Abstract
Co-translational protein targeting to the membrane is mediated by the signal recognition particle and its receptor (FtsY). Their homologous GTPase domains interact at the membrane and form a heterodimer in which both GTPases are activated. The prerequisite for protein targeting is the interaction of FtsY with phospholipids. However, the mechanism of FtsY regulation by phospholipids remained unclear. Here we show that the N terminus of FtsY (A domain) is natively unfolded in solution and define the complete membrane-targeting sequence. We show that the membrane-targeting sequence is highly dynamic in solution, independent of nucleotides and directly responds to the density of anionic phospholipids by a random coil-helix transition. This conformational switch is essential for tethering FtsY to membranes and activates the GTPase for its subsequent interaction with the signal recognition particle. Our results underline the dynamics of lipid-protein interactions and their importance in the regulation of protein targeting and translocation across biological membranes.
Keywords: Crystal Structure, Lipid-binding Protein, Membrane Lipids, Protein Targeting, Protein Translocation, FtsY, GTPase, SRP Receptor
Introduction
The biogenesis of most membrane proteins and many secretory proteins depends on the signal recognition particle (SRP)4 and the SRP receptor (SR). SRP binds to N-terminal signal sequences of nascent polypeptide chains at the ribosome (ribosome-nascent chain complexes (RNCs)) and acts as an adaptor between the ribosome and the membrane-embedded translocation channel (1–3). Interaction of SRP with SR (FtsY in bacteria and archaea and SRα in eukarya) specifies the target membrane and allows for the precise coordination of RNC release from SRP and its transfer to the translocation channel. Protein targeting critically depends on the two homologous GTPases present in SRP and SR forming a heterodimer (4). GTP binding to SRP and SR is required for heterodimer formation and GTP hydrolysis triggers the dissociation of the SRP-SR complex which resets the SRP system for a new round of translocation (5). Although FtsY does not contain a hydrophobic, transmembrane sequence, it was shown to be almost exclusively localized at the membrane (6). FtsY contains three domains: an N-terminal negatively charged A domain of unknown structure and function and the highly conserved N and G domains that form a structural and functional unit, the NG domain (7, 8) (see Fig. 1A). The A domain acts as negative regulator of the FtsY GTPase in a lipid-free environment (9) and was suggested to participate in membrane interaction of FtsY by its N-terminal region (10). However, the A domain is not essential in Escherichia coli as a truncation variant (termed NG+1) is functional in vivo (11). It was shown that the FtsY GTPase is activated by anionic phospholipids (9) and that the membrane interaction of FtsY is crucial for the release of RNCs from the SRP-FtsY complex (12, 13), which was confirmed recently (14). Membrane interaction of E. coli FtsY depends on a conserved motif at the N terminus of the N domain, referred to as membrane-targeting sequence (MTS) (15). Recently, strong genetic evidence was provided also for a functional interaction of FtsY with acidic lipids in vivo (16). However, the precise mechanism of how membrane lipids activate the FtsY GTPase and a detailed understanding of the consequences of FtsY-lipid interaction on SRP-mediated protein targeting are still unknown. Here we address the molecular mechanisms of the regulation of FtsY by anionic phospholipids.
FIGURE 1.
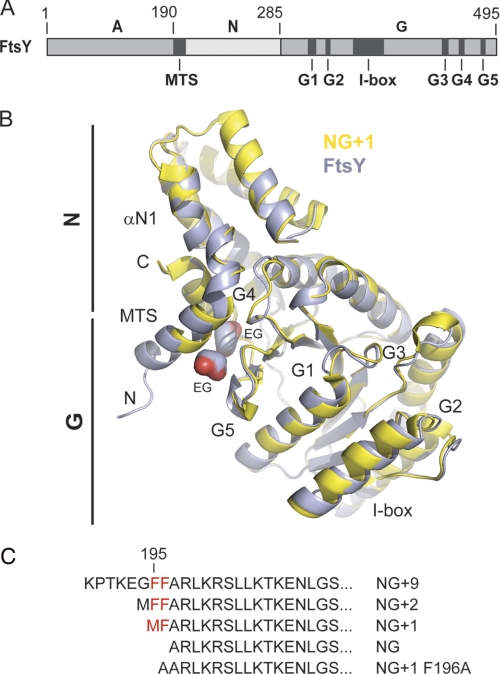
Crystal structure of FtsY allows definition of complete MTS. A, the domain architecture of the E. coli SRP receptor FtsY. The location of the MTS, the I-box, and the conserved G elements (G1–G5) are indicated. B, superposition of FtsY (blue) and NG+1 (yellow; Protein Data Bank code 2QY9) crystal structures. FtsY and NG+1 are shown in ribbon representation. The MTS present at the N terminus of αN1 is extended in the FtsY structure by two turns (residues 188–195). N and C termini of FtsY are indicated by “N” and “C,” respectively. Two ethylene glycol molecules associated to the hydrophobic face of the MTS are indicated by “EG.” C, N-terminal sequences from various FtsY variants used in this study. The conserved double phenylalanine motif is colored in red.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
E. coli BL21 (DE3) or C43 (DE3) were used for the overexpression of SRP and FtsY constructs. The purification was performed as described previously (15, 17).
Crystallization, Data Collection, and Structure Determination
Crystals were prepared by mixing 1 μl of fresh FtsY protein with 1 μl of reservoir solution containing 0.1 m Hepes (pH 7.0) and 30% Jeffamine ED-2001 (pH 7.0). All crystals were grown at 4 °C by the sitting drop vapor diffusion method. Crystals were flash frozen in liquid nitrogen directly or after the addition of 20% (v/v) ethylene glycol as a cryoprotectant. Data were collected at 100 K at the European Synchrotron Radiation Facility in France. Data were processed using iMOSFLM and SCALA (18). Structures were determined by molecular replacement using CCP4-implemented PHASER (18) with the NG domain of FtsY (Protein Data Bank code 1FTS) as a search model (7). The structures were manually built with Coot (19) and refined with Refmac 5 (20) and Phenix (21). Refinement statistics are given in supplemental Table S1. Figures were generated with PyMol (Schrödinger, LLC).
Amide Hydrogen 1H/2H Exchange (HX)-MS Experiments
Amide HX-MS experiments were performed similarly to those described earlier (22, 23). Amide HX was initiated by a 20-fold dilution of 100 pmol of either FtsY/NG/NG+1 (apo) or FtsY/NG/NG+1 with an excess of nucleotides into D2O buffer containing 20 mm Hepes (pD 7.5), 200 mm NaCl, 10 mm MgCl2, and 10 mm KCl at 30 °C. After various time intervals (10 s to 1 h), the exchange reaction was quenched by decreasing the temperature to 0 °C and the pH with ice-cold quench buffer (400 mm KH2PO4/H3PO4 (pH 2.2)). Quenched samples were injected into an HPLC-MS setup as described. The deuteron content of the peptic peptides covering the FtsY and the NG+1/NG constructs were determined from the centroid of the molecular ion isotope envelope. The deuteron content was calculated after adjustment for deuteron gain/loss during digestion and the HPLC-MS setup. For this adjustment, non-deuterated and fully deuterated protein samples were analyzed (24). Fully deuterated samples were prepared by three cycles of drying and resolubilization in D2O containing 6 m guanidinium hydrochloride. The 0% control was not treated with D2O.
CD Spectroscopy
Peptides were synthesized at the peptide synthesis unit of the German Cancer Research Center (DKFZ), Heidelberg, Germany. Far-UV CD spectra were measured with a Jasco J-810 spectroscope at 20 °C. 0.3 mm peptide and 6 mm large unilamellar vesicles (LUVs) were mixed in a buffer containing 5 mm NaH2PO4 (pH 7.4). For the measurements, a quartz cuvette with a 0.2-mm path length was used. The peptide:lipid molar ratio was 1:20. Spectra were the average of 10 acquired scans. The scan rate was 50 nm/min, the bandwidth was 1 nm, and the sensitivity was 100 millidegrees. The background signal was subtracted from the final spectra before expressing values as mean residue molar ellipticity.
Fluorescence Microscopy
The E. coli strain BL21 (DE3) (Novagen) was used for fluorescence microscopy studies. Freshly transformed cells were grown overnight on agar plates containing 1% glucose and the appropriate antibiotics. Cells were inoculated into LB medium containing 1% glucose and the appropriate antibiotics. After 3 h of growth at 37 °C, cells were collected by centrifugation, washed twice, and resuspended in fresh LB medium (A600 0.04). The cultures were incubated with shaking at 30 °C for 4 h (A600 0.6) before induction with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. After induction, cells were grown for additional 30 min to 1 h. Cells were sampled directly from the culture for microscopy or were fixed for 45 min at room temperature by the addition to the culture of 1.7% formaldehyde and 0.17% glutaraldehyde. Fixed cells were collected by low speed centrifugation and resuspended in 1× phosphate-buffered saline (PBS). Microscopic analysis was performed using an Axiovert 200M microscope (Zeiss).
Co-translational Protein Targeting Assay
The co-translational protein targeting assay was performed as described previously (25, 26). The model substrate was pre-prolactin (ppl). Briefly, pre-prolactin was first translated using wheat germ extract (Promega) in the presence of [35S]methionine followed by addition of 7-methyl-GTP. Pre-prolactin translocation across the microsomal membranes (2–4 eq of TKRM, high salt washed and partially trypsinized microsomal membranes) was then initiated by the addition of equimolar amounts (300 nm) of SRP and FtsY or FtsY variants. After an incubation time of 30 min at 25 °C, the reaction was stopped by TCA precipitation, and the translocation products were separated by SDS-PAGE and quantified with a phosphorimaging system (Fuji FLA7000).
GTPase Activity Assay
The GTPase assay was performed as described previously with slight modifications (27). 100 nm FtsY was incubated with 100 nm SRP in the presence of 10 μm LUVs (phosphatidylglycerol (PG)). The reaction was performed in a buffer containing 20 mm Tris (pH 7.5), 65 mm NaCl, 2 mm MgCl2, and 1% glycerol. The reaction was started by adding GTP with a final concentration of 10 μm supplemented with 2–10 μCi of radioactive γ-32P-labeled GTP (Amersham Biosciences). The reactions were incubated at 37 °C and stopped by transferring a 10 μl aliquot of the reaction into 400 μl of an ice-cold 12% charcoal slurry containing 10 mm KH2PO4 and 0.1 m HCl. The absorption mixture was vortexed and centrifuged for 10 min in a tabletop centrifuge at 14,000 rpm. 150 μl of supernatant were transferred into 3 ml of scintillation mixture (ReadySafe, Beckman) and counted for 1 min by Cerenkov counting.
RESULTS
Structural and Dynamic Properties of SRP Receptor FtsY
To understand the structural properties of the complete SRP receptor FtsY, we determined its crystal structure at 1.6-Å resolution (supplemental Table S1) (the atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 2YHS). Compared with the previously solved crystal structures of the NG and NG+1 variants of FtsY (7, 15), an additional eight residues (residues 188–195) were ordered at the N terminus. The MTS as part of helix αN1 is thereby extended by two turns (Fig. 1B), implicating a longer MTS than previously assumed (Fig. 1C). Unexpectedly, the major part of the A domain (residues 1–187) could not be assigned to the available electron density, indicating that the A domain is highly flexible and/or even unstructured. To gain further insights into the dynamic properties of FtsY in solution, we performed continuous labeling HX-MS (28–31). Deuteron incorporation allows analysis of the global stability and different conformational states of a protein (32). FtsY exchanged about 80% of its exchangeable amide hydrogens in D2O within 1 h (supplemental Fig. S1, A and B). The overall exchange characteristics indicate the presence of highly dynamic, solvent-accessible regions without extensive, stable hydrogen bonds typical for proteins with a low amount of secondary structure. To define the contributions of the A and NG domains to the global deuteron exchange properties of FtsY, the HX-MS experiment was repeated with the NG+1 variant. The difference between the NG+1 variant and FtsY of about 160 fast exchanging amide hydrogens could be exclusively attributed to the A domain. These results show that the A domain is highly dynamic. To identify regions in the A domain that are tightly folded in solution or are in a rapid unfolding-folding equilibrium, we analyzed deuteron incorporation into peptic peptides derived from FtsY (Fig. 2, A and B). Briefly, FtsY and the NG+1 variant were digested after the continuous labeling HX reaction under quench conditions when hydrogen exchange is slow (22). Peptide fragments with an average size of about eight amino acids were obtained and subsequently analyzed by liquid chromatography-electrospray ionization-mass spectrometry. All analyzed peptides of the A domain exhibited a high degree of solvent accessibility with complete deuteration within 10 s (Fig. 2A). These data provide strong evidence that the A domain is natively unfolded in solution. The absence of secondary structure elements was confirmed by 1H NMR experiments performed with an isolated A domain (residues 1–187) obtained from a proteolytically cleavable FtsY variant (supplemental Fig. S2).
FIGURE 2.
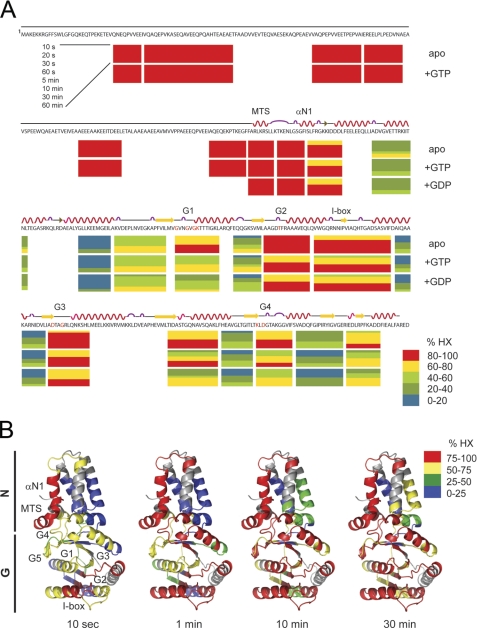
Analysis of dynamic properties of FtsY in solution by HX-MS. A, relative deuteron incorporation in FtsY for the nucleotide-free (apo), GDP-, and GTP-bound states. Each horizontal block represents an analyzed peptic fragment. Peptides are colored according to the relative deuteron incorporation as indicated. Secondary structure elements and the positions of the MTS, helix αN1, the G elements (G1–G5), and the I-box are marked above the amino acid sequence. B, fast and slow exchanging regions are mapped on the crystal structure shown in a ribbon representation. Segments corresponding to peptic fragments are colored according to the relative deuteron incorporation into the nucleotide-free protein after 10 s and 1, 10, and 30 min in D2O as indicated.
MTS Does Not Respond to Nucleotide-induced Conformational Changes in FtsY
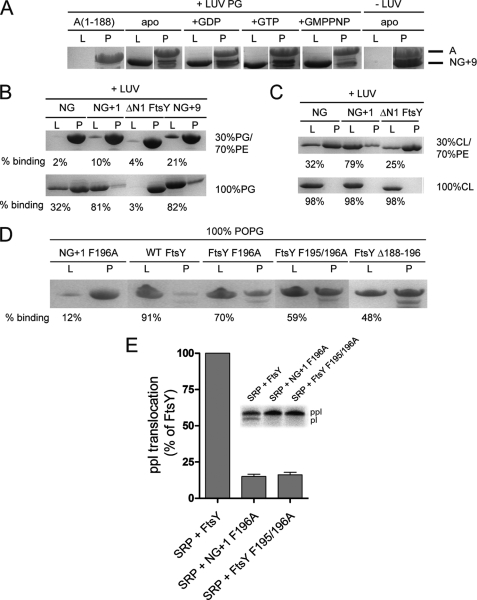
The kinetics of deuteron incorporation into peptides derived from the NG+1 variant overall correlates nicely with the crystal structures (7, 15) (Fig. 2B). However, although the MTS adopts an α-helix in the crystal structure that packs against the NG domain (Fig. 1B), in solution, the MTS is highly dynamic and solvent-accessible (Fig. 2, A and B). To understand this surprising observation, we asked whether the presence of nucleotides would influence the dynamic character of the MTS. We repeated the HX-MS experiments for FtsY, NG, and NG+1 preincubated with an excess of either GDP or GTP (Fig. 2A and supplemental Fig. S1, A and B). Comparison of the global exchange kinetics of nucleotide-free and GDP-, GTP-, or GMP-PNP-loaded states disclosed a similar behavior for all three analyzed variants (supplemental Fig. S1). Nucleotide interaction induced a significant reduction of deuteron incorporation, which suggests a more compact structure with reduced solvent accessibility and more amide hydrogens protected by hydrogen bonds. The reduction of deuteron incorporation was most prominent in the presence of GDP followed by GTP and to a lower extent in the presence of GMP-PNP. The observed conformational changes are exclusively localized in the N-G domain interface and in the G domain (Fig. 2A). Regions with a significant degree of nucleotide-induced protection include the G1, G2, and G4 elements; the switch 1 region; the N-terminal region of helix α5; and the C terminus containing β8 and the connecting loop to helix α7. Differences in the deuteration level between different nucleotides could be assigned primarily to the G1 element (supplemental Fig. S3, A and B). However, the dynamic character of the MTS did not change upon addition of nucleotides, indicating that the MTS secondary structure is not altered by nucleotides. To confirm that the MTS-dependent association of FtsY with membrane lipids is not influenced by nucleotides, we performed density gradient flotation analysis. FtsY was proteolytically cleaved, and the resulting A and NG+9 domains (supplemental Fig. S2) were tested for association with phosphatidylglycerol (Fig. 4A). The A domain was unable to stably interact with LUVs, whereas its counterpart NG+9 efficiently associated with LUVs.
FIGURE 4.
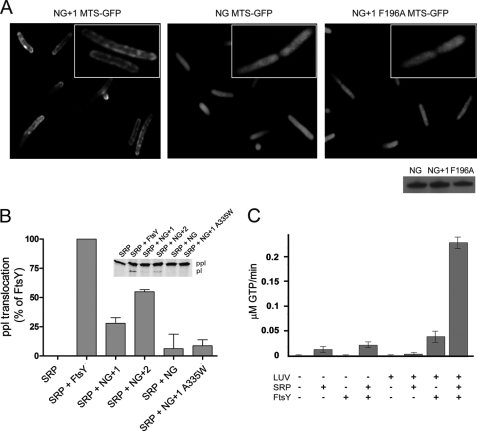
Lipid binding properties of FtsY variants. A, the nucleotide dependence of NG+9 and A domain binding to LUVs (PG) was investigated by density gradient flotation analysis. The LUV and pellet fractions are labeled with “L” and “P,” respectively. B and C, binding of NG, NG+1, NG+9, and ΔN1 FtsY to LUVs containing different molar ratios of PG, PE, and cardiolipin (CL). The ΔN1 FtsY lacks the complete MTS sequence and was used as a negative control. D, binding of full-length FtsY and different MTS variants to LUVs (PG). The amount of protein bound to LUVs is given as percentage of total protein. For lipid binding studies, 20 μg (5–10 μm) of proteins and 1.8 mm phospholipids in the absence or presence of 2 mm nucleotides were used. E, ability of FtsY, NG+1 F196A, and FtsY F195/196A to support membrane translocation of the SRP model substrate ppl. The inset shows the SDS-PAGE used to separate ppl and prolactin (pl). The efficiency of ppl translocation was quantified and is given in percent relative to FtsY. The error bars represent the standard deviation between three independent measurements. POPG, 1-palmitoyl-2-oleoyl-phosphatidylglycerol.
MTS Responds to Density of Anionic Phospholipids by Random Coil-Helix Transition
Next, we analyzed the secondary structure of a synthetic peptide representing the minimal MTS by circular dichroism (CD) spectroscopy. The NG+1 peptide exhibited a single minimum at 200 nm, typical for a random coil with no secondary structure present (Fig. 3A). This agrees with the observed HX-MS pattern of this region in the context of FtsY (Fig. 2A). To assess the ability of the peptide to adopt an α-helical structure, we used trifluoroethanol to artificially induce helix formation. The presence of trifluoroethanol induced a random coil-helix transition in the MTS peptide, showing its ability to form an α-helix in solution (Fig. 3A).
FIGURE 3.
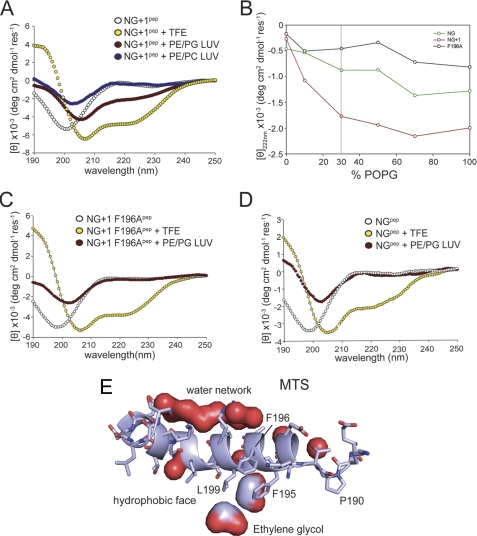
Anionic phospholipids induce random coil-helix transition of the MTS. Three different MTS peptides were analyzed by CD spectroscopy in the absence of LUVs (white) and the presence of plasma membrane-mimicking LUVs (70% PE and 30% PG; red) and LUVs composed entirely of zwitterionic phospholipids (70% PE and 30% phosphatidylcholine (PC); blue). Samples were treated with 83% trifluoroethanol (TFE) to induce random coil-helix transition (yellow). The MTS of NG+1 (NG+1pep; MFARLKRSLLKTKENLG) (A), MTS of the NG+1 F196A variant (NG+1 F196Apep; AARLKRSLLKTKENLG) (C), and MTS of the NG variant (NGpep; ARLKRSLLKTKENLG) (D) are shown. Only the in vivo functional MTS (NG+1 peptide) undergoes a random coil-helix transition upon interaction with anionic phospholipids. B, secondary structure transition of the three MTS variants is shown as a function of the concentration of anionic phospholipids. The physiologically relevant PG concentration is indicated by a vertical red line. POPG, 1-palmitoyl-2-oleoyl-phosphatidylglycerol. The mean residue ellipticity is shown in degrees cm2 dmol−1 residue−1 (deg cm2 dmol−1 res−1). E, close-up of the MTS as part of the FtsY structure. The MTS is shown in ribbon representation. The pronounced amphipathic character of the MTS is indicated by a network of bound water molecules at the hydrophilic side and the presence of two ethylene glycol molecules at the hydrophobic side.
We previously showed that the MTS is involved in the interaction of FtsY with membrane lipids (15). We therefore asked whether lipid interaction can induce a conformational change of the MTS. The peptide was analyzed in the presence of LUVs containing 70% phosphatidylethanolamine (PE) and 30% PG to mimic the composition of the E. coli inner membrane. Upon incubation with these LUVs, a change in the mean residual ellipticity was noticed at shorter wavelengths, which can be attributed to an increase in light scattering by LUVs. Importantly, the peptide showed an alteration of the CD spectrum characteristic for a random coil-helix transition with a red shift of the minimum at 200 nm to 208 nm and the minimum at 222 nm becoming more pronounced (Fig. 3A). In contrast, LUVs with zwitterionic lipids (PE/phosphatidylcholine) did not induce this transition, emphasizing the importance of anionic phospholipids (Fig. 3A). The maximal transition in secondary structure was dependent on the anionic phospholipid concentration in the LUVs (Fig. 3B). This shows that the MTS responds directly to the density of negative charges at the LUVs and suggests that the MTS is tuned for the lipid composition of the E. coli inner membrane.
We have shown previously that truncation of the first 196 residues or mutating Phe-196 to Ala completely abolishes receptor activity in vivo (15). Peptides derived either from the inactive NG variant or the NG+1 F196A mutant failed to undergo a random coil-helix transition in the presence of physiologically relevant amounts of anionic phospholipids (Fig. 3, C and D). The CD data clearly demonstrate that peptides harboring the inactive variants (NG and NG+1 F196A) lack secondary structure both in the absence and in the presence of lipids and are therefore unable to respond to lipids. Only when LUVs enriched in anionic phospholipids were used the binding of the NG variant was enhanced (Fig. 4, B and C), accompanied by partial folding (Fig. 3B). This provides a mechanistic and structural basis for the recently described stabilization and activation of the NG variant in E. coli cells enriched in anionic phospholipids (16).
To analyze the F196A variant in the context of FtsY, we performed lipid interaction studies with different FtsY variants (FtsY F196A, FtsY F195A/F196A, and FtsYΔ188–196; Fig. 4D). All three FtsY variants showed a severe reduction in their interaction with LUVs enriched in anionic phospholipids. These experiments suggest that besides the electrostatic component of the MTS-lipid interaction a hydrophobic component plays a major role. Support for this model comes from two ethylene glycol molecules that pack between the hydrophobic face of the amphipathic helix and the protein core in the crystal structure of FtsY (Fig. 3E). Ethylene glycol molecules could be viewed as indicators for the position of the glycerol moiety of the phospholipid head groups in the context of the lipid bilayer. Interestingly, ethylene glycol interacts with the conserved Phe-195 and Leu-199 residues, which are essential for FtsY interaction with phospholipids. By contrast, the hydrophilic face of the helix is involved in a continuous network of water molecules, indicating the helix orientation at the membrane.
Efficiency of Protein Translocation Requires Complete MTS
To address the membrane targeting properties of the MTS in vivo, three peptides (representing the MTS of NG, NG+1 F196A, and NG+1; Fig. 1C) were tested for their ability to target GFP to the E. coli membrane. N-terminally tagged GFP with the MTS of either NG or NG+1 F196A was uniformly distributed in the cytoplasm, showing its inability to interact with the membrane in vivo (Fig. 5A). In contrast, a GFP with the MTS of NG+1 resulted in a predominantly peripheral localization. These results indicate that the NG+1 MTS is necessary and sufficient for binding to the plasma membrane in vivo and represents the minimal functional MTS. The crystal structure of FtsY suggested that the MTS is longer than previously thought. To test the impact of MTS length on protein targeting, we used a well established protein translocation assay (25). An N-terminally extended NG+2 variant was compared with NG and NG+1 for its ability to support membrane insertion of ppl (Fig. 5B). When the translocation experiments were performed with the NG variant defective in lipid interaction, a diminished translocation activity was observed, in line with its inactivity in vivo (15). Next, we assessed the translocation efficiencies for NG+1 and NG+2 variants, which are competent for lipid interaction. NG+1 showed about 30% translocation efficiency with respect to FtsY, explaining its ability to complement for FtsY depletion in vivo. Extending the MTS further by only one additional phenylalanine residue (Phe-195; NG+2) increased the translocation efficiency to about 60% with respect to FtsY, indicating an enhanced lipid interaction. The diminished translocation efficiency observed for the NG+1 F196A and FtsY F195A/F196A variants (Fig. 4E) nicely correlates with the observed decrease in lipid interaction (Fig. 4D). Our results show that protein translocation depends on a functional MTS, and the efficiency of protein translocation correlates with the length of the MTS.
FIGURE 5.
MTS is functional in vivo and in vitro. A, fluorescence micrographs of the localization of the MTS of NG+1 (left), NG (middle), and NG+1 F196A (right) fused to the N terminus of GFP. The inset shows the magnification of a representative E. coli cell. The Western blot shows that all MTS-GFP fusions were expressed in E. coli at similar levels after 1 h of induction. B, ability of FtsY, NG+1, NG+2, and NG to support membrane translocation of the SRP model substrate ppl. The inset shows the SDS-PAGE used to separate ppl and prolactin (pl). The efficiency of ppl translocation was quantified and is given in percent relative to FtsY. The error bars represent the standard deviation between three independent measurements. An FtsY variant that contains a functional MTS but is defective in reciprocal GTPase activation in the context of SRP (NG+1 A335W variant) (26) was used as a control and shows a similar translocation defect as observed for NG. C, anionic phospholipids stimulate both the basal GTPase activity of FtsY and its complex assembly with SRP.
Lipid-induced Conformational Change Restricts Activity of FtsY to Membrane
After the correct localization of FtsY at the membrane, protein translocation requires the formation of the SRP-FtsY heterodimer in which both GTPases are activated (26). Only FtsY variants that contain at least a minimal MTS are functional in vivo and can be activated in GTP hydrolysis upon SRP interaction (13). In the crystal structure of the SRP-FtsY GTPase heterodimer, αN1 of the N domain is absent in both proteins (33, 34). Deletion of FtsY αN1 greatly facilitates SRP-FtsY complex formation and enhances basal GTP hydrolysis (35), similar to what we observed in the presence of phospholipids. We hypothesized that phospholipid interaction of the MTS results in a similar conformation of αN1 as seen in the SRP-FtsY heterodimer. We determined the rate of GTP hydrolysis in the SRP-FtsY complex in the presence and absence of phospholipids (Fig. 5C). Although FtsY was significantly stimulated upon lipid interaction, the stimulation of the SRP-FtsY complex by far exceeded the stimulation of the individual components. In summary, our data indicate that the phospholipid-induced structural change in the MTS of FtsY is required for efficient SRP-FtsY complex formation. The MTS-mediated lipid association represents a checkpoint in protein targeting and restricts the activity of FtsY to the membrane.
DISCUSSION
The SRP system works predominantly co-translationally and coordinates two cellular machines, a translating ribosome and the membrane-embedded SecYEG translocation channel. The ability of FtsY to bind to lipids is essential for its in vivo function. Here we address the dynamic properties of FtsY in solution and its response to lipids. Our previous work showed that FtsY-lipid interaction is mediated by an MTS present in αN1 of the FtsY N domain. However, mechanistic and structural insights were not available. Although the MTS adopts a helical conformation in the crystal structures of FtsY (Ref. 15 and this study), we show that the MTS is highly dynamic in solution. Previous studies showed that FtsY has a preference for anionic phospholipids (9), whereas other studies also suggested a role of zwitterionic lipids (36). The current study clarifies that only the interaction with anionic phospholipids triggers a random-coil helix transition of the MTS. This conformational change, which might enable FtsY to sense local lipid distributions in the membrane, depends on the concentration of anionic phospholipid head groups. The in vivo relevance of a functional interaction of FtsY with acidic lipids has been shown recently (16). The dominant negative phenotype of the inactive NG variant can be suppressed, and NG is stabilized by expression of PgsA, which increases the amount of anionic phospholipids (cardiolipin and phosphatidylglycerol) in E. coli. This supports the idea that the MTS of FtsY is tuned to the anionic phospholipid content of the E. coli membrane. Lipid segregation in bacterial membranes is well documented (37) and might restrict binding of FtsY to specific sites at the inner membrane. Indeed, anionic phospholipids have been found to be enriched at the SecYEG translocation channel (38). Therefore, the lipid-dependent conformational switch of the MTS may serve in the specific targeting of FtsY at the correct location. The function of a number of GTP- and ATP-binding proteins, e.g. MinD (39, 40) and RGS4 (41), also relies on specific membrane localization. In these cases, the MTS undergoes a similar random-coil helix transition in the presence of anionic phospholipids as shown here for FtsY. A lipid-dependent conformational change of the MTS seems to be a common theme used by different proteins for membrane targeting. Previous work showed that anionic phospholipids induce a conformational change in FtsY and stimulate its basal GTPase activity (9). Our data show that the structural transition of the MTS is essential for a lipid-induced conformational change within the NG domain, resulting in GTPase activation. The interaction of FtsY with lipids and the lipid-induced conformational change of the MTS are nucleotide-independent, indicating that lipid association of FtsY via its MTS is independent from its GTPase cycle. The recently shown predominant membrane association of FtsY supports this idea (6), while another study suggests that SRP facilitates the lipid association of FtsY (14).
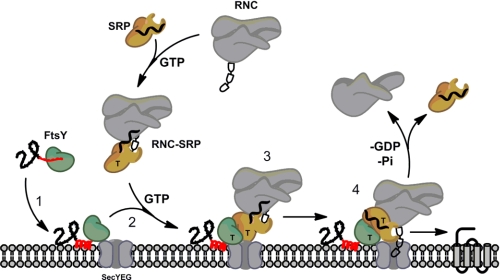
We suspect that the MTS-lipid interaction is a prerequisite for the GTP-dependent association of FtsY with the RNC-SRP complex. Indeed, preformed RNC-SRP-FtsY complexes do not allow efficient membrane interaction (6). Therefore, localization of FtsY at the membrane/SecYEG translocon needs to be ensured prior to its association with the RNC-SRP complex (Fig. 6). RNC-SRP interaction provides another checkpoint within the process (42) and could be viewed as the counterpart of the FtsY-lipid interaction. SRP-FtsY heterodimer formation depends on the presence of GTP in both catalytic half-sites and requires significant structural rearrangements that include a rotation of up to 30° of the N versus the G domain and a release of αN1 from the four-helix bundle of the N domain (33, 34). SRP was shown to undergo significant rearrangements upon RNC interaction that allow for efficient interaction with FtsY (42–44). Likewise, the lipid-induced structural transition of the MTS as part of αN1 triggers a conformational switch, which prepares FtsY for subsequent interaction with the RNC-SRP probably by detaching αN1. The presence of anionic phospholipids significantly enhanced SRP-FtsY complex formation. Furthermore, we show that a compromised MTS-lipid association leads to complete failure of protein translocation. Therefore, the described conformational switch constitutes a central element in the coordination of membrane lipid binding with the RNC-SRP-FtsY interaction.
FIGURE 6.
Scheme of SRP-mediated protein targeting. FtsY (green) is shown with the A domain (black) and the MTS (red). Prior to membrane interaction, the MTS is dynamic (red line) and undergoes a random coil-helix (red helix) transition upon interaction with anionic phospholipids, which are enriched at the SecYEG translocation channel (Step 1). This conformational switch allows subsequent interaction of FtsY with the RNC-SRP in a GTP-dependent manner (Step 2). Upon SRP-FtsY interaction, the two GTPases present in both proteins are activated (Step 3), and the cargo protein is inserted into the membrane (Step 4). GTP is indicated by T.
How FtsY is initially targeted to membrane remains unclear. An attractive model proposes a co-translational targeting of FtsY to the membrane (45, 46), which relies on the A domain. The natively unfolded character of the A domain may allow it to serve as a flexible linker associating ribosomes with the inner membrane. However, further experiments are needed to clarify whether the A domain interacts exclusively with lipids as suggested (10) or whether a yet undefined protein component contributes to its membrane interaction. In eukaryotes, the N terminus of the A domain (X2 domain) of SRα is permanently tethered to the transmembrane protein SRβ via a longin domain interaction (47–49). Taken together, our results underline the importance of membrane lipids in the spatiotemporal restriction of protein activity to membranes, and we envision that other membrane-associated protein complexes have adapted similar mechanisms.
Supplementary Material
Acknowledgments
We thank A. Hendricks for excellent technical assistance, T. Ruppert for support in the mass spectrometry facility at the ZMBH, Heidelberg, B. Simon (European Molecular Biology Laboratory) for NMR measurements Rüdiger Pipkorn (German Cancer Research Center (DKFZ)) for peptide synthesis, and J. Kopp and C. Siegmann from the Cluster of Excellence:CellNetworks crystallization platform for support in crystallization. We are grateful to K. te Kaat for the contribution in the beginning of the project, and we thank Eitan Bibi and Przemyslaw Grudnik for stimulating discussions. Data collection was performed at the European Synchrotron Radiation Facility in Grenoble, France.
This work was supported by the German-Israeli Foundation and Deutsche Forschungsgemeinschaft Grants GRK1188 and SFB638.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (code 2YHS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SRP
- signal recognition particle
- SR
- SRP receptor
- LUV
- large unilamellar vesicle
- MTS
- membrane-targeting sequence
- HX
- hydrogen 1H/2H exchange
- ppl
- pre-prolactin
- RNC
- ribosome-nascent chain complex
- PG
- phosphatidylglycerol
- GMP-PNP
- guanosine 5′-(β,γ-imido)triphosphate
- PE
- phosphatidylethanolamine.
REFERENCES
- 1. Facey S. J., Kuhn A. (2010) Cell. Mol. Life Sci. 67, 2343–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cross B. C., Sinning I., Luirink J., High S. (2009) Nat. Rev. Mol. Cell Biol. 10, 255–264 [DOI] [PubMed] [Google Scholar]
- 3. Grudnik P., Bange G., Sinning I. (2009) Biol. Chem. 390, 775–782 [DOI] [PubMed] [Google Scholar]
- 4. Wilson C., Connolly T., Morrison T., Gilmore R. (1988) J. Cell Biol. 107, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Powers T., Walter P. (1995) Science 269, 1422–1424 [DOI] [PubMed] [Google Scholar]
- 6. Mircheva M., Boy D., Weiche B., Hucke F., Graumann P., Koch H. G. (2009) BMC Biol. 7, 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Montoya G., Svensson C., Luirink J., Sinning I. (1997) Nature 385, 365–368 [DOI] [PubMed] [Google Scholar]
- 8. Freymann D. M., Keenan R. J., Stroud R. M., Walter P. (1997) Nature 385, 361–364 [DOI] [PubMed] [Google Scholar]
- 9. de Leeuw E., te Kaat K., Moser C., Menestrina G., Demel R., de Kruijff B., Oudega B., Luirink J., Sinning I. (2000) EMBO J. 19, 531–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weiche B., Bürk J., Angelini S., Schiltz E., Thumfart J. O., Koch H. G. (2008) J. Mol. Biol. 377, 761–773 [DOI] [PubMed] [Google Scholar]
- 11. Eitan A., Bibi E. (2004) J. Bacteriol. 186, 2492–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Valent Q. A., Scotti P. A., High S., de Gier J. W., von Heijne G., Lentzen G., Wintermeyer W., Oudega B., Luirink J. (1998) EMBO J. 17, 2504–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bahari L., Parlitz R., Eitan A., Stjepanovic G., Bochkareva E. S., Sinning I., Bibi E. (2007) J. Biol. Chem. 282, 32168–32175 [DOI] [PubMed] [Google Scholar]
- 14. Lam V. Q., Akopian D., Rome M., Henningsen D., Shan S. O. (2010) J. Cell Biol. 190, 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parlitz R., Eitan A., Stjepanovic G., Bahari L., Bange G., Bibi E., Sinning I. (2007) J. Biol. Chem. 282, 32176–32184 [DOI] [PubMed] [Google Scholar]
- 16. Erez E., Stjepanovic G., Zelazny A. M., Brugger B., Sinning I., Bibi E. (2010) J. Biol. Chem. 285, 40508–40514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Montoya G., Svensson C., Luirink J., Sinning I. (1997) Proteins 28, 285–288 [DOI] [PubMed] [Google Scholar]
- 18. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 21. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rist W., Graf C., Bukau B., Mayer M. P. (2006) J. Biol. Chem. 281, 16493–16501 [DOI] [PubMed] [Google Scholar]
- 23. Bozkurt G., Stjepanovic G., Vilardi F., Amlacher S., Wild K., Bange G., Favaloro V., Rippe K., Hurt E., Dobberstein B., Sinning I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21131–21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Z., Post C. B., Smith D. L. (1996) Biochemistry 35, 779–791 [DOI] [PubMed] [Google Scholar]
- 25. Powers T., Walter P. (1997) EMBO J. 16, 4880–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shan S. O., Chandrasekar S., Walter P. (2007) J. Cell Biol. 178, 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samuelsson T., Olsson M. (1993) Nucleic Acids Res. 21, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Engen J. R., Smith D. L. (2001) Anal. Chem. 73, 256A–265A [DOI] [PubMed] [Google Scholar]
- 29. Englander S. W. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 213–238 [DOI] [PubMed] [Google Scholar]
- 30. Englander S. W., Kallenbach N. R. (1983) Q. Rev. Biophys. 16, 521–655 [DOI] [PubMed] [Google Scholar]
- 31. Rist W., Jørgensen T. J., Roepstorff P., Bukau B., Mayer M. P. (2003) J. Biol. Chem. 278, 51415–51421 [DOI] [PubMed] [Google Scholar]
- 32. Wales T. E., Engen J. R. (2006) Mass Spectrom. Rev. 25, 158–170 [DOI] [PubMed] [Google Scholar]
- 33. Egea P. F., Shan S. O., Napetschnig J., Savage D. F., Walter P., Stroud R. M. (2004) Nature 427, 215–221 [DOI] [PubMed] [Google Scholar]
- 34. Focia P. J., Shepotinovskaya I. V., Seidler J. A., Freymann D. M. (2004) Science 303, 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neher S. B., Bradshaw N., Floor S. N., Gross J. D., Walter P. (2008) Nat. Struct. Mol. Biol. 15, 916–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Millman J. S., Qi H. Y., Vulcu F., Bernstein H. D., Andrews D. W. (2001) J. Biol. Chem. 276, 25982–25989 [DOI] [PubMed] [Google Scholar]
- 37. Vanounou S., Parola A. H., Fishov I. (2003) Mol. Microbiol. 49, 1067–1079 [DOI] [PubMed] [Google Scholar]
- 38. Gold V. A., Robson A., Bao H., Romantsov T., Duong F., Collinson I. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 10044–10049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mileykovskaya E., Fishov I., Fu X., Corbin B. D., Margolin W., Dowhan W. (2003) J. Biol. Chem. 278, 22193–22198 [DOI] [PubMed] [Google Scholar]
- 40. Szeto T. H., Rowland S. L., Habrukowich C. L., King G. F. (2003) J. Biol. Chem. 278, 40050–40056 [DOI] [PubMed] [Google Scholar]
- 41. Bernstein L. S., Grillo A. A., Loranger S. S., Linder M. E. (2000) J. Biol. Chem. 275, 18520–18526 [DOI] [PubMed] [Google Scholar]
- 42. Zhang X., Schaffitzel C., Ban N., Shan S. O. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1754–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wild K., Halic M., Sinning I., Beckmann R. (2004) Nat. Struct. Mol. Biol. 11, 1049–1053 [DOI] [PubMed] [Google Scholar]
- 44. Halic M., Blau M., Becker T., Mielke T., Pool M. R., Wild K., Sinning I., Beckmann R. (2006) Nature 444, 507–511 [DOI] [PubMed] [Google Scholar]
- 45. Herskovits A. A., Seluanov A., Rajsbaum R., ten Hagen-Jongman C. M., Henrichs T., Bochkareva E. S., Phillips G. J., Probst F. J., Nakae T., Ehrmann M., Luirink J., Bibi E. (2001) EMBO Rep. 2, 1040–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bibi E. (2011) Biochim. Biophys. Acta 1808, 841–850 [DOI] [PubMed] [Google Scholar]
- 47. Young J. C., Ursini J., Legate K. R., Miller J. D., Walter P., Andrews D. W. (1995) J. Biol. Chem. 270, 15650–15657 [DOI] [PubMed] [Google Scholar]
- 48. Schlenker O., Hendricks A., Sinning I., Wild K. (2006) J. Biol. Chem. 281, 8898–8906 [DOI] [PubMed] [Google Scholar]
- 49. Schwartz T., Blobel G. (2003) Cell 112, 793–803 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.