, mitochondria dysfunction, rather than impaired mobility, may be more important in rapid-onset motor neuron degeneration.
Supplementary Material
Acknowledgments
We thank X. Lin and Huaibin Cai (NIA, NIH) for generous help in training us in mouse pathology analysis and behavioral tests, and insightful discussion on the ALS mouse model. We also thank the following people: Q. Cai, Y. Chen, Y. Xie, R. Chia, K. Tsubota, and other members of the Sheng laboratory for technical help and critical comments; J. Tokita and M. Davis for editing the manuscript.
This work was supported, in whole or in part, by the Intramural Research Program of the NINDS, National Institutes of Health (to Z.-H. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movies S1–S4.
Animal care and use in this study were carried out in accordance with National Institutes of Health guidelines and was approved by the National Institutes of Health, NINDS/NIDCD Animal Care and Use Committee.
- ALS
- amyotrophic lateral sclerosis
- ANOVA
- analysis of variance
- DIV
- days in vitro
- DRG
- dorsal root ganglion
- GFAP
- glial fibrillary acidic protein
- Iba-1
- ionized calcium binding adaptor molecule 1
- SOD1
- copper/zinc superoxide dismutase 1
- SNPH
- syntaphilin.
REFERENCES
- 1. Hollenbeck P. J., Saxton W. M. (2005) J. Cell Sci. 118, 5411–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cai Q., Sheng Z. H. (2009) Neuron 61, 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Macaskill A. F., Rinholm J. E., Twelvetrees A. E., Arancibia-Carcamo I. L., Muir J., Fransson A., Aspenstrom P., Attwell D., Kittler J. T. (2009) Neuron 61, 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang X., Schwarz T. L. (2009) Cell 136, 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hirokawa N., Niwa S., Tanaka Y. (2010) Neuron 68, 610–638 [DOI] [PubMed] [Google Scholar]
- 6. Cleveland D. W., Rothstein J. D. (2001) Nat. Rev. Neurosci. 2, 806–819 [DOI] [PubMed] [Google Scholar]
- 7. Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X. (1993) Nature 362, 59–62 [DOI] [PubMed] [Google Scholar]
- 8. Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., Deng H. X. (1994) Science 264, 1772–1775 [DOI] [PubMed] [Google Scholar]
- 9. Sasaki S., Iwata M. (1996) Neurology 47, 535–540 [DOI] [PubMed] [Google Scholar]
- 10. De Vos K. J., Chapman A. L., Tennant M. E., Manser C., Tudor E. L., Lau K. F., Brownlees J., Ackerley S., Shaw P. J., McLoughlin D. M., Shaw C. E., Leigh P. N., Miller C. C., Grierson A. J. (2007) Hum. Mol. Genet. 16, 2720–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Magrané J., Hervias I., Henning M. S., Damiano M., Kawamata H., Manfredi G. (2009) Hum. Mol. Genet. 18, 4552–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kang J. S., Tian J. H., Pan P. Y., Zald P., Li C., Deng C., Sheng Z. H. (2008) Cell 132, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ilieva H. S., Yamanaka K., Malkmus S., Kakinohana O., Yaksh T., Marsala M., Cleveland D. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12599–12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kieran D., Hafezparast M., Bohnert S., Dick J. R., Martin J., Schiavo G., Fisher E. M., Greensmith L. (2005) J. Cell Biol. 169, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kong J., Xu Z. (1998) J. Neurosci. 18, 3241–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perlson E., Jeong G. B., Ross J. L., Dixit R., Wallace K. E., Kalb R. G., Holzbaur E. L. (2009) J. Neurosci. 29, 9903–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boillée S., Yamanaka K., Lobsiger C. S., Copeland N. G., Jenkins N. A., Kassiotis G., Kollias G., Cleveland D. W. (2006) Science 312, 1389–1392 [DOI] [PubMed] [Google Scholar]
- 18. Yamanaka K., Chun S. J., Boillee S., Fujimori-Tonou N., Yamashita H., Gutmann D. H., Takahashi R., Misawa H., Cleveland D. W. (2008) Nat. Neurosci. 11, 251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bruijn L. I., Houseweart M. K., Kato S., Anderson K. L., Anderson S. D., Ohama E., Reaume A. G., Scott R. W., Cleveland D. W. (1998) Science 281, 1851–1854 [DOI] [PubMed] [Google Scholar]
- 20. Clement A. M., Nguyen M. D., Roberts E. A., Garcia M. L., Boillée S., Rule M., McMahon A. P., Doucette W., Siwek D., Ferrante R. J., Brown R. H., Jr., Julien J. P., Goldstein L. S., Cleveland D. W. (2003) Science 302, 113–117 [DOI] [PubMed] [Google Scholar]
- 21. Nagai M., Re D. B., Nagata T., Chalazonitis A., Jessell T. M., Wichterle H., Przedborski S. (2007) Nat. Neurosci. 10, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guégan C., Vila M., Rosoklija G., Hays A. P., Przedborski S. (2001) J. Neurosci. 21, 6569–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li M., Ona V. O., Guégan C., Chen M., Jackson-Lewis V., Andrews L. J., Olszewski A. J., Stieg P. E., Lee J. P., Przedborski S., Friedlander R. M. (2000) Science 288, 335–339 [DOI] [PubMed] [Google Scholar]
- 24. Bruijn L. I., Miller T. M., Cleveland D. W. (2004) Annu. Rev. Neurosci. 27, 723–749 [DOI] [PubMed] [Google Scholar]
- 25. Pasinelli P., Brown R. H. (2006) Nat. Rev. Neurosci. 7, 710–723 [DOI] [PubMed] [Google Scholar]
- 26. Liu J., Lillo C., Jonsson P. A., Vande Velde C., Ward C. M., Miller T. M., Subramaniam J. R., Rothstein J. D., Marklund S., Andersen P. M., Brännström T., Gredal O., Wong P. C., Williams D. S., Cleveland D. W. (2004) Neuron 43, 5–17 [DOI] [PubMed] [Google Scholar]
- 27. Vijayvergiya C., Beal M. F., Buck J., Manfredi G. (2005) J. Neurosci. 25, 2463–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vande Velde C., Miller T. M., Cashman N. R., Cleveland D. W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dal Canto M. C., Gurney M. E. (1994) Am. J. Pathol. 145, 1271–1279 [PMC free article] [PubMed] [Google Scholar]
- 30. Wong P. C., Pardo C. A., Borchelt D. R., Lee M. K., Copeland N. G., Jenkins N. A., Sisodia S. S., Cleveland D. W., Price D. L. (1995) Neuron 14, 1105–1116 [DOI] [PubMed] [Google Scholar]
- 31. Vielhaber S., Winkler K., Kirches E., Kunz D., Büchner M., Feistner H., Elger C. E., Ludolph A. C., Riepe M. W., Kunz W. S. (1999) J. Neurol. Sci. 169, 133–139 [DOI] [PubMed] [Google Scholar]
- 32. Echaniz-Laguna A., Zoll J., Ribera F., Tranchant C., Warter J. M., Lonsdorfer J., Lampert E. (2002) Ann. Neurol. 52, 623–627 [DOI] [PubMed] [Google Scholar]
- 33. Mattiazzi M., D'Aurelio M., Gajewski C. D., Martushova K., Kiaei M., Beal M. F., Manfredi G. (2002) J. Biol. Chem. 277, 29626–29633 [DOI] [PubMed] [Google Scholar]
- 34. Wiedemann F. R., Manfredi G., Mawrin C., Beal M. F., Schon E. A. (2002) J. Neurochem. 80, 616–625 [DOI] [PubMed] [Google Scholar]
- 35. Dupuis L., di Scala F., Rene F., de Tapia M., Oudart H., Pradat P. F., Meininger V., Loeffler J. P. (2003) FASEB J. 17, 2091–2093 [DOI] [PubMed] [Google Scholar]
- 36. Higgins C. M., Jung C., Xu Z. (2003) BMC Neurosci. 4, 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Damiano M., Starkov A. A., Petri S., Kipiani K., Kiaei M., Mattiazzi M., Flint B. M., Manfredi G. (2006) J. Neurochem. 96, 1349–1361 [DOI] [PubMed] [Google Scholar]
- 38. Sasaki S., Iwata M. (2007) J. Neuropathol. Exp. Neurol. 66, 10–16 [DOI] [PubMed] [Google Scholar]
- 39. Nguyen K. T., García-Chacón L. E., Barrett J. N., Barrett E. F., David G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2007–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Israelson A., Arbel N., Da Cruz S., Ilieva H., Yamanaka K., Shoshan-Barmatz V., Cleveland D. W. (2010) Neuron 67, 575–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Warita H., Itoyama Y., Abe K. (1999) Brain. Res. 819, 120–131 [DOI] [PubMed] [Google Scholar]
- 42. Williamson T. L., Cleveland D. W. (1999) Nat. Neurosci. 2, 50–56 [DOI] [PubMed] [Google Scholar]
- 43. Hafezparast M., Klocke R., Ruhrberg C., Marquardt A., Ahmad-Annuar A., Bowen S., Lalli G., Witherden A. S., Hummerich H., Nicholson S., Morgan P. J., Oozageer R., Priestley J. V., Averill S., King V. R., Ball S., Peters J., Toda T., Yamamoto A., Hiraoka Y., Augustin M., Korthaus D., Wattler S., Wabnitz P., Dickneite C., Lampel S., Boehme F., Peraus G., Popp A., Rudelius M., Schlegel J., Fuchs H., Hrabe de Angelis M., Schiavo G., Shima D. T., Russ A. P., Stumm G., Martin J. E., Fisher E. M. (2003) Science 300, 808–812 [DOI] [PubMed] [Google Scholar]
- 44. Ligon L. A., LaMonte B. H., Wallace K. E., Weber N., Kalb R. G., Holzbaur E. L. (2005) Neuroreport 16, 533–536 [DOI] [PubMed] [Google Scholar]
- 45. Morfini G. A., Burns M., Binder L. I., Kanaan N. M., LaPointe N., Bosco D. A., Brown R. H., Jr., Brown H., Tiwari A., Hayward L., Edgar J., Nave K. A., Garberrn J., Atagi Y., Song Y., Pigino G., Brady S. T. (2009) J. Neurosci. 29, 12776–12786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bosco D. A., Morfini G., Karabacak N. M., Song Y., Gros-Louis F., Pasinelli P., Goolsby H., Fontaine B. A., Lemay N., McKenna-Yasek D., Frosch M. P., Agar J. N., Julien J. P., Brady S. T., Brown R. H., Jr. (2010) Nature Neurosci. 13, 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guipponi M., Li Q. X., Hyde L., Beissbarth T., Smyth G. K., Masters C. L., Scott H. S. (2010) J. Mol. Neurosci. 41, 172–182 [DOI] [PubMed] [Google Scholar]
- 48. Tateno M., Kato S., Sakurai T., Nukina N., Takahashi R., Araki T. (2009) Hum. Mol. Genet. 18, 942–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang F., Ström A. L., Fukada K., Lee S., Hayward L. J., Zhu H. (2007) J. Biol. Chem. 282, 16691–16699 [DOI] [PubMed] [Google Scholar]
- 50. Chen H., Chan D. C. (2009) Hum. Mol. Genet. 18, 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chang D. T., Reynolds I. J. (2006) Neuroscience 141, 727–736 [DOI] [PubMed] [Google Scholar]
- 52. Teuchert M., Fischer D., Schwalenstoecker B., Habisch H. J., Böckers T. M., Ludolph A. C. (2006) Exp. Neurol. 198, 271–274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.