Abstract
Angiogenesis requires concomitant remodeling of cell junctions and migration, as exemplified by recent observations of extensive endothelial cell movement along growing blood vessels. We report that a protein complex that regulates cell junctions is required for VEGF-driven directional migration and for angiogenesis in vivo. The complex consists of RhoA and Syx, a RhoA guanine exchange factor cross-linked by the Crumbs polarity protein Mupp1 to angiomotin, a phosphatidylinositol-binding protein. The Syx-associated complex translocates to the leading edge of migrating cells by membrane trafficking that requires the tight junction recycling GTPase Rab13. In turn, Rab13 associates with Grb2, targeting Syx and RhoA to Tyr1175-phosphorylated VEGFR2 at the leading edge. Rab13 knockdown in zebrafish impeded sprouting of intersegmental vessels and diminished the directionality of their tip cells. These results indicate that endothelial cell mobility in sprouting vessels is facilitated by shuttling the same protein complex from disassembling junctions to the leading edges of cells.
Keywords: Cell Migration, Membrane Trafficking, Rho, Trafficking, Zebra Fish
Introduction
Several of the major organs in living organisms: the vascular and lymphatic systems, the lung airways, and the ureteric system, to name a few, consist of a tubular network. The development of such networks requires recursive remodeling of the junctions between the mural cells, concurrently with their collective directional migration in response to chemotactic factors (1). A highly dynamic behavior of mural cells has been observed in several in vivo and in vitro model systems. Epithelial cells of Drosophila tracheal branches undergo extensive rearrangement (2), requiring remodeling of adherens junctions (3). Similarly, mural endothelial cells (ECs)5 move extensively during intersegmental vessel (ISV) sprouting in the zebrafish (4, 5), during the expansion of the retinal vasculature in the mouse, and during vessel sprouting from embryoid bodies (5). Junction remodeling between epithelial and ECs requires endocytosis and trafficking of junction transmembrane proteins (6). The recycling of vertebrate adherens and tight junctions is mediated by the GTPases Rab8 and Rab13, respectively (7).
Recent studies have shown that endocytosis and trafficking play an important role in facilitating directional cell migration (8). In mouse neurons, directional migration required Rab5- and Rab11-dependent trafficking of the adherens junction protein cadherin (9), similar to epithelial cells in the Drosophila tracheal system (10). The directional migration of border cells during Drosophila oogenesis, guided by EGF and PDGF/VEGF receptor tyrosine kinases, required proteins that mediate receptor tyrosine kinase endocytosis (11) and Rab11 (12). The coupling between trafficking and cell migration was highlighted in a recent study showing that the activity of Rac, a Rho GTPase essential for cell migration (13), required Rab5-dependent trafficking (14). Rac was activated on early endosomes, likely by the guanine exchange factor Tiam1 residing on the same endosome. Thus, Rab5-dependent trafficking regulated the spatial activity pattern of Rac and facilitated directional cell migration by determining where lamellipodia formed.
RhoA participates both in the regulation of cell junctions (15) and in driving cell migration (13), but the mechanism that regulates the cellular localization of RhoA so as to enable its dual functions is unknown. The spatial pattern of Rho GTPase activity in the cell is determined primarily by the localization of Rho guanine exchange factors, which are often targeted to specific cellular locations via the binding of their carboxyl terminus PDZ-binding motif to PDZ domain-containing proteins (16). In accordance, we had found that the localization the RhoA-specific guanine exchange factor Syx (named alternatively Plekhg5 or Tech) and the spatial activity pattern of RhoA depended on the PDZ-binding motif of Syx (17). Syx binds via this motif the PDZ domain of the adaptor protein synectin (named alternatively Gipc1) (17) and PDZ domains 10 and 13 of multi-PDZ domain protein 1 (Mupp1) (18). Syx is required for cell migration (17), for angiogenesis in the zebrafish and the mouse (19), and for the regulation of cell junctions.6 Here we report that the functional duality of Syx and RhoA as regulators of both cell junctions and migration is conferred by their cotrafficking from EC junctions to the leading edge.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Affinity purified anti-Syx was raised in chicken (Aves Labs) as described (17) and used at 10 mg/ml. Amot antiserum was raised in rabbit (Dr. Earl Poptic, Hybridoma Core Facility, Lerner Research Institute, Cleveland Clinic Foundation) against the 24 most carboxyl-terminal amino acids, as in Ernkvist et al. (21) and used at a 1:1600 dilution. Antiserum to Mupp1 (a gift of Dr. Ronald Javier, Baylor College of Medicine) (22) was used at 1:100 dilution. Antibodies to Rab13 (Millipore or Santa Cruz Biotechnology), RhoA (Abcam), Grb2 (Santa Cruz Biotechnology), Tyr(P)1175 VEGFR2 (GenScript), and phalloidin-Alexa647 (Invitrogen) were used according to the manufacturer's instructions. Conjugated secondary antibodies were from Invitrogen (Alexa 488, 568, and 647), from Jackson ImmunoResearch (FITC), and from Bio-Rad (peroxidase). Validated lentiviral shRNA targeting mouse rab13 (CCGGGCCAAGAACGATTCAAGACAACTCGAGTTGTCTTGAATCGTTCTTGGCTTTTTG) and negative control lentiviral shRNA (SHC002V) were purchased from Sigma.
Cell Isolation and Culture
ECs from syx+/+ and syx−/− hearts of adult mice of congenic C57/BL6 background were isolated as described (23) and maintained up to passage 5 in DMEM (Lonza Biowhittaker), 20% FBS (Fisher), 1% glutamate (Invitrogen), 1% nonessential amino acids (Invitrogen), 20 μg/ml gentamycin (Sigma), and 100 μg/ml EC mitogen supplement (Biomedical Technologies). The polyoma middle T-immortalized amot+/+ and amot−/− ECs and their growth conditions were described previously (21).
Immunofluorescence
ECs were grown on fibronectin (R & D Systems)-coated round (12-mm diameter) glass coverslips. Two days before immunolabeling, ECs were starved for the first day in 0.5% and in serum-free DMEM on the second day. In part of the experiments, ECs were incubated in starvation medium at a uniform VEGF-A164 (R & D Systems) concentration of 20 ng/ml on ice for 30 min. At the start of the experiment (0 min), the medium was replaced with fresh starvation medium prewarmed to 37 °C, and the cells were placed in 37 °C at 5% CO2 for the durations indicated under “Results.” In Dunn chamber experiments (see below), the coverslip was placed on top of the chamber and incubated as above for 30 min. ECs were washed immediately after the coverslip was dismounted by ice-cold PBS, fixed in 4% paraformaldehyde/PBS for 30 min, and permeabilized in 10% Triton X-100/PBS for 10 min. ECs labeled by anti-ZO1 were fixed and permeabilized by methanol at −20 °C for 5 min. ECs immunolabeled by anti-Syx underwent antigen retrieval by 1 min of immersion in boiling 10 mm sodium citrate, pH 8.6, containing 0.05% Tween 20, instead of permeabilization. ECs were blocked for 40 min at room temperature by 10% PBS serum of the host species of the secondary antibody. After immunolabeling by primary antibodies diluted in PBS overnight at 4 °C, ECs were washed with PBS and incubated with fluorescently tagged secondary antibodies diluted in PBS for 30 min at room temperature. The coverslips were mounted on glass slides with Prolong Gold (Invitrogen), a DAPI-containing medium.
Microscopy
ECs were imaged a Leica TCS-NT confocal microscope by ×63 (1.4 NA) or ×40 (1.0 NA) objectives. Coverslips dismounted from Dunn chamber were imaged only around the region that was directly above the bridge separating the two wells of the chamber, so as to record only ECs subjected to the 0–20 ng/ml VEGF-A164 gradient.
FRET Imaging
The adenovirus-encoded FRET RhoA probe (24) subcloned into pAd/CMV/V5-DEST (Invitrogen) was coexpressed with lentivirus-encoded rab13-targeted or control nontargeted shRNA (Mission shRNA; Sigma) 48 h ahead of the experiment. The coverslips were mounted on top of a Dunn chamber and imaged either with or without a 0–100 ng/ml VEGF-A164 linear gradient for 5 min at 37 °C on Leica TCS-NT confocal microscope after paraformaldehyde fixation. The VEGF concentration and timing followed conditions of peak FRET-detected RhoA activity in previous studies (24). FRET efficiency was calculated by dividing the background-subtracted yellow fluorescent protein emission by the background-subtracted cyan fluorescent protein emission, using custom-written macros in Image-Pro 7.0 (Media Cybernetics). A total of 40 ECs treated by nontargeting or by rab13-targeted shRNA were imaged in each group. The direction of the FRET pattern polarization was determined by eye.
Quantification of Colocalization and of Polarization of Cellular Proteins
Colocalization was quantified by CoolLocalizer (Cytolight, Stockholm, Sweden), based on Pearson's correlation coefficient (25). The polarization of the colocalization pattern was determined by measuring the colocalization level in each of six 60-degree sectors. The center of the first sector was aligned in the gradient direction and designated as 0 degrees. Colocalization was calculated in 10 ECs from at least two experiments and presented as a radial graph of the means and standard deviations of the colocalization level in each sector. The significance of the difference (p) was determined by a two-sample t test of the sectors with the highest and the second highest colocalization levels.
Immunoprecipitation
Confluent ECs grown in a 35-mm dish were lysed in 250 μl of lysis solution (Cytoskeleton) and incubated with 1 μg of anti-Rab13 (Santa Cruz Biotechnology) overnight at 4 °C. Antibody-bound complexes were precipitated with protein G-Sepharose beads (Santa Cruz Biotechnology) and eluted by boiling the thoroughly washed beads in SDS sample buffer. Eluted proteins were resolved on 12% SDS-PAGE gel (Bio-Rad), transferred to a PVDF membrane (Pierce) at 25 V for 1 h, and detected by anti-Grb2 (Santa Cruz) followed by peroxidase-conjugated goat anti-mouse IgG (Bio-Rad). Immunoblotted bands were detected by exposure to BioMax XAR film (Kodak).
Dunn Chamber Chemotaxis Assay
The Dunn chamber and its assembly were described in detail before (26). Approximately 104 primary mouse heart ECs were seeded on fibronectin-coated square glass coverslips and starved overnight prior to the assay. Both concentric wells of the chamber were filled with starvation medium (DMEM with 0.5% BSA), and a coverslip seeded with ECs was inverted onto the chamber in an offset position leaving a narrow slit at one edge for refilling the outer well. The coverslip was sealed in place using a 60 °C-melted wax mixture (paraffin, beeswax, Vaseline, 1:1:1) around the edges, except for the filling slit. To set up a linear radially directed VEGF-A164 gradient, the medium in the outer well was drained and replaced by starvation medium containing 0.5% BSA and 20 ng/ml VEGF-A164. The slit was then sealed by hot wax mixture. For control experiments in which ECs were subjected to uniform concentrations of chemoattractant, both wells were filled with medium containing VEGF (20 ng/ml). Migrating ECs were imaged on Leica DM IRB inverted microscope equipped with CoolSnap HQ cooled CCD camera (Photometrics) on a motorized stage (Prior Scientific) prewarmed to 37 °C. Stage positions were preprogrammed by MetaMorph 6.3r2 software (Molecular Devices). The Dunn chamber assembly was preincubated for 30 min to let its temperature equilibrate and a linear gradient to form between the outer and inner wells. The coverslip region laying directly above the circular ridge separating the outer and inner wells was imaged every 5 min for 1 h. The trajectories of migrating ECs were tracked manually, and their X-Y positions were input into Image-Pro for path plotting and analysis. The probability of the null hypothesis (that the migration of the 20–50 ECs sampled in each experiment was not directional) was determined by Rayleigh test for unimodal clustering (27). The 95% confidence interval of the mean direction was calculated using a Batschelet chart (27).
Zebrafish Protocols
Zebrafish were maintained, and the embryos were imaged as described (28). Morpholino antisense oligonucleotides (MOs) were designed and purchased from by GeneTools. The MO1 sequence spans the rab13 ATG codon: GAAGTCGTACTTCTTTGCCATTGTT. MO2 targets the rab13 exon 2-intron 2 border: AGAATCTTTGTTTACCAGACTTGCA. A 5-base pair mismatch of MO2 with the sequence AGAATGTTTCTTTAGCACACTTCCA served as negative control.
RESULTS
syx−/− ECs Fail to Follow a VEGF-A165 Gradient
To investigate the mechanism underlying the angiogenic defects we detected previously in the syx−/− mouse and in the zebrafish syx morphant (19), we compared the directional response of syx+/+ and syx−/− ECs induced by VEGF-A164. An angiogenesis paradigm generalized from observations of vascular development in the retina (29) posits that sprouting vessels follow a VEGF gradient. To simulate the in vivo conditions, we subjected ECs to a VEGF-A164 gradient (0–20 ng/ml) in a Dunn chemotaxis chamber (27). The basal migration distances of syx+/+ and syx−/− ECs in the absence of VEGF-A were similarly low (Fig. 1A). Both cell types migrated in response to a uniform VEGF-A164 concentration, although the distances traveled by syx−/− ECs were shorter than those traveled by syx+/+ ECs (Fig. 1B), similar to our previous observations (19). The responses of the two cell types to a VEGF-A164 gradient were drastically different from each other: syx+/+ EC followed the direction of the gradient, whereas syx−/− ECs failed to respond in a directional manner (Fig. 1C). The difference between the responses of syx+/+ EC and syx−/− ECs cannot be accounted for by a lower VEGFR2 expression level in the latter (supplemental Fig. S1). These results indicate that Syx is required for EC directional migration.
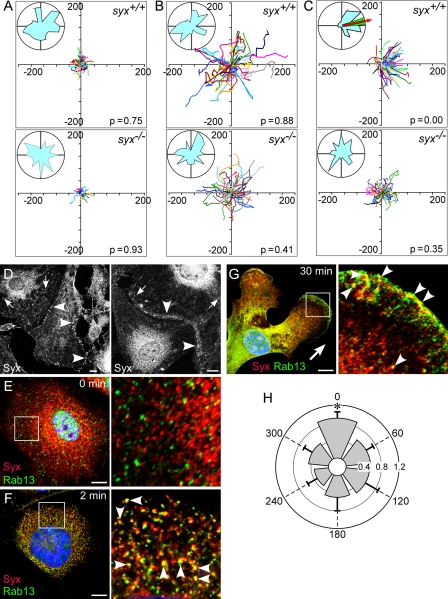
FIGURE 1.
Syx was required for directional migration in response to VEGF and colocalized with Rab13. A–C, the migration of syx+/+ and syx−/− ECs did not have a predominant direction in the absence (A) or presence (B) of a uniform concentration of VEGF-A164 (20 ng/ml). Under a 0–20 ng/ml VEGF-A164 gradient (C), syx+/+ ECs migrated preferentially (red arrow in circular histogram represents mean direction; the green region represents 95% confidence interval) along the gradient direction (the x axis), whereas syx−/− ECs did not. x and y axis units are μm; total time, 240 min; p, probability of absence of a predominant direction, based on Raleigh test, n = 50–100. D, endogenous Syx was localized mainly to the cell periphery and cell junctions in confluent cells (arrowheads) and partially to cytoplasmic punctae (arrows). E, Syx and Rab13 appeared as cytoplasmic punctae in quiescent ECs. F, Syx and Rab13 colocalized within 2 min of VEGF-A164 stimulation (20 ng/m). G, under a VEGF-A164 gradient, Syx and Rab13 colocalized at the leading edge facing the gradient direction (arrow). H, quantification of the angular distribution of Syx and Rab13 colocalization in ECs subjected to a VEGF-A164 gradient (0–20 ng/m). The gradient direction is 0 degrees (mean ± S.D., n = 10, p < 0.007). Bars, 10 μm.
Syx Colocalizes with Rab13 on Endocytic Vesicles
We have recently found that Syx is required for the maintenance of endothelial and epithelial cell junctions.6 We had found earlier that Syx was localized to the leading edge of migrating ECs and that it was required for EC migration (17, 19). Syx associates through the PDZ adaptor protein synectin with myosin VI (17), an actin-based molecular motor involved in membrane traffic (30). This association implies that Syx could traffic, possibly facilitating its translocation from cell junctions to the leading edge and enabling its involvement in the regulation of both cell junctions and migration. In quiescent ECs, Syx was localized primarily at cell junctions and in the cytoplasm (Fig. 1D). Immunofluorescence revealed that stimulation by VEGF-A164 induced the appearance of Syx primarily in cytoplasmic punctae (Figs. 1F and 2B). We used standard endocytic markers to test whether these punctae represented endocytic vesicles. The Syx-containing punctae did not colocalize, however, with clathrin heavy chain, AP3, Rab5, EEA1, Rab4, and Rab11 (data not shown). Because Syx is localized to cell junctions of confluent cells, we immunolabeled VEGF-treated ECs for Rab13, a GTPase known to be involved in recycling the tight junction transmembrane proteins occludin (31) and claudin-1 (32). Although Syx and Rab13 did not colocalize in quiescent ECs (Fig. 1E), treatment with a uniform VEGF-A164 concentration induced extensive colocalization throughout the cytoplasm within the first 2 min (Fig. 1F). This is in agreement with previous studies reporting that in unpolarized cells, such as the ECs shown in Fig. 1 (D–G), Rab13 is present exclusively in cytoplasmic vesicles (33). When placed under a VEGF-A164 gradient (0–20 ng/ml) in a Dunn chamber, ECs formed lamellipodia and polarized in the gradient direction (Fig. 1G). Strikingly, Rab13 and Syx colocalized at the leading edge within 30 min. The extent of colocalization between Rab13 and Syx was highest in the 60-degree sector facing the gradient direction (Fig. 1H), indicating that the VEGF-A164 gradient polarized the colocalization pattern of Syx and Rab13. The localization at the leading edge suggests that similar to Syx, Rab13 has a multifaceted role that includes trafficking of junction proteins to the leading edge of migrating cells.
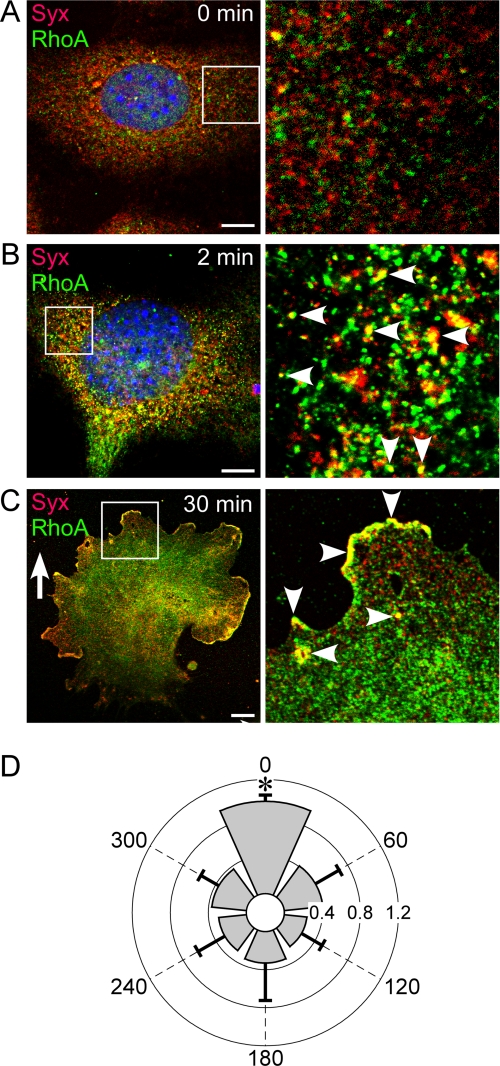
FIGURE 2.
Syx and RhoA colocalized in response to VEGF. A, there was little colocalization of Syx and RhoA in quiescent ECs. B, Syx and RhoA colocalized on endocytic vesicles within 2 min of stimulation by 20 ng/ml VEGF-A164. C, Syx and RhoA polarized in the direction of the VEGF-A164 gradient (0–20 ng/m) and colocalized on endocytic vesicles. D, quantification of the extent and angular distribution of Syx and RhoA colocalization in ECs subjected to a VEGF-A164 gradient (0–20 ng/m). The gradient direction is 0 degrees (mean ± S.D., n = 10; p < 0.0001). Bars, 10 μm.
RhoA Colocalizes with Syx on Endocytic Vesicles
Because Syx is a RhoA-specific GTPase (18, 34), we sought to determine the localization of its effector. There was little colocalization between Syx and RhoA in quiescent ECs (Fig. 2A). In contrast, a uniform VEGF-A164 concentration (20 ng/ml) induced extensive colocalization of Syx and RhoA on endocytic vesicles within 2 min (Fig. 2B). The localization of Syx and RhoA in ECs under VEGF-A164 gradient was polarized in the gradient direction, mainly at the leading edge (Fig. 2C). The extent of colocalized Syx and RhoA in the frontal 60-degree sector was more than 2-fold higher than in all other 60-degree sectors (Fig. 2D).
Rab13 Is Required for Directional Migration, for Syx and RhoA Trafficking, and for the Polarization of Activated RhoA
To investigate the dependence of EC directional migration and Syx trafficking on Rab13, we knocked down rab13 by shRNA (supplemental Fig. S2). The migration of Rab13-deficient ECs in response to a VEGF-A164 gradient was not directional, similar to that of syx−/− ECs (Fig. 3A), suggesting that Rab13-dependent trafficking of Syx and RhoA is required for maintaining directional migration. In agreement, Syx and RhoA did not colocalize in Rab13-deficient ECs neither under uniform VEGF-A164 (Fig. 3B) nor under a VEGF-A164 gradient (Fig. 3C). These results show that Rab13 is required for VEGF-guided migration, as well as for Syx and RhoA trafficking. In addition to its role in tight junction recycling, Rab13 mediates traffic between the trans-Golgi network and recycling endosomes (35). Because all of the Syx-associated proteins are cytoplasmic rather than transmembrane receptors, they are not expected to traffic via the trans-Golgi network. It is unlikely, therefore, that the directional migration and trafficking defects caused by rab13 silencing resulted from impairment of Rab13-dependent traffic from the trans-Golgi network to recycling endosomes.
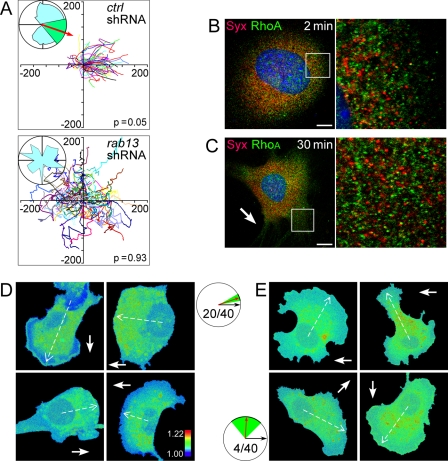
FIGURE 3.
Rab13 was required for directional migration and Syx trafficking. A, ECs in which rab13 had been silenced did not migrate predominantly along the VEGF-A164 0–20 ng/ml gradient (represented by the X+ direction). B and C, rab13 silencing prevented colocalization of Syx and RhoA on endocytic vesicles in VEGF-A164-treated (20 ng/ml) (B) ECs and in ECs subjected to a 0–20 ng/ml VEGF-A164 gradient. (C), Syx and RhoA traffic was normal in ECs transfected by control shRNA (data not shown). D and E, FRET efficiency images of ECs (four cells of 40 in each group) transfected by nontargeting (D) or by rab13-targeted shRNA (E). A solid arrow denotes the direction of the VEGF gradient; a dashed arrow denotes the mean direction of FRET pattern polarization, if such a pattern was manifest. The circular histograms show the average direction (red arrow) and its standard deviation (green sector) for each group, relative to the gradient direction (black arrow). The FRET pattern was polarized in 20 of 40 cells transfected by nontargeting shRNA at a mean direction of 18.4 ± 10.5 degrees, whereas it was polarized only in four of 40 cells transfected by rab13-targeted shRNA at a mean direction of 87 ± 41.8 degrees. Scale bars, 10 μm.
The dependence of Syx and RhoA trafficking on Rab13 suggests that the spatial pattern of RhoA activity may similarly depend on Rab13-mediated trafficking. To test this premise, we used a FRET probe consisting of the Rho-binding domain of rhotekin, cyan fluorescent protein, yellow fluorescent protein, and RhoA (24). The probe detects RhoA-specific guanine exchange factor activity. In 50% of ECs transfected by nontargeting shRNA and stimulated by VEGF-A164 in a Dunn chamber, the FRET spatial pattern was polarized at a mean direction of 18.4 ± 10.5 degrees relative to the gradient direction (Fig. 3D). Active RhoA was localized to either cytoplasmic punctae or along the leading edge, similar to RhoA localization (Fig. 2C) and to the RhoA activity pattern reported by previous FRET studies (36, 37). The FRET pattern was polarized only in 10% of ECs transfected with rab13-targeted shRNA, at a mean direction of 87 ± 41.8 degrees (Fig. 3E), i.e. without a directional preference. These results confirm the dependence of the spatial pattern of RhoA activity on Rab13.
Angiomotin Is Required for the Recruitment of Syx to Endocytic Vesicles
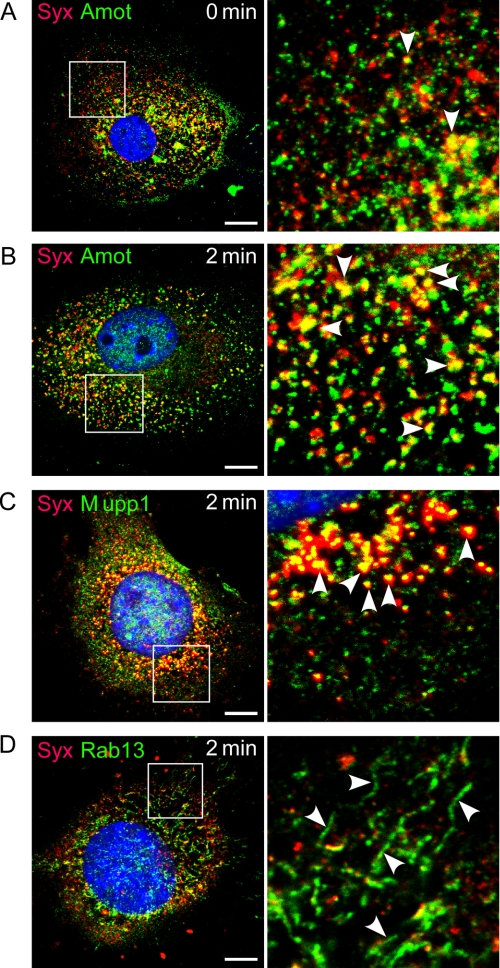
In a recent study, we proposed a molecular mechanism to account for the VEGF-induced removal of Syx from tight junctions.6 Similar to other junctional proteins (9, 31), the removal of Syx from cell junctions is likely mediated by Rab13-dependent vesicle trafficking, but the recruitment mechanism of Syx to vesicles is unknown. It is unlikely that Syx is recruited to vesicles by its pleckstrin homology domain, because such domains favor binding to phosphatidylinositol 3,4,5-phosphate, whereas the prevalent phosphoinositide-phosphate in endosomal membranes is phosphatidylinositol 3-phosphate (38). Mupp1, which binds Syx directly (18, 39), cross-links Syx to Amot (39, 40). Because Amot has a lipid-binding domain that favors mono- over di- or tri-phosphorylated phosphatidylinositols (41), it could recruit Mupp1 and Syx to vesicles. The colocalization of Amot with Syx (Fig. 4, A and B) and with Rab13 (supplemental Fig. S3) increased after treatment with VEGF-A164. Furthermore, Mupp1 also colocalized with vesicular Syx (Fig. 4C), suggesting the presence of a Syx-Mupp1-Amot complex on endocytic vesicles. The recruitment of the Syx-Mupp1-Amot complex to vesicles likely occurs before the VEGF-induced decrease in binding of Syx to Mupp1.6 To test whether Amot targets Syx to endocytic vesicles, we imaged Syx in quiescent and VEGF-A164-treated amot−/− and amot+/+ ECs. In the absence of Amot, the localizations of Syx and Rab13 were markedly different from those in amot+/+ ECs (Fig. 4D). Their level of colocalization in amot−/− was lower than in amot+/+ ECs, and most of the Rab13 cellular population in amot−/− ECs was associated with tubular structures that reached a length of 5 μm rather than with round vesicles. These results indicate that Amot is required not only for the recruitment of Syx to endocytic vesicles but also for structuring the intracellular membranes that support Rab13-dependent trafficking. Synectin did not appear to be involved in the recruitment of Syx to endocytic vesicles in response to VEGF, because the colocalization of the two proteins was very low (data not shown).
FIGURE 4.
Amot colocalized with Syx and was required for Syx trafficking. A and B, Amot colocalized with Syx in quiescent (A) and VEGF-A164-stimulated (20 ng/m) ECs (B). C, Syx colocalized also with Mupp1 in similarly stimulated ECs. D, knockdown of Amot reduced the colocalization of Syx and Rab13 and caused appearance of Rab13-associated tubules (arrowheads). Scale bars, 10 μm.
Rab13 and Syx Are Targeted to the Leading Edge of VEGF-A-treated ECs by Grb2
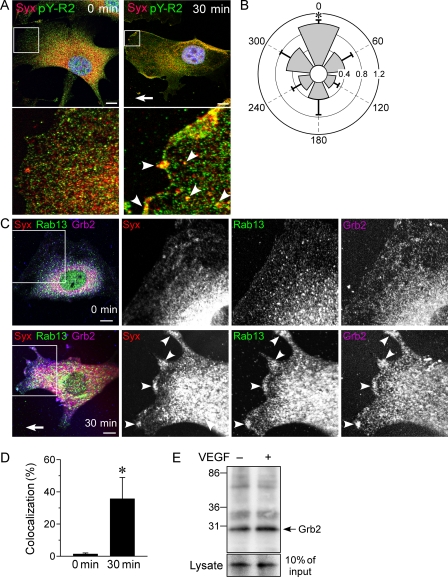
The translocation of Syx and Rab13 to the leading edge of ECs subjected to a VEGF gradient (Fig. 1F) implies the presence of a targeting motif at that location. Because VEGF induces endocytosis of VEGFR2 at the leading edge (42), the cytoplasmic domain of the internalized receptor could conceivably recruit Syx-carrying vesicles. In agreement, we found that Syx colocalized with Tyr1175-phosphorylated VEGFR2 at the leading edge (Fig. 5A). The extent of colocalization in the 60-degree sector facing the VEGF gradient was ∼2-fold higher than in any other 60-degree sector (Fig. 5B). We next sought to identify the protein that docks the Syx-carrying vesicles to Tyr(P)-VEGFR2 at the leading edge. Because Rab13 is not known to bind VEGFR2 directly, we searched for other proteins that could dock Syx-carrying vesicles to Tyr(P)-VEGFR2. Rab13 binds the cell junction protein molecule interacting with CasL-like 2 (MICAL-L2) (43). MICAL-L2 binds, in turn, actinin-4 (60), which interacts with Grb2 (61). Grb2 binds the adaptor protein growth factor receptor binding protein 2 (Grb2) (44). Grb2 has the capacity to recruit other proteins to activated VEGFR2 because its Src homology 2 domain binds phosphorylated tyrosines in the cytoplasmic domains of several receptor tyrosine kinases, including VEGFR2 (45). In agreement, Rab13, Syx, and Grb2 colocalized in the frontal lamellipodia of VEGF-stimulated ECs but were absent from the leading edge of untreated, spontaneously polarized ECs (Fig. 5, C and D). Importantly, Rab13 and Grb2 coimmunoprecipitated from EC lysates (Fig. 5E), showing that Rab13-carrying vesicles can be targeted to Tyr(P)-VEGFR2 at the leading edge by a Rab13 and Grb2-containing complex.
FIGURE 5.
Targeting of Syx to Tyr(P)1175-VEGFR2 by Grb2. A, Syx localization was not polarized in quiescent ECs, and the Tyr(P)1175-VEGFR2 signal at the cell edges was low. In ECs subjected to a VEGF-A164 gradient (0–20 ng/m), Syx colocalized with Tyr(P)1175-VEGFR2 (pY-R2) at the leading edge. B, quantification of the extent and angular distribution of Syx and Tyr(P)1175-VEGFR2 colocalization in ECs subjected to a VEGF-A164 gradient (0–20 ng/m). The gradient direction is 0 degrees (mean ± S.D., n = 10, p < 0.0001). C, Syx, Rab13, and Grb2 were absent from the leading edge of untreated spontaneously polarized ECs. A VEGF-A164 gradient (0–20 ng/m) induced colocalization of the three proteins in frontal lamellipodia (arrowheads). D, quantification of the colocalization of Grb2 with Rab13 within a distance of ∼5 μm from the leading edge of untreated ECs and of ECs subjected to a 0–20 ng/m VEGF-A164 gradient (mean ± S.D., n = 10, p < 3 × 10−7). E, the extent of Rab13 coimmunoprecipitated with Grb2 (molecular mass = 25.2 kDa) was higher in ECs treated by VEGF-A164 (20 ng/ml, 5 min). Scale bars, 10 μm.
Rab13 Is Required for ISV Growth in the Zebrafish
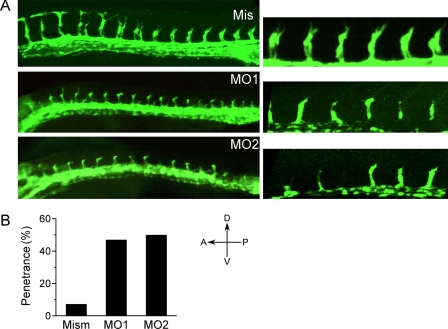
The zebrafish Rab13 ortholog has 78% sequence identity to mouse Rab13, and its mRNA is expressed in vessels of the trunk (46). To study the function of Rab13 in ISV sprouting, we knocked rab13 down in transgenic Tg(flk1:EGFP) zebrafish. Embryos were injected at the one-cell stage by MO1 or MO2, targeting the start codon and the exon 2-intron 2 splicing site, respectively. By 28 h postfertilization, part of the ISVs of embryos injected by nontargeting control MO were fully grown and started coalescing into the dorsal anastomotic longitudinal vessel (Fig. 6B). In zebrafish embryos injected by either MO1 or MO2, however, ISVs stopped growing approximately midway between the dorsal aorta and the would-be location of the unformed dorsal anastomotic longitudinal vessel (Fig. 6B). The lengths of the ISVs and the shape of their tips were variable (right-hand panels in Fig. 6A), unlike the growth-arrested ISVs in syx and amot morphants that ended uniformly in a round tip (19, 47). These observations conform to a scenario in which the loss of directional migration caused by Rab13 depletion randomizes the shape of the tips cells of sprouting ISVs. The penetrance of the ISV defects induced by MO1 and MO2 was 47 and 50%, respectively, versus 7% of the embryos injected by the control MO (Fig. 6B). The loss of tip directionality is consistent with the directional defect in cell migration caused also by rab13 knockdown (Fig. 3A). The absence of dorsal aorta defects suggests that Rab13 is specifically required for angiogenesis and is not involved in vasculogenesis.
FIGURE 6.
rab13 knockdown caused ISV defects. A, images of the vasculature in the trunk of Tg(flk1:EGFP) embryos injected by 12 ng each of a five-base mismatch control MO (Mis) or MOs targeting the ATG codon (MO1) and the exon 2-intron 2 splicing site (MO2). The embryos were fixed at 28 h postfertilization. B, quantification of the penetrance of the mismatch MO, of MO1, and of MO2 in groups of 44, 75, and 80 embryos, respectively.
DISCUSSION
Accumulating evidence indicates that membrane traffic is tightly integrated into cell migration (8, 48). Whereas it is appreciated that directional cell migration requires trafficking of adhesion and growth factor receptors (8), including that of VEGFR2 (42, 49, 50), the dependence of directional migration on trafficking of cytoskeletal and motility proteins has come to light only recently. Our finding that Rab13-dependent trafficking of Syx and RhoA is required for directional cell migration resonates with a recent report on the requirement of Rab5-dependent trafficking of Rac and Tiam1 for hepatocyte growth factor-stimulated migration of mouse embryo fibroblasts (14). Interestingly, our results suggest that Rac and RhoA traffic along separate pathways. This previously unknown difference between Rac and RhoA indicates that the cellular targeting of Rac and RhoA, and hence their spatial activity patterns, are determined by different mechanisms. Although the targeting mechanism of Rac is unknown, our data describe how Grb2 and Tyr-phosphorylated VEGFR2 target RhoA to the leading edge of migrating cells. Grb2 binds to Tyr(P) not only of VEGFR2 (45) but also of EGFR, PDGFR (51), and FGF receptor (52). Consequently, the Grb2-dependent targeting of RhoA to the leading edge of migrating cells may be a general mechanism shared by several growth factors and present in multiple cell types. Depletion of Grb2 has indeed been shown to decrease the EGF-induced migration of fibroblasts and macrophages (53).
Based on in vitro experiments with synthetic lipid membranes, a recent study attributed liposome tubulation activity to a region in the amino terminus of Amot (41). Although our findings also indicate that Amot structures intracellular membranes, we observed that amot−/− ECs contained numerous Rab13-associated tubular structures (Fig. 5D). This indicates that Amot may cooperate with dynamin in vesicle fission, similar to amphiphysin (54), rather than form tubules. Because other membrane-molding proteins bind Rho and Rab GTPases (55), it is possible that Amot targets RhoA and Rab13 to endocytic vesicles, in addition to recruiting Mupp1/Patj (41) and Syx (this study). The multifunctionality of Amot, including its involvement in the regulation of Cdc42 via the GTPase-activating protein Rich1 (56), highlights Amot as pivotal adaptor protein in the intersection between trafficking, cell junctions, and cell migration. Whereas the colocalization of Amot and Rab13 on perinuclear endosomes was observed before (41), we find that Amot and Rab13 colocalize extensively on endocytic vesicles throughout the cytoplasm. Unlike the study of Heller et al. (41), we did not observe colocalization of Amot and Rab11. However, Amot-Rab11 colocalization occurred only in unpolarized epithelial cells, whereas we focused on ECs. We previously reported that the polarization of active RhoA in migrating ECs required the Amot-Mupp1/Patj-Syx complex (39). The dependence of Syx recruitment to endocytic vesicles on Amot, which we report here, explains that finding.
The colocalization of the Crumbs complex protein Mupp1 with Syx on endocytic vesicles in VEGF-stimulated ECs could account for the presence of the Mupp1 paralog Pals1-associated tight junction protein (Patj) at the leading edge of migrating epithelial cells and for its involvement in directional cell migration (57). Similarly, the role of Amot in Rab13-dependent trafficking of Syx and RhoA may explain how Amot regulates both cell migration and cell junctions (58). The involvement of Patj/Mupp1 and Amot in the regulation of directional cell migration conveys a common theme whereby junctional proteins are required also for cell migration, a seemingly counterintuitive notion. However, the functional versatility of these proteins and their alternate localization at cell junctions and at the leading edge are probably essential for conferring the dynamic cell remodeling inherent to growing vessels, where the same cell regions may alternate between being a leading edge and forming junctions. Amot and Syx had already been shown to be required for vascular development in the mouse and zebrafish (19, 47).
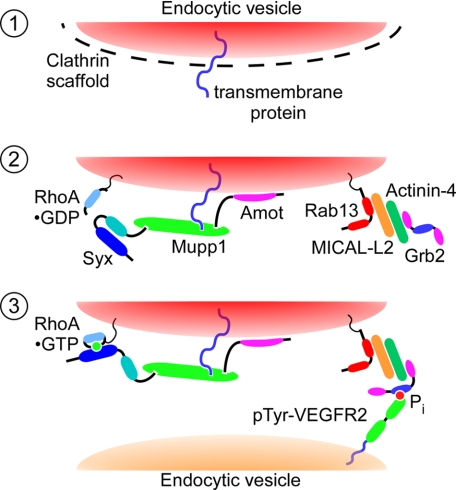
Our findings suggest a distinct scenario for the regulation of directional cell migration by Rab13-dependent trafficking of Syx and its effector, RhoA (Fig. 7): 1) the binding of VEGF-A to VEGFR2 initiates signals, still unidentified, that trigger endocytosis of junctional transmembrane receptors, likely in a clathrin-dependent manner (59), and Amot recruits the Syx-Mupp1-RhoA complex to endocytic vesicles harboring junctional transmembrane receptors; 2) Rab13 associates with and mediates the trafficking of these vesicles; it recruits Grb2 through binding MICAL-L2 and actinin-4; and 3) Grb2 targets these vesicles to the Tyr-phosphorylated cytoplasmic domain of internalized VEGFR2; Syx activates RhoA residing on the same vesicle, near the leading edge of migrating cells. In this manner, Rab13-mediated trafficking of the Syx-associated protein complex could sustain movement of mural ECs in sprouting vessels.
FIGURE 7.
Hypothetical mechanism of Syx and RhoA trafficking from cell junctions to the leading edge. In step 1, VEGF-A binding to VEGFR2 triggers clathrin-dependent endocytosis of junctional transmembrane proteins. Amot targets the Syx-associated complex, containing also Mupp1 and RhoA, to the cytoplasmic leaflet of uncoated endocytic vesicles enclosing the endocytosed junctional transmembrane proteins (step 2). Rab13 associates with and mediates the trafficking of these vesicles. It recruits Grb2 (step 3), which targets the Rab13-associated vesicles to Tyr-phosphorylated VEGFR2. Syx activates RhoA cotrafficking on the same vesicles, at the leading edge of the migrating cell.
Supplementary Material
Acknowledgments
We thank Dr. Earl Poptic (Lerner Research Institute of the Cleveland Clinic Foundation) for raising polyclonal antibodies. We thank Dr. Lars Holmgren (Karolinska Institute, Sweden) for sharing polyoma middle T-immortalized amot+/+ and amot−/− ECs, and Dr. Ronald Javier (Baylor College of Medicine) and Dr. Elior Peles (Weizman Institute of Science, Israel) for providing antisera to Mupp1. We thank Dr. Klaus Hahn (University of North Carolina) for sharing the RhoA FRET probe.
This work was supported, in whole or in part, by National Institutes of Health Predoctoral Fellowships F30-HL-094020 and T32-GM-80079 (to C. M. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Siu P. Ngok, Rory Geyer, Sudesh Agrawal, Antonios Kourtidis, Laura J. Lewis-Tuffin, Karen L. Moodie, Deborah Huveldt, Chuanshen Wu, Miaoliang Liu, Ruth Marx, Jay M. Baraban, Peter Storz, Arie Horowitz, and Panos Z. Anastasiadis, unpublished results.
- EC
- endothelial cell
- ISV
- intersegmental vessel
- MO
- morpholino antisense oligonucleotide
- VEGFR
- VEGF receptor.
REFERENCES
- 1. Rørth P. (2009) Annu. Rev. Cell. Dev. Biol. 25, 407–429 [DOI] [PubMed] [Google Scholar]
- 2. Affolter M., Caussinus E. (2008) Development 135, 2055–2064 [DOI] [PubMed] [Google Scholar]
- 3. Cheshire A. M., Kerman B. E., Zipfel W. R., Spector A. A., Andrew D. J. (2008) Dev. Dyn. 237, 2874–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blum Y., Belting H. G., Ellertsdottir E., Herwig L., Lüders F., Affolter M. (2008) Dev. Biol. 316, 312–322 [DOI] [PubMed] [Google Scholar]
- 5. Jakobsson L., Franco C. A., Bentley K., Collins R. T., Ponsioen B., Aspalter I. M., Rosewell I., Busse M., Thurston G., Medvinsky A., Schulte-Merker S., Gerhardt H. (2010) Nat. Cell. Biol. 12, 943–953 [DOI] [PubMed] [Google Scholar]
- 6. Yap A. S., Crampton M. S., Hardin J. (2007) Curr. Opin. Cell. Biol. 19, 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamamura R., Nishimura N., Nakatsuji H., Arase S., Sasaki T. (2008) Mol. Biol. Cell 19, 971–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ulrich F., Heisenberg C. P. (2009) Traffic 10, 811–818 [DOI] [PubMed] [Google Scholar]
- 9. Kawauchi T., Sekine K., Shikanai M., Chihama K., Tomita K., Kubo K., Nakajima K., Nabeshima Y., Hoshino M. (2010) Neuron 67, 588–602 [DOI] [PubMed] [Google Scholar]
- 10. Shaye D. D., Casanova J., Llimargas M. (2008) Nat. Cell. Biol. 10, 964–970 [DOI] [PubMed] [Google Scholar]
- 11. Jékely G., Sung H. H., Luque C. M., Rørth P. (2005) Dev. Cell. 9, 197–207 [DOI] [PubMed] [Google Scholar]
- 12. Janssens K., Sung H. H., Rørth P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 7323–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raftopoulou M., Hall A. (2004) Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 14. Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P. P. (2008) Cell 134, 135–147 [DOI] [PubMed] [Google Scholar]
- 15. Jou T. S., Schneeberger E. E., Nelson W. J. (1998) J. Cell. Biol. 142, 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. García-Mata R., Burridge K. (2007) Trends Cell. Biol. 17, 36–43 [DOI] [PubMed] [Google Scholar]
- 17. Liu M., Horowitz A. (2006) Mol. Biol. Cell 17, 1880–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Estévez M. A., Henderson J. A., Ahn D., Zhu X. R., Poschmann G., Lübbert H., Marx R., Baraban J. M. (2008) J. Neurochem. 106, 1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garnaas M. K., Moodie K. L., Liu M. L., Samant G. V., Li K., Marx R., Baraban J. M., Horowitz A., Ramchandran R. (2008) Circ. Res. 103, 710–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deleted in proof.
- 21. Ernkvist M., Aase K., Ukomadu C., Wohlschlegel J., Blackman R., Veitonmäki N., Bratt A., Dutta A., Holmgren L. (2006) FEBS J. 273, 2000–2011 [DOI] [PubMed] [Google Scholar]
- 22. Lee S. S., Glaunsinger B., Mantovani F., Banks L., Javier R. T. (2000) J. Virol. 74, 9680–9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chittenden T. W., Claes F., Lanahan A. A., Autiero M., Palac R. T., Tkachenko E. V., Elfenbein A., Ruiz de Almodovar C., Dedkov E., Tomanek R., Li W., Westmore M., Singh J. P., Horowitz A., Mulligan-Kehoe M. J., Moodie K. L., Zhuang Z. W., Carmeliet P., Simons M. (2006) Dev. Cell. 10, 783–795 [DOI] [PubMed] [Google Scholar]
- 24. Pertz O., Hodgson L., Klemke R. L., Hahn K. M. (2006) Nature 440, 1069–1072 [DOI] [PubMed] [Google Scholar]
- 25. Manders E. M., Stap J., Brakenhoff G. J., van Driel R., Aten J. A. (1992) J. Cell. Sci. 103, 857–862 [DOI] [PubMed] [Google Scholar]
- 26. Wells C. M., Ridley A. J. (2005) Methods Mol. Biol. 294, 31–41 [DOI] [PubMed] [Google Scholar]
- 27. Zicha D., Dunn G., Jones G. (1997) Methods Mol. Biol. 75, 449–457 [DOI] [PubMed] [Google Scholar]
- 28. Xie J., Farage E., Sugimoto M., Anand-Apte B. (2010) BMC Dev. Biol. 10, 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerhardt H., Golding M., Fruttiger M., Ruhrberg C., Lundkvist A., Abramsson A., Jeltsch M., Mitchell C., Alitalo K., Shima D., Betsholtz C. (2003) J. Cell. Biol. 161, 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aschenbrenner L., Lee T., Hasson T. (2003) Mol. Biol. Cell 14, 2728–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morimoto S., Nishimura N., Terai T., Manabe S., Yamamoto Y., Shinahara W., Miyake H., Tashiro S., Shimada M., Sasaki T. (2005) J. Biol. Chem. 280, 2220–2228 [DOI] [PubMed] [Google Scholar]
- 32. Marzesco A. M., Dunia I., Pandjaitan R., Recouvreur M., Dauzonne D., Benedetti E. L., Louvard D., Zahraoui A. (2002) Mol. Biol. Cell 13, 1819–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zahraoui A., Joberty G., Arpin M., Fontaine J. J., Hellio R., Tavitian A., Louvard D. (1994) J. Cell. Biol. 124, 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marx R., Henderson J., Wang J., Baraban J. M. (2005) J. Neurochem. 92, 850–858 [DOI] [PubMed] [Google Scholar]
- 35. Nokes R. L., Fields I. C., Collins R. N., Fölsch H. (2008) J. Cell. Biol. 182, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kurokawa K., Matsuda M. (2005) Mol. Biol. Cell 16, 4294–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M., Danuser G. (2009) Nature 461, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindmo K., Stenmark H. (2006) J. Cell. Sci. 119, 605–614 [DOI] [PubMed] [Google Scholar]
- 39. Ernkvist M., Luna Persson N., Audebert S., Lecine P., Sinha I., Liu M., Schlueter M., Horowitz A., Aase K., Weide T., Borg J. P., Majumdar A., Holmgren L. (2009) Blood 113, 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugihara-Mizuno Y., Adachi M., Kobayashi Y., Hamazaki Y., Nishimura M., Imai T., Furuse M., Tsukita S. (2007) Genes Cells 12, 473–486 [DOI] [PubMed] [Google Scholar]
- 41. Heller B., Adu-Gyamfi E., Smith-Kinnaman W., Babbey C., Vora M., Xue Y., Bittman R., Stahelin R. V., Wells C. D. (2010) J. Biol. Chem. 285, 12308–12320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sawamiphak S., Seidel S., Essmann C. L., Wilkinson G. A., Pitulescu M. E., Acker T., Acker-Palmer A. (2010) Nature 465, 487–491 [DOI] [PubMed] [Google Scholar]
- 43. Terai T., Nishimura N., Kanda I., Yasui N., Sasaki T. (2006) Mol. Biol. Cell 17, 2465–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suzuki T., Nakamoto T., Ogawa S., Seo S., Matsumura T., Tachibana K., Morimoto C., Hirai H. (2002) J. Biol. Chem. 277, 14933–14941 [DOI] [PubMed] [Google Scholar]
- 45. D'Angelo G., Martini J. F., Iiri T., Fantl W. J., Martial J., Weiner R. I. (1999) Mol. Endocrinol. 13, 692–704 [DOI] [PubMed] [Google Scholar]
- 46. Thisse B., Pflumio S., Fürthauer M., Loppin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X. Q., Thisse C. (2001) Direct Data Submission to The Zebrafish Model Organism Database [Google Scholar]
- 47. Aase K., Ernkvist M., Ebarasi L., Jakobsson L., Majumdar A., Yi C., Birot O., Ming Y., Kvanta A., Edholm D., Aspenström P., Kissil J., Claesson-Welsh L., Shimono A., Holmgren L. (2007) Genes Dev. 21, 2055–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Polo S., Di Fiore P. P. (2006) Cell 124, 897–900 [DOI] [PubMed] [Google Scholar]
- 49. Lanahan A. A., Hermans K., Claes F., Kerley-Hamilton J. S., Zhuang Z. W., Giordano F. J., Carmeliet P., Simons M. (2010) Dev. Cell 18, 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Y., Nakayama M., Pitulescu M. E., Schmidt T. S., Bochenek M. L., Sakakibara A., Adams S., Davy A., Deutsch U., Lüthi U., Barberis A., Benjamin L. E., Mäkinen T., Nobes C. D., Adams R. H. (2010) Nature 465, 483–486 [DOI] [PubMed] [Google Scholar]
- 51. Lowenstein E. J., Daly R. J., Batzer A. G., Li W., Margolis B., Lammers R., Ullrich A., Skolnik E. Y., Bar-Sagi D., Schlessinger J. (1992) Cell. 70, 431–442 [DOI] [PubMed] [Google Scholar]
- 52. Kanai M., Göke M., Tsunekawa S., Podolsky D. K. (1997) J. Biol. Chem. 272, 6621–6628 [DOI] [PubMed] [Google Scholar]
- 53. Knapek K., Frondorf K., Post J., Short S., Cox D., Gomez-Cambronero J. (2010) Mol. Cell. Biol. 30, 4492–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takei K., Slepnev V. I., Haucke V., De Camilli P. (1999) Nat. Cell. Biol. 1, 33–39 [DOI] [PubMed] [Google Scholar]
- 55. Habermann B. (2004) EMBO Rep. 5, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wells C. D., Fawcett J. P., Traweger A., Yamanaka Y., Goudreault M., Elder K., Kulkarni S., Gish G., Virag C., Lim C., Colwill K., Starostine A., Metalnikov P., Pawson T. (2006) Cell 125, 535–548 [DOI] [PubMed] [Google Scholar]
- 57. Shin K., Wang Q., Margolis B. (2007) EMBO Rep. 8, 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bratt A., Birot O., Sinha I., Veitonmäki N., Aase K., Ernkvist M., Holmgren L. (2005) J. Biol. Chem. 280, 34859–34869 [DOI] [PubMed] [Google Scholar]
- 59. Ivanov A. I., Nusrat A., Parkos C. A. (2004) Mol. Biol. Cell 15, 176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nakatsuji H., Nishimuri N., Yamamura R., Kanayama H. O., Sasaki T. (2008) Mol. Cell Biol. 28, 3324–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. (2003) Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.