Abstract
In adult muscles and under normal physiological conditions, satellite cells are found in a quiescent state but can be induced to enter the cell cycle by signals resulting from exercise, injury-induced muscle regeneration, or specific disease states. Once activated, satellite cells proliferate, self-renew, and differentiate to form myofibers. In the present study, we found that the zinc finger-containing factor Teashirt-3 (TSHZ3) was expressed in quiescent satellite cells of adult mouse skeletal muscles. We showed that following treatment with cardiotoxin TSHZ3 was strongly expressed in satellite cells of regenerating muscles. Moreover, immunohistochemical analysis indicated that TSHZ3 was expressed in both quiescent and activated satellite cells on intact myofibers in culture. TSHZ3 expression was maintained in myoblasts but disappeared with myotube formation. In C2C12 myoblasts, we showed that overexpression of Tshz3 impaired myogenic differentiation and promoted the down-regulation of myogenin (Myog) and up-regulation of paired-box factor 7 (Pax7). Moreover, knockdown experiments revealed a selective effect of Tshz3 on Myog regulation, and transcriptional reporter experiments indicated that TSHZ3 repressed Myog promoter. We identified the BRG1-associated factor 57 (BAF57), a subunit of the SWI/SNF complex, as a partner of TSHZ3. We showed that TSHZ3 cooperated with BAF57 to repress MYOD-dependent Myog expression. These results suggest a novel mechanism for transcriptional repression by TSHZ3 in which TSHZ3 and BAF57 cooperate to modulate MyoD activity on the Myog promoter to regulate skeletal muscle differentiation.
Keywords: Development, Differentiation, Gene Regulation, Mouse, Skeletal Muscle, BAF57, MyoD, Teashirt-3, Muscle Satellite Cells, Myoblast Differentiation
Introduction
Genetic studies in mice have indicated that muscle-regulatory factors (MRFs;3 MYOD, MYF5, Myogenin, and MRF4) orchestrate myogenesis (1–4). During embryogenesis, MRFs are critical for entry of skeletal muscle satellite progenitor cells (SCs) into the myogenic program (5). These progenitors are characterized by Pax3/Pax7 expression and the lack of MRFs (PAX3/7+MRF−) (6–10). The up-regulation of MRFs in the progenitors triggers the onset of the myogenic program. Between E16.5 and E18.5, PAX3/7+MRF− progenitors located underneath the basal lamina have been proposed to constitute the SC population responsible for postnatal muscle growth and regeneration (9).
In adult muscles, SCs are found in a quiescent state but can be induced to enter the cell cycle by signals resulting from exercise, injury-induced muscle regeneration, or specific disease states. Once activated, SCs proliferate, self-renew, and differentiate into mature muscle cells (5, 11–14). During skeletal muscle regeneration, the induction of MRFs is critical for SC differentiation. In particular, binding of MYOD to cis-regulatory regions establishes a specific and temporally ordered gene expression program, which allows SCs to progress efficiently through the myogenic precursor cell developmental program (4, 15–17).
MYOD-dependent gene activation is associated with chromatin remodeling in the regulatory regions of muscle-specific genes (18, 19). The modulation of MYOD activity affects the balance between proliferation and differentiation of activated SCs (17, 20). MyoD is expressed in myoblasts long before activation of its target genes both in vivo and in tissue culture systems, thereby providing evidence for the existence of mechanisms by which premature activity of MYOD is prevented on muscle-specific promoters and in particular on the myogenin (Myog) promoter (21, 22). Among the mechanisms that control Myog expression, chromatin-remodeling enzymes play an important role (23). Indeed, the activation of Myog expression requires the coordinated action of MRFs and chromatin-remodeling enzymes (24, 25). SWI/SNF chromatin-remodeling complexes employ energy derived from ATP hydrolysis to induce nucleosome movement along the DNA. These complexes can physically interact with histone deacetylases to repress Myog transcription (23, 25, 26). As chromatin remodeling activities are crucial for regulating gene expression, it is important to identify transcription factors and regulatory proteins involved in targeting and/or modulating chromatin modification complexes.
In Drosophila, the zinc finger protein Tsh represses developmental pathways in combination with HOX proteins (27, 28). Tsh function is also required for the development of imaginal discs (29–33). In the eye disc, Tsh functions as part of a complex with the homeoproteins Homothorax and Eyeless to prevent the expression of the downstream genes sine oculis (so), eyes absent (eya), and dachshund (dac) (29). The highly conserved vertebrate homologues Six1–6, Eya1–4, and Dach1–2/Ski/Sno, respectively, are co-expressed in multiple organs and participate in the organogenesis of kidney and skeletal muscle during embryogenesis (34, 35).
We previously generated and characterized Tshz1- and Tshz3-deficient mice (36–42). Mice deficient for Tshz3 develop to term but have congenital hydronephrosis, fail to breathe, and die at birth (37, 38). Tshz3 inactivation results in failure of neuronal and smooth muscle differentiation, indicating that Tshz3 has a role in the control of cellular differentiation (37, 38). In mouse, Tshz3 regulates myocardin (Myocd) expression, and the absence of Tshz3 precludes the activation of smooth muscle genes by Myocd (37). In human, TSHZ3 is part of a powerful gene-silencing complex that represses expression of caspase-4 (43). TSHZ proteins have been shown to interact with Class I histone deacetylases and members of the C-terminal binding protein family that function as co-repressors (41, 43, 44). However, the mechanisms whereby TSHZ factors function as transcriptional repressors and their role in cell differentiation have not been fully elucidated.
Here we report that TSHZ3 is expressed in developing skeletal muscle during late embryonic development and in quiescent and activated SCs in adult muscles. To determine the function of Tshz3 during myogenesis, we performed overexpression and knockdown experiments in the C2C12 myoblast cell line. These experiments revealed that forced expression of Tshz3 impaired differentiation through a selective effect of Tshz3 on Myog regulation. In vitro experiments indicated that TSHZ3 repressed MYOD-dependent activation of the Myog promoter. This property depended in part on BRG1-associated factor 57 (BAF57), a subunit of the SWI/SNF complex and a partner of TSHZ3. Together, these results suggest novel mechanisms that modulate MYOD activity to regulate skeletal muscle differentiation.
EXPERIMENTAL PROCEDURES
Mice
All mouse lines used in this study were maintained on a CD1 background. The Tshz3+/lacZ allele was described (37). Cardiotoxin-induced regeneration was performed as described (45). The cardiotoxin-injected and non-injected contralateral muscles were analyzed 5 days after injection. Mice received a 5-bromo-2-deoxyuridine (BrdU) single pulse (1 mg/10 g of body weight) 5 days after cardiotoxin injection followed by a 2-h chase. Care was taken to minimize the number of animals used, and experimental protocols were approved by the French Ethics Committee.
Preparation of Satellite Cells and Primary Cultures
SCs were prepared as described (46). Myofibers were cultivated for 5 days on Matrigel. After immunostaining, the number of cells expressing only one or a combination of markers was counted. The percentage of cells of each category was evaluated from the total number of DAPI-stained nuclei. We counted >1500 nuclei from five independent experiments.
Cell Lines
The C2C12 cell line was provided by Dr. S. Tajbakhsh (Pasteur Institute, Paris, France). The stable C2C12-derived cell line (C2i-Myog) that expresses a doxycycline (Dox)-inducible cDNA encoding FLAG-tagged MYOG was generated as follows. The Tet repressor-expressing myoblast cell line C2i was generated by electroporation of the plasmid pPyCAGIP-TetR into C2C12 cells and selection with puromycin until individual clones formed. Individual clones were screened by immunofluorescence to ensure that they retained the potential to differentiate (expression of Myog and Myh3 while forming multinucleated myotubes) upon serum withdrawal. C2i-Myog cells were generated by electroporation of the plasmid pCDNA5/TO-FL-Myog into C2i cells and selecting for hygromycin until individual clones formed. Individual clones were then screened for Dox-inducible Myogenin expression as well as differentiation. C2C12 cells lines were cultivated following the American Type Culture Collection protocol (ATCC number CRL-1772). To avoid differentiation, cells were kept in subconfluent (<60–70%) conditions. To induce differentiation, C2C12 cells grown to 80% confluence were then cultivated in differentiation medium (98% DMEM, 2% FBS).
Cell Transfection, Plasmids, and siRNA
C2C12 cells were transfected using AmaxaTM Nucleofector kit V (Lonza) or Lipofectamine 2000 reagent (Invitrogen). pCAβ-CMV MyoD (47), pcDNA3.1-HA-Tshz3 and pCX-Tshz3 clones were produced by subcloning a 3.4-kb PCR-generated fragment covering the complete HA-Tshz3 open reading frame, respectively, into the pcDNA3.1(+) (Invitrogen) and pCX-MCS2 (a gift from Dr. X. Morin, École Normale Supérieure, Paris, France) plasmids. The pcDNA3-VP16/Tshz3 fusion plasmid was made by subcloning a PCR-generated VP16 domain into a mouse Tshz3 expression plasmid. We used a VP16 plasmid provided by Dr. P. Lemaire (Institut de Biologie du Développement de Marseille Luminy, Marseille, France) as template and the primers VP16F (5′-CCAAGCTTCATGGCCCCCCCGACCGATGTCAGC-3′) and VP16R (5′-CGTGGATCCCCCACCGTACTCGTCAAT-3′). The VP16 PCR fragment was digested by HindIII and BamHI and subcloned; the resulting clones were verified by sequencing. pSG5-FLAG-BAF57 was a gift from B. Belandia (48), pCX-eGFP was provided with the Amaxa Nucleofector kit, and pCMV-dsRed2 has been described (49). 60–70% of the C2C12 cells were transfected as estimated by co-transfection with pCX-eGFP or pCMV-dsRed2. Silencer® Select predesigned siRNAs against Tshz3 (Ambion identification numbers s110169, s110170, and s110171) and smarce1/BAF57 (Ambion identification numbers s81270, s81271, and s81272) and a negative control siRNA (Ambion number 1) were used.
Immunohistochemical Analysis
The primary antibodies used were: guinea pig anti-TSHZ3 (1:2500) and guinea pig anti-LBX1 (1:2000) (Dr. A. Garratt, Max Delbrück Center for Molecular Medicine, Berlin, Germany); mouse anti-PAX7 (1:5) and mouse anti-myosin heavy chain (MyHC) (1:10) (MF20, Developmental Studies Hybridoma Bank); rabbit anti-PAX7 (1:100) (Euromedex); mouse anti-Myogenin (1:200) and rabbit anti-MYOD (1:200) (Santa Cruz Biotechnology); rabbit anti-laminin (1:500) (Sigma); rabbit anti-β-galactosidase (1:1000) (Cappel); and rat anti-BrdU (1:200) (Abcam). The secondary antibodies used were: Alexa Fluor 546 or 488-goat anti-rabbit, Alexa Fluor 555 or 488-IgG1 goat anti-mouse, Alexa-Fluor 488-goat anti-rat (Molecular Probes), and Cy5-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories). Secondary antibodies were diluted 1:1000 except for Cy5-conjugated donkey anti-mouse IgG, which was diluted 1:100. Muscles were fixed in 4% paraformaldehyde for 30 min, embedded in a gelatin/sucrose solution, and sectioned using a Cryostat (Leica CM 3050S). Cultured cells were fixed in 1% paraformaldehyde for 10 min. Cryosections and cultured cells were washed with 1% Triton X-100, PBS for 60 or 15 min, respectively. Samples were then processed as described (50, 51).
mRNA Extraction and Quantitative Real Time PCRs (RT-qPCR)
Total RNA from C2C12 cells was prepared using TRIsure reagent (Bioline). RNA samples were treated with RQ1 RNase-free DNase I (Promega), and first strand cDNA was synthesized using superscript II RT (Invitrogen). RT-qPCR conditions were as follows: 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s (iCycler iQTM5, Bio-Rad) using SYBR® GreenER RT-qPCR reagents (Invitrogen). Analyses were performed in triplicate from three independent experiments. Transcript levels were first normalized to two housekeeping genes, Gapdh and muscular phosphofructokinase (PFK), and then normalized to their respective control group: Relative quantity (RQ) = 2−ΔΔCt and ΔΔCt = (Ct exp − Ct PFK) − (Ct control − Ct PFK). Control experiments were performed by transfecting empty expression plasmid or control siRNA (mock). As these controls did not produce significant variation in the expression level of genes analyzed for overexpression and knockdown experiments, only one control condition is shown. Primer sequences used were: PFK (52), Gapdh (53), Myog (54), F-Tshz3 (5′-GCGCGCAGCAGCCTATGTTTC-3′) and R-Tshz3 (5′-TCAGCCATCCGGTCACTCGTC-3′), F-Pax7 (5′-GGCACAGAGGACCAAGCTC-3′) and R-Pax7 (5′-GCACGCCGGTTACTGAAC-3′), F-MyoD (5′-TCCTTTCGAAGCCGTTCTT-3′) and R-MyoD (5′-CTTTCTTTGGGGCTGGATCT-3′), F-MRF4 (5′-ATGGTACCCTATCCCCTTGC-3′) and R-MRF4 (5′-TAGCTGCTTTCCGACGATCT-3′), and F-BAF57 (5′-CCAGCTCCTGCAACACAA-3′) and R-BAF57 (5′-GTTGTTGTAGGCGAGATGACTG-3′).
Luciferase Assays
Myog proximal promoter linked to the firefly luciferase transgene was provided by Dr. H. Ohto (55). 24 or 48 h after transfection, both luciferase and Renilla activities (Dual-Luciferase reporter assay, Promega) were determined on a Tristar LB941 luminometer (Berthold Technologies). The experiments were performed in duplicate, and the reported results represent at least three independent experiments.
Yeast Two-hybrid Screening
The screen and data analysis were performed by Hybrigenics (Paris, France). The bait construct N-LexA-TSHZ3-C fusion (full-length Tshz3 cloned into the pB27 vector) was transformed into Saccharomyces cerevisiae strain L40 GAL4 (56). The screen was performed as described (57); a mouse embryo brain (RP2) cDNA library transformed into the Y187 yeast strain and containing 10 million independent fragments was used for mating, and 56.4 million interactions were tested. After selection, 39 positive clones were analyzed and compared with GenBankTM using BLASTN. A predicted biological score was used to assess the reliability of each interaction as described (58). The predicted biological scores have been shown to positively correlate with the biological significance of interactions (57). The clones matched BAF57 (GenBank accession number NM_02618.4).
GST Pulldown Assay
The N-terminal (N-Tshz3; amino acids 1–472) and C-terminal (C-Tshz3; amino acids 473–1081) Tshz3 fragments were generated by PCR from pGEX-Tshz3 (41) and subcloned into pGEX-4T2 (GE Healthcare). pGEX-BAF57, pGEX-N-BAF57, and pGEX-C-BAF57 were provided by Dr. B. Belandia (48). The pulldown assay was performed as described (41).
Co-immunoprecipitation and Western Blotting
C2C12 cells were seeded in 10-cm dishes and either transfected with HA-tagged or FLAG-tagged bait expression constructs. After 48 h, cells were broken open in lysis buffer (50 mm, Tris-HCl at pH 7.6, 200 mm NaCl, 1 mm EDTA, 0.25% sodium deoxycholate, 1 mm PMSF, 1 mm Na3VO4) containing protease inhibitors (Roche Applied Science). Cellular debris were precipitated by centrifugation at 13,000 rpm for 20 min at 4 °C; the supernatant was incubated with FLAG-agarose M2 beads (Sigma) prewashed in lysis buffer. Samples were incubated for 3 h at 4 °C on a rotator. Proteins eluted from the beads were analyzed by Western blot with rat anti-HA (3F10, Roche Applied Science) or mouse anti-FLAG M2 (F1804, Sigma) and anti-β-actin (AC-74, Sigma) antibodies, and 5% of input was loaded as a control.
RESULTS
TSHZ3 Is Expressed in Adult Satellite Cells
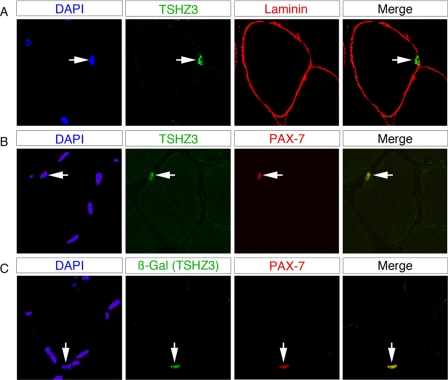
We reported previously that expression of the Tshz3lacZ allele recapitulated the expression pattern of endogenous Tshz3 (37). X-Gal staining performed on Tshz3+/lacZ mice revealed that Tshz3 was expressed in adult skeletal muscles (supplemental Fig. 1, A and A′). To identify the cell types expressing Tshz3 in skeletal muscles, immunohistochemical analysis were carried out on wild type (WT) and Tshz3+/lacZ mice. TSHZ3+ cells were found underneath the basal lamina and in close proximity to the muscle fiber (Fig. 1A) and were PAX7-positive (PAX7+) (Fig. 1B). In the gastrocnemius from Tshz3+/lacZ adult mice, β-galactosidase (β-gal)-positive cells were also found in a typical satellite position and were PAX7+ (Fig. 1C). This observation suggests that in adult skeletal muscles TSHZ3 is expressed in quiescent muscle SCs. It has been suggested that the PAX+ resident muscle progenitor cells present in embryonic and fetal muscle later constitute the satellite cell population (8, 9). We investigated the distribution of TSHZ3 in WT and Tshz3+/lacZ fetal muscles. At E18.5, X-Gal and immunohistochemical stainings showed that TSHZ3 was expressed in fetal muscles and that almost all PAX7+ cells expressed TSHZ3 (supplemental Fig. 2, A–H). These results suggest that at late stages of embryogenesis TSHZ3 is expressed in cells that are determined to become progenitor cells responsible for postnatal muscle growth and regeneration.
FIGURE 1.
TSHZ3 expression in adult satellite cells. Confocal optical views of transverse sections through gastrocnemius skeletal muscle taken from a 6-week-old wild type and Tshz3+/lacZ mice are shown. TSHZ3-positive cells are positioned under the basal lamina as shown by immunolabeling for laminin (A). TSHZ3 (B) or β-gal (C) labeling co-localized with the PAX7 satellite cell markers. Corresponding DAPI staining is shown with the labeled nucleus indicated by an arrow.
Tshz3 homozygous mutants (Tshz3lacZ/lacZ) die of an inability to breathe at birth (38) and display a bilateral hydronephrosis and proximal hydroureter (37). Apart from the renal defect, Tshz3 mutants did not display any significant anatomical defect compared with their littermate controls. To investigate whether Tshz3 was required in developing muscle to set aside SCs, we analyzed the expression of PAX7 in Tshz3lacZ/lacZ muscles and quantified the number of PAX7+ cells in WT and Tshz3lacZ/lacZ muscles at E18.5 (supplemental Fig. 2, I–M). In Tshz3lacZ/lacZ muscles, we found that the number of PAX7+ cells was not significantly different compared with WT littermate control muscles. Together, these results support that TSHZ3 is a new marker of SCs and that at late embryonic stages Tshz3 is dispensable for the emergence of the PAX7+ cells that later will constitute the satellite cell population.
TSHZ3 Is Expressed in Activated Satellite Cells during Adult Muscle Regeneration
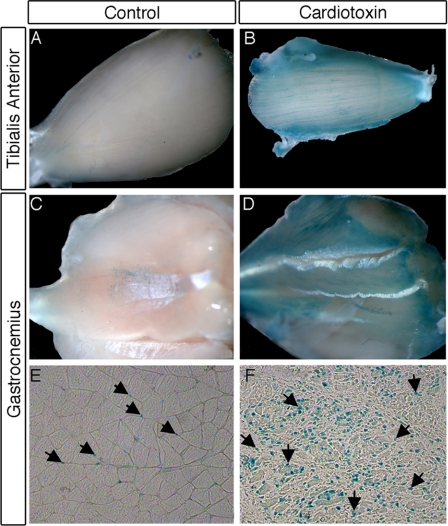
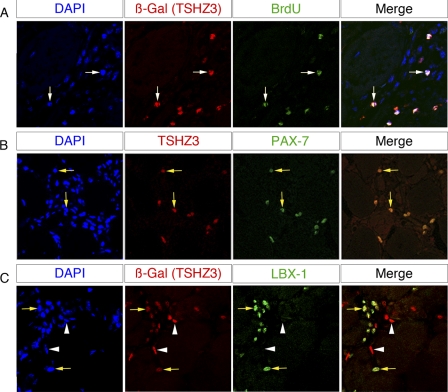
To investigate whether Tshz3 is expressed in activated SCs, muscle regeneration was induced in hind limb muscles by the administration of cardiotoxin (CTX). In response to CTX-induced injury, SCs undergo activation and proliferation expansion with the formation of newly regenerated myotubes (59–61). CTX was injected into hind limb muscles of 6-week-old WT CD1 and Tshz3+/lacZ mice. Examination of Tshz3+/lacZ injured muscles 5 days after injection revealed a significant increase of TSHZ3+ cells in the regenerating muscle compared with the contralateral non-damaged muscle (Fig. 2). To further characterize the TSHZ3+ cells present in a CTX-injured muscle (day 5), we performed immunofluorescence for detection of BrdU incorporation to mark proliferative cells after a single pulse and a 2-h chase. We also stained the same sections for PAX7 and LBX1, a marker of activated SCs (62). All β-gal-positive cells were positive for BrdU (Fig. 3A). In addition, all PAX7+ cells expressed TSHZ3 (Fig. 3B). On sections immunostained for β-gal (TSHZ3) and LBX1, we found that all LBX1+ cells were positive for β-gal (Fig. 3C, arrow). These results indicate that in adult skeletal muscle TSHZ3 is expressed in activated SCs during the course of CTX-induced muscle regeneration in addition to expression in quiescent SCs.
FIGURE 2.
Number of β-Gal-positive cells increased significantly in muscles of adult Tshz3+/lacZ mice after cardiotoxin-induced muscle damage. X-Gal-stained tibialis anterior (A and B) and gastrocnemius muscles from Tshz3+/lacZ mouse (A–D) are shown. The tibialis anterior and the gastrocnemius of one hind limb serve as a control (A and C), and the other hind limb was injected with cardiotoxin (B and D). Using X-Gal to localize the β-galactosidase activity on cryosections revealed the expansion of the Tshz3+ cell population. Fewer cells with β-galactosidase activity were found in control muscle (arrows; E) compared with regenerating muscle (arrows; F).
FIGURE 3.
TSHZ3 expression in activated adult satellite cells. Confocal optical views of transverse sections through 6-week-old Tshz3+/lacZ cardiotoxin-injected gastrocnemius are shown. The cardiotoxin-injected gastrocnemius muscle was taken for analysis 5 days after injection. 2 h before sacrifice, mice received a BrdU single pulse. Transverse sections were stained for β-gal/BrdU (A), TSHZ3/PAX7 (B), or TSHZ3/LBX1 (C). β-Gal+/BrdU+ cells are indicated by white arrows in A. All the PAX7-positive cells were TSHZ3-positive (B, yellow arrows). β-Gal+ cells co-expressed LBX1, a specific marker of activated SCs (C, yellow arrows). Note that β-gal+ LBX1− cells could correspond to quiescent SCs or to non-myogenic cells (C, arrowheads).
In vivo, X-Gal staining on the diaphragm muscle of Tshz3+/lacZ adult mice suggested that TSHZ3+ cells could contribute to the regeneration of adult skeletal muscle under physiological conditions (supplemental Fig. 1, A and A′). As a result of the elevated half-life of the β-gal, nuclei of Tshz3+/lacZ SCs incorporated in the newly formed muscle fibers (blue fibers) are likely to be still β-gal+ after incorporation. Together, these results indicate that TSHZ3+ cells could contribute to normal and induced muscle regeneration.
TSHZ3 Expression in SCs and Their Progeny Isolated from Myofibers
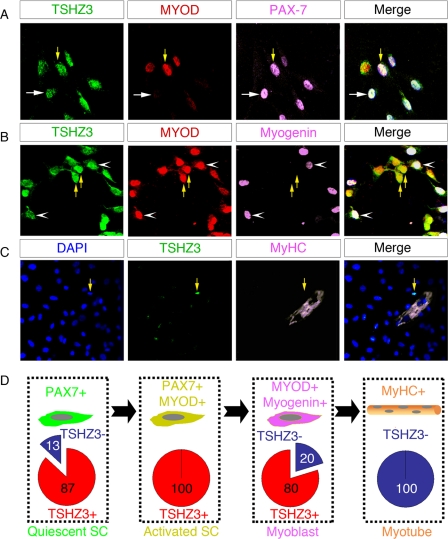
To further investigate the expression of Tshz3 in SCs, myofibers isolated from the gastrocnemius and extensor digitorum longus muscles of adult mice were cultured in suspension and on Matrigel. Culture of myofibers provides an excellent model to study the progression of activated SCs through the different steps of the myogenic program (62–65). TSHZ3+ cell populations were characterized by immunostaining after 5 days of culture when SCs have adopted different fates as revealed by the sequential expression of PAX7, MYOD, MYOG, and MyHC (Fig. 4). We analyzed and quantified the expression of TSHZ3 in four cell populations expressing, respectively, PAX7 only, PAX7 and MYOD, MYOD and MYOG, and the myosin heavy chain only (Fig. 4D and supplemental Fig. 3). This analysis revealed that 87% of the quiescent SCs (PAX7+/MYOD−) (Fig. 4, A, white arrow, and D) and 100% of the activated SCs (PAX7+/MYOD+) were TSHZ3+ (Fig. 4, A, yellow arrows, and D). Among the TSHZ3+/MYOD+ population, some cells did not express MYOG (Fig. 4B, yellow arrows). Finally, 80% of myoblasts (PAX7−/MYOD+/MYOG+) expressed TSHZ3 (Fig. 4, B and D). In some sporadic cases, TSHZ3 was detected in isolated MyHC-positive myocytes (data not shown) but never in fusing myotubes (Fig. 4, C and D).
FIGURE 4.
TSHZ3 expression in satellite cells and their progeny isolated from myofibers. SCs freshly isolated from extensor digitorum longus muscles were cultured in growth medium for 5 days and then immunostained for TSHZ3/MYOD/PAX7 (A), TSHZ3/MYOD/MYOG (B), and TSHZ3/MyHC (C). The number of TSHZ3+ cells that expressed, only PAX7, PAX7 and MYOD, MYOD and MYOG, and MyHC, respectively, was evaluated (see supplemental Fig. 3 for details). The mean percentage of TSHZ3+ cells in each category is reported in red (D). Notice that 87% of cells expressing only PAX7 also expressed TSHZ3 and that all PAX7+/MYOD+ cells expressed TSHZ3 (A, yellow arrows). The undifferentiated satellite PAX7+/MYOD− cells are indicated by white arrows in A. Some TSHZ3+/MYOD+ cells (B, yellow arrows) expressed MYOG (B, white arrowheads).
Together, these results support the conclusion that Tshz3 is expressed in both quiescent and activated SCs. In addition, the presence of TSHZ3/MYOD/MYOG-positive cells indicates that Tshz3 expression is maintained at the onset of SC differentiation. Finally, the absence of TSHZ3 in myotubes suggests that Tshz3 expression is down-regulated during myogenic differentiation.
Forced Expression and Down-regulation of Tshz3 Affect Myogenic Differentiation
To further investigate the role of Tshz3 in myogenesis, we used C2C12 myoblasts that have the potential to differentiate into myotubes following serum deprivation in an MRF-dependent manner (66). Analysis of Tshz3 expression in undifferentiated and differentiating C2C12 by RT-qPCR revealed a gradual reduction of Tshz3 expression during the myogenic differentiation process (supplemental Fig. 4A). Because Tshz3 expression was down-regulated in C2C12 differentiating myoblasts and absent in newly formed myotubes, we decided to investigate the effect of modulation of Tshz3 expression on myogenic differentiation.
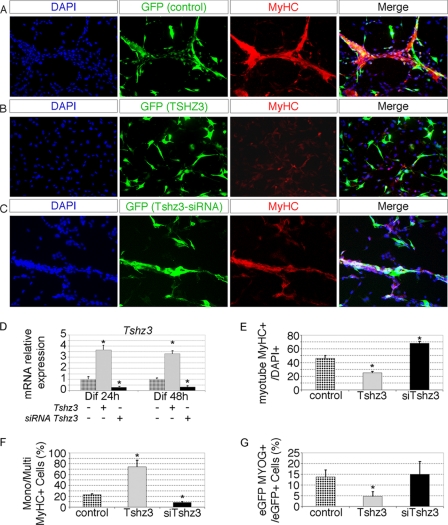
First, we conducted Tshz3 overexpression experiments by transfecting C2C12 cells with a Tshz3-expressing vector construct, pCX-Tshz3. The expression level of Tshz3 in overexpressing conditions increased the total expression of this gene as determined using RT-qPCR (Fig. 5D). Transfected C2C12 cells were morphologically normal and proliferated at the same rate as control cells when cultured in cell growth medium (supplemental Fig. 4C). After 48 h in differentiation medium, whereas control C2C12 cells differentiated into multinuclear myotubes as revealed by labeling with MyHC antibody (Fig. 5A), C2C12 cells overexpressing Tshz3 did not form mature myotubes, and most of them remained mononucleated myoblasts (Fig. 5B). Quantification indicated that overexpression of Tshz3 in C2C12 myoblasts significantly reduced the number of myotubes (Fig. 5E). About 70% of MyHC+ cells were mononucleated after Tshz3 overexpression, whereas in control experiments, only 20% of the MyHC+ cells were mononucleated (Fig. 5F). These results indicate that overexpression of Tshz3 reduced myotube formation.
FIGURE 5.
Forced expression and down-regulation of Tshz3 affect myogenic differentiation. C2C12 cells were transfected with pcDNA3 (control) (A), pcDNA3-HA-Tshz3 (B), and siRNAs targeting Tshz3 (C). To detect successfully transfected cells, pCX-eGFP was co-transfected. After 48 h of differentiation (Dif 48h), cells were fixed and immunostained with an anti-MyHC antibody. Cell nuclei were counterstained with DAPI. Representative fields are shown (A–C). D, the expression level of Tshz3 was tested in overexpression and knockdown conditions by RT-qPCR. E and F, bar graphs indicating the effects of Tshz3 overexpression and after knockdown of Tshz3 from the experiments in A–C. For each condition, >7000 nuclei were counted from randomly chosen fields. E, when cells were induced to differentiate by serum withdrawal, Tshz3 overexpression led to a reduction in myotube number, whereas the number of MyHC+ cells increased after Tshz3 knockdown. Data are expressed as the ratio of MyHC+ cells with two or more nuclei to total DAPI+ nuclei. F, quantification of the mononucleated MyHC+ cells. Tshz3 overexpression and knockdown, respectively, led to a significant increase of mononucleated (Mono) MyHC+ cells and of multinucleated (Multi) MyHC+ cells compared with control cells. Data are expressed as the ratio of mononucleated MyHC+ to total MyHC+ cells. G, quantification of MYOG+ cells. Data are presented as the ratio of eGFP+, MYOG+ cells to total eGFP+ cells. >1000 eGFP+ cells were scored. Tshz3 overexpression led to a significant reduction of MYOG+ cells. D–G, data are presented as mean values ± S.E. from three independent experiments. * indicates statistical significance, p < 0.05 (Kruskal and Wallis test), and errors bars represent the standard deviation.
Second, we performed a Tshz3 knockdown experiment by transfecting C2C12 cells with siRNA specific to Tshz3 (siTshz3). We showed that siTshz3 severely decreased the endogenous expression level of Tshz3 and did not affect cell proliferation (Fig. 5D and supplemental Fig. 4C). The ability of the siTshz3 to reduce Tshz3 expression was also tested in C2C12 cells overexpressing an HA-tagged version of TSHZ3. In these conditions, the data confirm that Tshz3 expression was severely decreased and that HA-TSHZ3 protein expression became undetectable after transfection with siTshz3 (supplemental Fig. 4, D–F).
In siTshz3-transfected cells, myotube formation was visible within 2 days as in non-transfected cells (Fig. 5C). However, quantification revealed that the number of myotubes was higher (×1.5-fold) and fusion improved (×2.5-fold) in siTshz3-transfected cells compared with control conditions (Fig. 5, E and F). These results show that the level of expression of Tshz3 was critical for the progression of myogenic differentiation.
To further investigate the effects of the modulation of Tshz3 expression, Myog expression was used to monitor the progression of differentiation of the C2C12 myoblasts. We quantified the number of MYOG+ cells after Tshz3 overexpression in the GFP-transfected population. When Tshz3 was overexpressed, only 5 ± 2.0% of the GFP+ cells expressed MYOG (p = 0.000059) compared with 13 ± 3.1% in the control condition (Fig. 5G). In contrast, when siTshz3 was transfected, 15 ± 5.9% of the GFP+ cells expressed MYOG (Fig. 5G). Together, these results show that Tshz3 affected the expression of myogenin, and as a consequence, modulation of Tshz3 expression levels perturbed the fusion of myoblasts.
Tshz3 Regulates Myogenin Expression Level
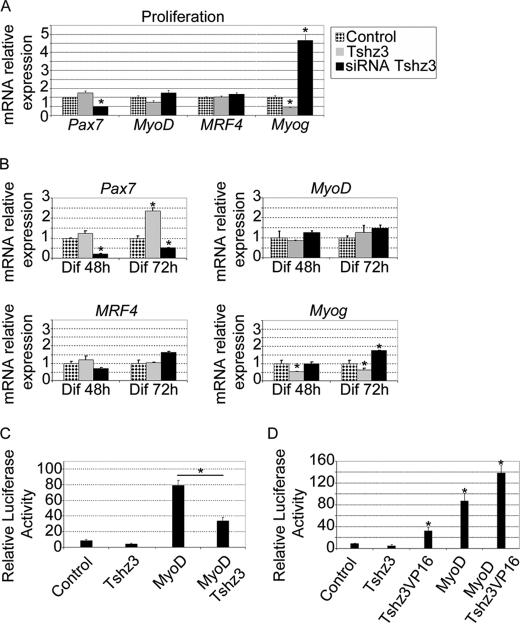
To investigate the effect of Tshz3 on the myogenic program, the expression levels of Pax7 and MRF genes were analyzed by RT-qPCR in C2C12 cells transfected with pCX-Tshz3 expression vector or siTshz3 and maintained either in proliferation or differentiation medium (Fig. 6).
FIGURE 6.
Tshz3-dependent modulation of myogenic regulatory factor expression. Quantitative real time PCR analysis of Pax7, MyoD, MRF4, and Myog mRNA levels in transfected C2C12 cells is shown. The analysis was performed with cells maintained in proliferation medium (A) or cultured in differentiation (Dif) medium for 2 and 3 days (B). Cells were transfected either with a vector allowing overexpression of TSHZ3 (gray bars) or with siRNA specific for Tshz3 (black bars). Transcript levels were first normalized to PFK internal loading control and then normalized to the expression level obtained in control conditions. Data presented are the mean ± S.E. of three samples obtained from three independent experiments. * indicates statistical significance, p < 0.05 (Kruskal and Wallis test), and errors bars represent the standard deviation.
After transfection in proliferation medium, forced expression of Tshz3 induced a significant decrease in Myog expression levels and an increase in the Pax7 expression levels (Fig. 6A). Conversely, transfection with siTshz3 induced a strong increase (5-fold) in Myog expression levels and a significant decline of Pax7 expression (Fig. 6A). These effects of Tshz3 overexpression on Myog and Pax7 expression levels were also found when cells were cultured for 48 h in the differentiation medium (Fig. 6B). Overexpression or down-regulation of Tshz3 did not significantly affect MyoD and MRF4 expression levels (Fig. 6, A and B). Thus, our data show that in C2C12 cells variation of Tshz3 expression levels significantly affected the expression of Pax7 and Myog and suggest that variations in levels of Myog expression are not mediated by altered MyoD expression.
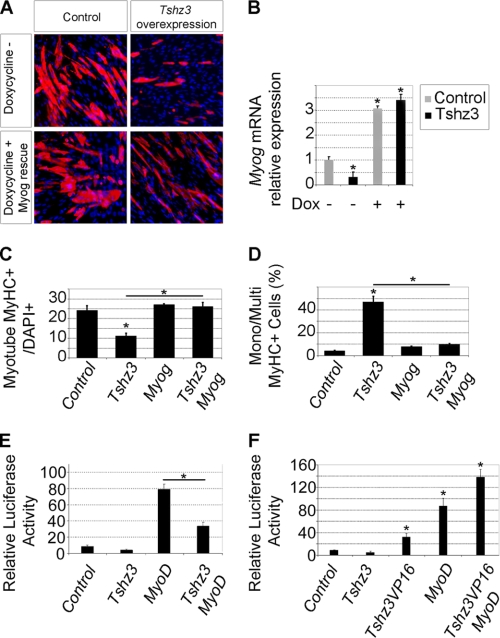
As MYOG is a key regulator of genes in the myogenic terminal differentiation, we hypothesized that Tshz3 might regulate myogenic differentiation by modulating the expression of Myog. To test this, we examined whether forced expression of Myog could rescue defective differentiation in Tshz3-overexpressing C2C12 cells. A stable C2C12-derived cell line (C2i-Myog) expressing a Dox-inducible cDNA encoding FLAG-tagged MYOG was transfected with a Tshz3-expressing vector construct and then cultivated for 48 h in differentiation medium with or without Dox. In the absence of Dox, overexpression of Tshz3 dramatically decreased the efficiency of muscle differentiation, and the expression of Myog was significantly reduced (Fig. 7, A and B). Conversely, in doxycycline-treated C2i-Myog cells, the expression of Myog was efficiently stimulated, and overexpression of Tshz3 did not affect terminal differentiation (Fig. 7, A–D).
FIGURE 7.
Tshz3-specific function on Myog expression. A and B, Myog expression rescues the myogenic differentiation in Tshz3-overexpressing C2i-Myog cells. C2i-Myog cells were transfected with either pBluescript (Control) or pcDNA3-HA-Tshz3 (Tshz3 overexpression). Myog expression was induced by addition of Dox in the differentiation medium. After 48 h in differentiation medium with or without Dox, cells were fixed and immunostained with an anti-MyHC antibody (cell nuclei were counterstained with DAPI, and representative fields are shown) (A), and the endogenous Myog mRNA levels were analyzed by RT-qPCR (B). In the absence of Dox (−), the analysis revealed a reduction of Myog expression in Tshz3-overexpressing C2i-Myog cells compared with control. In the presence of Dox (+), Myog expression increased about 3-fold in both control and Tshz3-overexpressing C2i-Myog cells. C and D, bar graphs indicating the effects of Tshz3 overexpression and after Myog rescue from the experiments in A. For each condition, >7000 nuclei were counted from randomly chosen fields. C, when cells were induced to differentiate by serum withdrawal, Tshz3 overexpression led to a reduction in myotube number, whereas the number of MyHC+ cells was rescued after Myog induction. Data are expressed as the ratio of MyHC+ cells with two or more nuclei to total DAPI+ nuclei. D, quantification of the mononucleated MyHC+. Tshz3 overexpression led to an increase of mononucleated (Mono) MyHC+ cells, whereas Myog induction restored the formation of multinucleated (Multi) MyHC+ cells compared with control cells. Data are expressed as the ratio of mononucleated MyHC+ to total MyHC+ cells. E and F, analysis of the effect of TSHZ3 on the Myog promoter. For the Dual-Luciferase assay, pGL3MG-185 was co-transfected with vectors expressing MyoD in the absence or presence of Tshz3 or with a construct that encodes TSHZ3 fused to the transactivation domain VP16 (TSHZ3-VP16) into C2C12 myoblasts. E, TSHZ3 significantly represses MYOD-dependent activation of the Myog promoter. Data are presented as mean values ± S.E. from five independent experiments. Myog promoter activity is presented as relative light units (×103). F, TSHZ3-VP16 significantly activates luciferase expression, whereas TSHZ3 shows no significant effect. TSHZ3-VP16 cooperates with MYOD to activate Myog expression. Data are presented as mean values ± S.E. from at least three independent experiments. * indicates statistical significance, p < 0.05 (Kruskal and Wallis test), and errors bars represent the standard deviation.
Thus, the induction of Myog expression in Tshz3-overexpressing C2C12 cells was able to restore the differentiation. This result highlights a TSHZ3-specific role in the control of Myog expression.
TSHZ3 Represses Stimulated Activity of Myogenin Promoter
To determine whether TSHZ3 could modulate the MYOD-dependent activation of Myog transcription, we performed transient transfection and luciferase assays in C2C12 cells using the proximal promoter (−185 to +49) of the Myog gene fused to the luciferase reporter gene (pGL3MG-185; Ref. 55). This regulatory region has been defined as a muscle-specific regulatory region and contains a number of transcription factor binding sites that are critical for activation of Myog transcription during differentiation (24, 55, 67, 68). MYOD along with various cofactors plays a critical role in the timing- and cell type-specific expression of Myog (24, 69). As shown in Fig. 7E, MYOD significantly activated transcription from the promoter containing the Myog enhancer. In contrast, the co-expression of TSHZ3 with MYOD resulted in a significant reduction of the MYOD-dependent activation of the Myog promoter (Fig. 7E). TSHZ3 alone did not affect the basal level of transcription (Fig. 7, E and F). These results demonstrate that TSHZ3 repressed Myog expression and suggest that TSHZ3 modulates MYOD-dependent activation of Myog.
The mechanisms by which TSHZ3 interferes with MYOD activity are not known; TSHZ3 might prevent MYOD from binding to (or hinder the accessibility of MYOD to) the Myog promoter, or TSHZ3 could prevent DNA-bound MYOD from activating transcription. To distinguish between these possibilities, C2C12 cells were transfected with a construct that encodes TSHZ3 fused to the transactivation domain VP16 (TSHZ3-VP16). This manipulation converted TSHZ3 to an activator that could induce expression of the Myog promoter (Fig. 7F). Moreover, TSHZ3-VP16 enhanced MYOD-dependent activation of Myog expression (Fig. 7F). These findings suggest that TSHZ3 acts as a specific negative regulator of Myog. Taken together, our data show that TSHZ3 acted on the proximal promoter of Myog and that TSHZ3 reduced the MYOD-dependent transcriptional activation.
TSHZ3 Interacts with SWI/SNF Chromatin-remodeling Subunit BAF57/SMARCE1
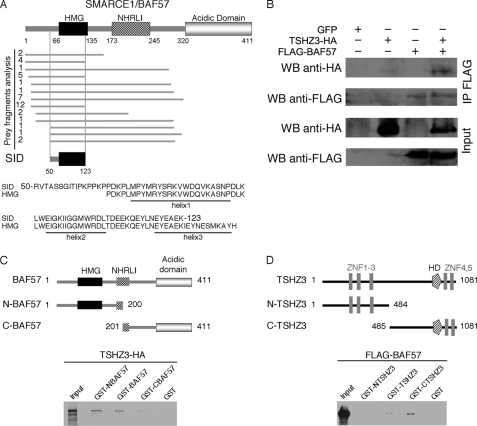
Data obtained in invertebrate and vertebrate showed that TSHZ-mediated regulation depends on interaction with different partners (42, 43, 70, 71). Furthermore, the regulation of Myog expression involves the formation of a multiprotein complex (72, 73). Therefore, to identify protein binding partners of TSHZ3, we performed a yeast two-hybrid screen using full-length TSHZ3 as bait. We identified 84 independent clones among which 40 (47.6%) clones correspond to the mouse SMARCE1/BAF57, a subunit of the SWI/SNF complex (Fig. 8A).
FIGURE 8.
TSHZ3 interacts with BAF57. A, sequence analysis of the 40 BAF57 clones obtained from the two-hybrid screens revealed that the selected interaction domain (SID) corresponds to amino acids 50–123 of BAF57 and contains helices 1 and 2 of the HMG domain. B, TSHZ3 associates with BAF57 in C2C12 cells. C2C12 cells were co-transfected with expression plasmids for HA-tagged TSHZ3 and FLAG-tagged BAF57. Whole cell extracts were immunoprecipitated (IP) using a FLAG antibody and probed for the presence of HA-TSHZ3 by Western blot (WB). Input represents 10% of the total cell extract. C, GST pulldown assay confirmed that TSHZ3 binds preferentially to the N-terminal domain of BAF57. D, BAF57 binds to the C-terminal domain of TSHZ3. NHRLI, a conserved region named after a group of conserved amino.
BAF57 contains DNA binding capability through its high mobility group (HMG) motif, and the analysis of the interacting clones indicated that part of the HMG domain belongs to the selected interaction domain (Fig. 8A). To strengthen these results, we assessed TSHZ3 interaction with BAF57 in vitro by glutathione S-transferase (GST) pulldown assays and in vivo in C2C12 cells by means of co-immunoprecipitation assays (Fig. 8, B–D). Different GST-TSHZ3 and GST-BAF57 fusion proteins produced in bacteria were immobilized on agarose-glutathione beads, and tagged recombinant forms of FLAG-BAF57 and HA-TSHZ3 were detected by Western blotting using anti-HA and anti-FLAG antibodies, respectively. Consistent with the result of the two-hybrid screen, GST pulldown experiments showed that TSHZ3 interacted with the full-length BAF57 and the N-terminal part of BAF57, which contains the HMG domain (Fig. 8C). GST pulldown assays performed to map the region of TSHZ3 that binds BAF57 showed that BAF57 specifically interacts with the C-terminal domain of TSHZ3 that includes the region related to the homeodomain and two zinc finger domains (Fig. 8D).
TSHZ3-mediated Repression of Myogenin Depends in Part on BAF57
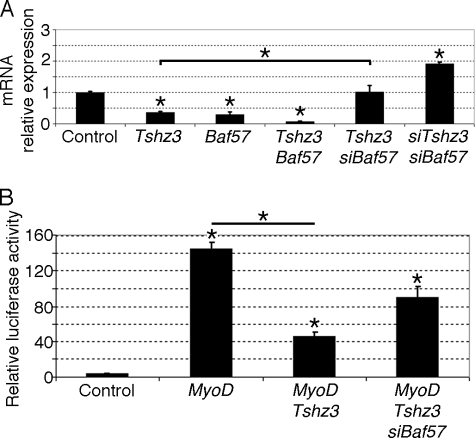
Mammalian SWI/SNF complexes are ATP-dependent chromatin-remodeling enzymes that have been implicated in the regulation of gene expression. Interestingly, a role for SWI/SNF chromatin-remodeling enzymes in MYOD-mediated activation of muscle-specific genes has been established, and it has been shown that MYOD could initiate chromatin remodeling at the Myog locus (18, 19, 25). In undifferentiated C2C12 cells, we showed that BAF57 was expressed and co-localized with TSHZ3 (supplemental Fig. 4, A and B). Analysis of the expression level of BAF57 by RT-qPCR in undifferentiated and differentiating C2C12 cells showed a gradual reduction of BAF57 expression during the myogenic differentiation process (supplemental Fig. 4A). Both the expression pattern and profile allow us to speculate that interaction between TSHZ3 and BAF57 might affect SWI/SNF-dependent muscle-specific gene expression. To test this hypothesis, we first compared the effects of overexpression of Tshz3 and BAF57 on Myog expression in C2C12 myoblasts maintained in proliferation medium for 48 h. We observed that forced expression of either Tshz3 or BAF57 alone led to a significant reduction in the levels of Myog mRNA (Fig. 9A). Interestingly, co-transfection of Tshz3 and BAF57 strongly reduced the expression of Myog (Fig. 9A). To determine whether BAF57 could contribute to TSHZ3-mediated repression of Myog, we used siRNA (siBAF57) to knock down BAF57. We showed that siBAF57 significantly reduced the TSHZ3-mediated repression of Myog, suggesting that endogenous BAF57 is required for the TSHZ3-dependent repression of MYOD activation of Myog (Fig. 9A). In addition, we observed that knockdown of both Tshz3 and BAF57 led to an increase in the levels of Myog mRNA (Fig. 9A). Efficient knockdown was confirmed by RT-qPCR or immunoblotting and did not affect cell proliferation (supplemental Fig. 4, B and G–I). These results suggest that in C2C12 myoblasts TSHZ3 and BAF57 cooperate to down-regulate Myog.
FIGURE 9.
TSHZ3 and BAF57 regulate Myog expression. A, RT-qPCR was used to examine the level of expression of Myog in control and transfected C2C12 cells. Transfected C2C12 cells were maintained for 48 h in proliferation medium. Transcript levels were first normalized to PFK internal loading control and then normalized to the expression level obtained in control conditions. In C2C12 myoblasts, TSHZ3 and BAF57 cooperate to down-regulate Myog. Data presented are the mean ± S.E. of four samples obtained from three independent experiments. B, Dual-Luciferase assays were used to analyze the regulation of Myog. C2C12 cells were harvested 24 h after transfection. pGL3MG-185 was co-transfected with MyoD or Tshz3 or in combination with siRNA targeting specific regions within Baf57. The depletion of BAF57 in C2C12 led to a reduction of TSHZ3-mediated repression of the Myog promoter. Data are presented as mean values ± S.E. from three independent experiments. * indicates statistical significance, p < 0.05 (Kruskal and Wallis test), and errors bars represent the standard deviation.
To examine whether endogenous BAF57 contributes to TSHZ3-mediated repression of the Myog promoter, we used siBAF57 to knock down BAF57 and examined activation of the pGL3MG-185 reporter in C2C12 cells transfected with MyoD and Tshz3 expression vectors. These analyses revealed that targeting of BAF57 improved the activity of Myog promoter, suggesting that TSHZ3 was less efficient to repress the Myog promoter (Fig. 9B). These experiments demonstrate that in C2C12 cells TSHZ3 and BAF57 cooperate to repress the MYOD-mediated activation of the Myog promoter.
DISCUSSION
TSHZ3 Is Expressed in Undifferentiated Myogenic Progenitors
Here we demonstrated that, in vivo, Tshz3 is expressed in quiescent and activated SCs. Indeed, TSHZ3 protein was detected in quiescent skeletal muscles of 6-week-old mice and in regenerating muscle tissue after CTX injection as well as in primary cultured SCs isolated from myofibers. During regeneration, we found that TSHZ3 was expressed in a pool of LBX1+ cells from which muscle fibers can be generated during adulthood in response to damage. In C2C12 myoblasts, Tshz3 expression was progressively down-regulated as cells differentiated into mature myofibers. Overexpression of Tshz3 in C2C12 myoblasts cells inhibited myogenic differentiation, decreased Myog expression, and led to retention of a high level of Pax7 expression. In contrast, down-regulation of endogenous Tshz3 expression in C2C12 cells enhanced Myog expression. Together, our data suggest that Tshz3 plays an important role during myogenesis likely by controlling the commitment of myoblasts and repressing or delaying the skeletal muscle differentiation program. Accordingly, in transfected C2C12 cells that express Tshz3, the expression of Pax7 was maintained at high levels. Thus, Tshz3 may have multiple roles; it may facilitate the maintenance of the cells in an undifferentiated state, potentially facilitating their self-renewal. For example, during regeneration, TSHZ3 could facilitate the expression of genes like Pax7 that keep the cell poised to respond to differentiation signals.
So far, examination of Tshz3 mutant embryos has not revealed any defect in skeletal muscle, suggesting that Tshz3 function is not required for embryonic myogenesis or for production of satellite cells during late stages of embryogenesis. Tshz3 may be co-expressed with other Tshz genes during muscle development, and it is possible that these proteins can substitute for each other. In vitro data indicate that the three TSHZ proteins behave similarly (41, 43), and it is possible that compensatory Tshz2 function might explain why we failed to see significant abnormalities in skeletal muscles of Tshz3-deficient mice. Of note, an analysis that provided novel insight into the transcriptome of quiescent and activated satellite cells in vivo demonstrated that both Tshz2 and Tshz3 are expressed in quiescent and activated SCs (74). Immunohistochemical stainings for TSHZ2 in fetal muscles suggest that, like TSHZ3, TSHZ2 is detected in cells that are fated to become muscle progenitor cells (data not shown). As Tshz3 mutant mice die at birth, in the future, conditional inactivation will allow investigation of Tshz3 function during the regeneration process.
TSHZ3 Interacts with BAF57 to Control Myogenin Promoter
During myogenesis, the SWI/SNF complex is thought to facilitate transcriptional activation of approximately one-third of the MYOD target genes (25). Here we identified BAF57 as an interacting factor for TSHZ3. BAF57 is a component of mammalian SWI/SNF complexes and contains DNA binding capability through its HMG domain. However, complexes with mutations in the HMG domain of BAF57 can still bind DNA (75). Thus, the role for the BAF57 HMG domain within the mammalian BAF complex remains poorly understood. We found that TSHZ3 interacts specifically with the HMG domain of BAF57. To understand the role of the interaction of TSHZ3 with BAF57, we focused on Myog expression in C2C12 cells. Overexpression of either TSHZ3 or BAF57 led to a significant reduction of the transcriptional activation of myogenin. When expressed together, they repressed Myog transcription to a greater extent. In addition, Myog expression was significantly increased when we used siRNA to knock down both BAF57 and Tshz3 in C2C12 cells. The depletion of only BAF57 partially blocked TSHZ3-mediated repression of the Myog promoter. These data suggest that the ability of TSHZ3 to down-regulate Myog expression and inhibit myogenic differentiation depends on interaction with BAF57.
SWI/SNF is a chromatin-remodeling complex containing a central catalytic subunit (either Brahma (BRM) or Brahma-related gene-1 (BRG1)) as well as several variable BAFs (76, 77). SWI/SNF is essential for MYOD-mediated muscle differentiation (18, 25, 78) and is required at different steps during the activation of the Myog promoter (24). A greater understanding of how individual BAF subunits contribute to the different steps, however, is needed. Recent studies have suggested roles for BAF subunits as a modulator of SWI/SNF nucleosome remodeling activity or as a determinant of SWI/SNF specificity, facilitating the recruitment to specific transcription factors (79, 80). Through its interaction with BAF57, TSHZ3 could be one of the factors that contributes both to the heterogeneity and functional specificity of SWI/SNF complexes.
Several hypotheses can be envisaged to explain how TSHZ3 affects/represses Myog transcription. The ability of MYOD to interact with BRG1 (25, 81) and to potentially target SWI/SNF chromatin-remodeling enzymes to the Myog locus appears to be required for the activation of the locus. TSHZ3, by interacting with BAF57, may antagonize the activity of SWI/SNF enzymes and participate in the maintenance of a repressive chromatin environment. It has been reported that the stable association of MYOD with the Myog promoter is dependent on functional SWI/SNF enzymes and immediately precedes a significant increase in Myog gene expression (25). It is also possible that TSHZ3 affects the potential of the SWI/SNF enzymes to facilitate activator binding to chromatin. In addition, TSHZ3 may also control other chromatin remodeling activities, in particular histone acetylation. It has been shown that TSHZ3 can directly interact with histone deacetylases and participate in repressor complexes via a direct association with the promoter region of CASP4 (43). TSHZ3 may thus recruit histone deacetylase and interfere with histone acetylation, which is induced after MYOD recruitment to the Myog promoter (25).
Further studies are required to elucidate the molecular mechanism by which TSHZ3 modulates expression of the Myog locus as well as the exact order of recruitment of MYOD, SWI/SNF, and TSHZ3 to the Myog promoter. However, the identification of TSHZ3 as a BAF57-interacting protein provides new mechanistic insight into how TSHZ3 is able to function as a transcriptional repressor.
In addition to a role in differentiation, SWI/SNF complexes are required for stem cell self-renewal and the maintenance of a pluripotent state (80, 82). Several reports showed that SWI/SNF complexes participate in the MYOD-dependent differentiation program. However, in muscle stem cells it is not known whether SWI/SNF complexes have a dual role as in other studied stem cells. Our studies suggest that the SWI/SNF complex together with TSHZ3 may control the chromatin structure to maintain muscle stem cells in an undifferentiated state.
In conclusion, we report the ability of TSHZ3 to impact basic helix-loop-helix-dependent transcription, and our data provide new insight into the mechanisms involved in early myogenesis. We suggest that during myogenesis Tshz3 controls the transition from undifferentiated myogenic progenitors to differentiated myoblasts by modulating SWI/SNF activity and antagonizing its ability to promote myogenesis.
Supplementary Material
Acknowledgments
We thank B. Gayraud-Morel and S. Tajbakhsh (Pasteur Institute, Paris, France) for advice on muscle fiber dissection and culture conditions. We thank M. Buckingham, R. Dono, R. Kelly, P. Maire, D. Montarras, A. Woolf, and S. Zaffran for helpful discussions.
This work was supported in part by Association Française contre les Myopathies (AFM) Grants 12545 and 13013 (to F. L.) and Canadian Institutes of Health Research Grant MOP-93777 (to F. J. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- MRF
- muscle-regulatory factor
- TSHZ
- Teashirt
- MYOG
- Myogenin
- SC
- satellite progenitor cell
- E
- embryonic day
- Dox
- doxycycline
- MyHC
- myosin heavy chain
- RT-qPCR
- quantitative real time PCR
- PFK
- phosphofructokinase
- CTX
- cardiotoxin
- HMG
- high mobility group
- BAF
- BRG1-associated factor
- eGFP
- enhanced GFP.
REFERENCES
- 1. Hasty P., Bradley A., Morris J. H., Edmondson D. G., Venuti J. M., Olson E. N., Klein W. H. (1993) Nature 364, 501–506 [DOI] [PubMed] [Google Scholar]
- 2. Kassar-Duchossoy L., Gayraud-Morel B., Gomès D., Rocancourt D., Buckingham M., Shinin V., Tajbakhsh S. (2004) Nature 431, 466–471 [DOI] [PubMed] [Google Scholar]
- 3. Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., Nabeshima Y. (1993) Nature 364, 532–535 [DOI] [PubMed] [Google Scholar]
- 4. Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. (1993) Cell 75, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 5. Kuang S., Gillespie M. A., Rudnicki M. A. (2008) Cell Stem Cell 2, 22–31 [DOI] [PubMed] [Google Scholar]
- 6. Ben-Yair R., Kalcheim C. (2005) Development 132, 689–701 [DOI] [PubMed] [Google Scholar]
- 7. Gros J., Manceau M., Thomé V., Marcelle C. (2005) Nature 435, 954–958 [DOI] [PubMed] [Google Scholar]
- 8. Kassar-Duchossoy L., Giacone E., Gayraud-Morel B., Jory A., Gomès D., Tajbakhsh S. (2005) Genes Dev. 19, 1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Relaix F., Rocancourt D., Mansouri A., Buckingham M. (2005) Nature 435, 948–953 [DOI] [PubMed] [Google Scholar]
- 10. Schienda J., Engleka K. A., Jun S., Hansen M. S., Epstein J. A., Tabin C. J., Kunkel L. M., Kardon G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collins C. A., Olsen I., Zammit P. S., Heslop L., Petrie A., Partridge T. A., Morgan J. E. (2005) Cell 122, 289–301 [DOI] [PubMed] [Google Scholar]
- 12. Kuang S., Kuroda K., Le Grand F., Rudnicki M. A. (2007) Cell 129, 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M. (2005) Science 309, 2064–2067 [DOI] [PubMed] [Google Scholar]
- 14. Sherwood R. I., Christensen J. L., Conboy I. M., Conboy M. J., Rando T. A., Weissman I. L., Wagers A. J. (2004) Cell 119, 543–554 [DOI] [PubMed] [Google Scholar]
- 15. Bergstrom D. A., Penn B. H., Strand A., Perry R. L., Rudnicki M. A., Tapscott S. J. (2002) Mol. Cell 9, 587–600 [DOI] [PubMed] [Google Scholar]
- 16. Megeney L. A., Kablar B., Garrett K., Anderson J. E., Rudnicki M. A. (1996) Genes Dev. 10, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 17. Sabourin L. A., Girgis-Gabardo A., Seale P., Asakura A., Rudnicki M. A. (1999) J. Cell Biol. 144, 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de la Serna I. L., Carlson K. A., Imbalzano A. N. (2001) Nat. Genet. 27, 187–190 [DOI] [PubMed] [Google Scholar]
- 19. Gerber A. N., Klesert T. R., Bergstrom D. A., Tapscott S. J. (1997) Genes Dev. 11, 436–450 [DOI] [PubMed] [Google Scholar]
- 20. Cornelison D. D., Olwin B. B., Rudnicki M. A., Wold B. J. (2000) Dev. Biol. 224, 122–137 [DOI] [PubMed] [Google Scholar]
- 21. Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B. D. (2005) Genes Dev. 19, 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mal A., Harter M. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sartorelli V., Caretti G. (2005) Curr. Opin. Genet. Dev. 15, 528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berkes C. A., Bergstrom D. A., Penn B. H., Seaver K. J., Knoepfler P. S., Tapscott S. J. (2004) Mol. Cell 14, 465–477 [DOI] [PubMed] [Google Scholar]
- 25. de la Serna I. L., Ohkawa Y., Berkes C. A., Bergstrom D. A., Dacwag C. S., Tapscott S. J., Imbalzano A. N. (2005) Mol. Cell. Biol. 25, 3997–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nielsen A. L., Oulad-Abdelghani M., Ortiz J. A., Remboutsika E., Chambon P., Losson R. (2001) Mol. Cell 7, 729–739 [DOI] [PubMed] [Google Scholar]
- 27. Fasano L., Röder L., Coré N., Alexandre E., Vola C., Jacq B., Kerridge S. (1991) Cell 64, 63–79 [DOI] [PubMed] [Google Scholar]
- 28. Röder L., Vola C., Kerridge S. (1992) Development 115, 1017–1033 [DOI] [PubMed] [Google Scholar]
- 29. Bessa J., Gebelein B., Pichaud F., Casares F., Mann R. S. (2002) Genes Dev. 16, 2415–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Erkner A., Gallet A., Angelats C., Fasano L., Kerridge S. (1999) Dev. Biol. 215, 221–232 [DOI] [PubMed] [Google Scholar]
- 31. Pan D., Rubin G. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15508–15512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soanes K. H., MacKay J. O., Core N., Heslip T., Kerridge S., Bell J. B. (2001) Mech. Dev. 105, 145–151 [DOI] [PubMed] [Google Scholar]
- 33. Wu J., Cohen S. M. (2000) Mech. Dev. 94, 47–56 [DOI] [PubMed] [Google Scholar]
- 34. Brodbeck S., Englert C. (2004) Pediatr. Nephrol. 19, 249–255 [DOI] [PubMed] [Google Scholar]
- 35. Relaix F., Buckingham M. (1999) Genes Dev. 13, 3171–3178 [DOI] [PubMed] [Google Scholar]
- 36. Caubit X., Coré N., Boned A., Kerridge S., Djabali M., Fasano L. (2000) Mech. Dev. 91, 445–448 [DOI] [PubMed] [Google Scholar]
- 37. Caubit X., Lye C. M., Martin E., Coré N., Long D. A., Vola C., Jenkins D., Garratt A. N., Skaer H., Woolf A. S., Fasano L. (2008) Development 135, 3301–3310 [DOI] [PubMed] [Google Scholar]
- 38. Caubit X., Thoby-Brisson M., Voituron N., Filippi P., Bévengut M., Faralli H., Zanella S., Fortin G., Hilaire G., Fasano L. (2010) J. Neurosci. 30, 9465–9476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Caubit X., Tiveron M. C., Cremer H., Fasano L. (2005) J. Comp. Neurol. 486, 76–88 [DOI] [PubMed] [Google Scholar]
- 40. Coré N., Caubit X., Metchat A., Boned A., Djabali M., Fasano L. (2007) Dev. Biol. 308, 407–420 [DOI] [PubMed] [Google Scholar]
- 41. Manfroid I., Caubit X., Kerridge S., Fasano L. (2004) Development 131, 1065–1073 [DOI] [PubMed] [Google Scholar]
- 42. Onai T., Matsuo-Takasaki M., Inomata H., Aramaki T., Matsumura M., Yakura R., Sasai N., Sasai Y. (2007) EMBO J. 26, 2350–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kajiwara Y., Akram A., Katsel P., Haroutunian V., Schmeidler J., Beecham G., Haines J. L., Pericak-Vance M. A., Buxbaum J. D. (2009) PLoS One 4, e5071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saller E., Kelley A., Bienz M. (2002) Genes Dev. 16, 1828–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cooper R. N., Tajbakhsh S., Mouly V., Cossu G., Buckingham M., Butler-Browne G. S. (1999) J. Cell Sci. 112, 2895–2901 [DOI] [PubMed] [Google Scholar]
- 46. Allen R. E., Temm-Grove C. J., Sheehan S. M., Rice G. (1997) Methods Cell Biol. 52, 155–176 [DOI] [PubMed] [Google Scholar]
- 47. Delfini M. C., Duprez D. (2004) Development 131, 713–723 [DOI] [PubMed] [Google Scholar]
- 48. García-Pedrero J. M., Kiskinis E., Parker M. G., Belandia B. (2006) J. Biol. Chem. 281, 22656–22664 [DOI] [PubMed] [Google Scholar]
- 49. Aziz A., Miyake T., Engleka K. A., Epstein J. A., McDermott J. C. (2009) Dev. Biol. 332, 116–130 [DOI] [PubMed] [Google Scholar]
- 50. Brown J. K., Pemberton A. D., Wright S. H., Miller H. R. (2004) J. Histochem. Cytochem. 52, 1219–1230 [DOI] [PubMed] [Google Scholar]
- 51. Lu Q. L., Partridge T. A. (1998) J. Histochem. Cytochem. 46, 977–984 [DOI] [PubMed] [Google Scholar]
- 52. Pelé M., Tiret L., Kessler J. L., Blot S., Panthier J. J. (2005) Hum. Mol. Genet. 14, 1417–1427 [DOI] [PubMed] [Google Scholar]
- 53. Iwanaga Y., Kihara Y., Takenaka H., Kita T. (2006) J. Mol. Cell. Cardiol. 41, 798–806 [DOI] [PubMed] [Google Scholar]
- 54. Oustanina S., Hause G., Braun T. (2004) EMBO J. 23, 3430–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ohto H., Kamada S., Tago K., Tominaga S. I., Ozaki H., Sato S., Kawakami K. (1999) Mol. Cell. Biol. 19, 6815–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fromont-Racine M., Rain J. C., Legrain P. (1997) Nat. Genet. 16, 277–282 [DOI] [PubMed] [Google Scholar]
- 57. Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., Legrain P. (2001) Nature 409, 211–215 [DOI] [PubMed] [Google Scholar]
- 58. Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C., Jacq B., Arpin M., Bellaiche Y., Bellusci S., Benaroch P., Bornens M., Chanet R., Chavrier P., Delattre O., Doye V., Fehon R., Faye G., Galli T., Girault J. A., Goud B., de Gunzburg J., Johannes L., Junier M. P., Mirouse V., Mukherjee A., Papadopoulo D., Perez F., Plessis A., Rossé C., Saule S., Stoppa-Lyonnet D., Vincent A., White M., Legrain P., Wojcik J., Camonis J., Daviet L. (2005) Genome Res. 15, 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Anderson J. E. (1998) Biochem. Cell Biol. 76, 13–26 [PubMed] [Google Scholar]
- 60. Goetsch S. C., Hawke T. J., Gallardo T. D., Richardson J. A., Garry D. J. (2003) Physiol. Genomics 14, 261–271 [DOI] [PubMed] [Google Scholar]
- 61. Hawke T. J., Atkinson D. J., Kanatous S. B., Van der Ven P. F., Goetsch S. C., Garry D. J. (2007) Am. J. Physiol. Cell Physiol. 293, C1636–C1644 [DOI] [PubMed] [Google Scholar]
- 62. Watanabe S., Kondo S., Hayasaka M., Hanaoka K. (2007) J. Cell Sci. 120, 4178–4187 [DOI] [PubMed] [Google Scholar]
- 63. Allen D. L., Yasui W., Tanaka T., Ohira Y., Nagaoka S., Sekiguchi C., Hinds W. E., Roy R. R., Edgerton V. R. (1996) J. Appl. Physiol. 81, 145–151 [DOI] [PubMed] [Google Scholar]
- 64. Yablonka-Reuveni Z., Rivera A. J. (1994) Dev. Biol. 164, 588–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zammit P. S., Golding J. P., Nagata Y., Hudon V., Partridge T. A., Beauchamp J. R. (2004) J. Cell Biol. 166, 347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yamaguchi A. (1995) Semin. Cell Biol. 6, 165–173 [DOI] [PubMed] [Google Scholar]
- 67. Edmondson D. G., Cheng T. C., Cserjesi P., Chakraborty T., Olson E. N. (1992) Mol. Cell. Biol. 12, 3665–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Spitz F., Demignon J., Porteu A., Kahn A., Concordet J. P., Daegelen D., Maire P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14220–14225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tapscott S. J. (2005) Development 132, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 70. Gallet A., Angelats C., Kerridge S., Thérond P. P. (2000) Development 127, 5509–5522 [DOI] [PubMed] [Google Scholar]
- 71. Taghli-Lamallem O., Gallet A., Leroy F., Malapert P., Vola C., Kerridge S., Fasano L. (2007) Dev. Biol. 307, 142–151 [DOI] [PubMed] [Google Scholar]
- 72. Albini S., Puri P. L. (2010) Exp. Cell Res. 316, 3073–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aziz A., Liu Q. C., Dilworth F. J. (2010) Epigenetics 5, 691–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pallafacchina G., François S., Regnault B., Czarny B., Dive V., Cumano A., Montarras D., Buckingham M. (2010) Stem Cell Res. 4, 77–91 [DOI] [PubMed] [Google Scholar]
- 75. Wang W., Chi T., Xue Y., Zhou S., Kuo A., Crabtree G. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mohrmann L., Verrijzer C. P. (2005) Biochim. Biophys. Acta 1681, 59–73 [DOI] [PubMed] [Google Scholar]
- 77. Sif S., Saurin A. J., Imbalzano A. N., Kingston R. E. (2001) Genes Dev. 15, 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. de la Serna I. L., Roy K., Carlson K. A., Imbalzano A. N. (2001) J. Biol. Chem. 276, 41486–41491 [DOI] [PubMed] [Google Scholar]
- 79. de la Serna I. L., Ohkawa Y., Imbalzano A. N. (2006) Nat. Rev. Genet. 7, 461–473 [DOI] [PubMed] [Google Scholar]
- 80. Saladi S. V., de la Serna I. L. (2010) Stem Cell Rev. 6, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Simone C., Forcales S. V., Hill D. A., Imbalzano A. N., Latella L., Puri P. L. (2004) Nat. Genet. 36, 738–743 [DOI] [PubMed] [Google Scholar]
- 82. Lessard J., Wu J. I., Ranish J. A., Wan M., Winslow M. M., Staahl B. T., Wu H., Aebersold R., Graef I. A., Crabtree G. R. (2007) Neuron 55, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.