Abstract
The regulation of gene expression in response to environmental signals and metabolic imbalances is a key step in maintaining cellular homeostasis. BTB and CNC homology 1 (BACH1) is a heme-binding transcription factor repressing the transcription from a subset of MAF recognition elements at low intracellular heme levels. Upon heme binding, BACH1 is released from the MAF recognition elements, resulting in increased expression of antioxidant response genes. To systematically address the gene regulatory networks involving BACH1, we combined chromatin immunoprecipitation sequencing analysis of BACH1 target genes in HEK 293 cells with knockdown of BACH1 using three independent types of small interfering RNAs followed by transcriptome profiling using microarrays. The 59 BACH1 target genes identified by chromatin immunoprecipitation sequencing were found highly enriched in genes showing expression changes after BACH1 knockdown, demonstrating the impact of BACH1 repression on transcription. In addition to known and new BACH1 targets involved in heme degradation (HMOX1, FTL, FTH1, ME1, and SLC48A1) and redox regulation (GCLC, GCLM, and SLC7A11), we also discovered BACH1 target genes affecting cell cycle and apoptosis pathways (ITPR2, CALM1, SQSTM1, TFE3, EWSR1, CDK6, BCL2L11, and MAFG) as well as subcellular transport processes (CLSTN1, PSAP, MAPT, and vault RNA). The newly identified impact of BACH1 on genes involved in neurodegenerative processes and proliferation provides an interesting basis for future dissection of BACH1-mediated gene repression in neurodegeneration and virus-induced cancerogenesis.
Keywords: Cell Cycle, Chromatin Immunoprecipitation (ChiP), Gene Regulation, Heme, Hemostasis, Oxidative Stress, siRNA, Transcription, Transcription Regulation, Transcription Repressor
Introduction
The coupling of key metabolic activities and gene expression is central to the coordination of cellular homeostasis. The transcriptional repressor BTB and CNC homology 1 (BACH1) is a heme-binding protein belonging to the cap‘n’collar type of basic leucine zipper factors and constitutes a major link between the cellular heme level, the redox state, and the transcriptional response. BACH1 was identified as a key player in the physiological regulation of oxidative stress, where it acts as a repressor of its main target, HMOX1 (1), the rate-limiting enzyme of the heme degradation pathway that cleaves heme to form ferrous iron and biliverdin, which is subsequently converted to bilirubin, a potent radical scavenger, and carbon monoxide.
Although heme is essential to life, it can exert toxic effects through its ability to catalyze the formation of reactive oxygen species, whose action is counterbalanced by HMOX1. The regulation of HMOX1 expression involves a direct sensing of heme levels by BACH1, an analogy with the lac repressor sensitivity to lactose, generating a feedback loop whereby the substrate effects a repressor-activator antagonism (2). Heme induces export of BACH1 from the nucleus into the cytoplasm (3), where BACH1 can be sequestered at microtubules (4). Finally, BACH1 is ubiquitinated and degraded (5). Other BACH1 targets include the NAD(P)H menadione oxidoreductase 1 (6), the ferritin heavy and light chains, as well as thioredoxin reductase 1 (7) and the glutamate-cysteine ligase catalytic and modifier subunits (8). In the mouse, Bach1 can bind the p53 apoptosis effector Perp and the cyclin-dependent kinase inhibitor 1A together with p53, resulting in negative regulation of oxidative stress-induced cellular senescence (9). Several lines of evidence suggest the involvement of BACH1 in neurodegenerative diseases and cancer. Among other proteins involved in the heme degradation pathway, HMOX1 and the biliverdin reductases A and B were proposed as serum biomarkers for the early detection of Alzheimer disease (10). Furthermore, BACH1 expression was down-regulated by microRNAs in viral infection-associated cancerogenesis (11–13).
At low heme levels, BACH1 forms heterodimers with small MAF proteins and functions as a competitive repressor at MAF recognition elements (MAREs).2 In this, BACH1 competes with nuclear factor erythroid-derived 2-like 2 (NFE2L2, also termed NRF2), which acts as a transcriptional activator at MAREs together with small MAFs (14). NFE2L2 controls the expression of genes associated with antioxidant response elements encoding, among others, a number of phase II detoxification enzymes. NFE2L2 is recognized as master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes in response to oxidative stress. The binding of NFE2L2 to antioxidant response elements is regulated through competition with Kelch-like ECH-associated protein 1 (KEAP1) and BACH1 (8, 15, 16). However, the full subset of MARE-driven genes regulated by BACH1 has not been characterized systematically, although BACH1 knockdown by arsenite identified a small set of potential genes (17).
To address the global gene regulatory networks involving BACH1, we performed chromatin immunoprecipitation followed by high throughput deep sequencing (ChIP-seq) as well as knockdown of the BACH1 transcription factor in HEK 293 cells using three independent small interfering RNAs. The integration of the genome-wide data sets allowed the identification of transcriptional regulatory networks targeted by BACH1 to outline associated metabolic and signal transduction pathways.
EXPERIMENTAL PROCEDURES
Additional information on the experimental procedures can be found in the supplemental material.
Chromatin Immunoprecipitation, Library Construction, and Sequencing
Chromatin immunoprecipitation from HEK 293 cells was performed as described previously (18) using a well characterized goat polyclonal BACH1 antibody (sc-14700X, Santa Cruz Biotechnology). The input and ChIP DNA were used for preparation of sequencing libraries according to the manufacturer's instructions for the Illumina Genome Analyzer, with some modifications. After PCR preamplification, size selection was performed by agarose gel electrophoresis, excision, and purification of DNA fragments in the range of 150–250 bp. The purified libraries were sequenced on an Illumina 1G Genome Analyzer.
Genomic Alignment of Reads and Peak Calling
Sequencing reads were aligned to the human genome (UCSC hg18) using the Eland algorithm allowing up to two mismatches without insertions or deletions, resulting in 8,153,703 ChIP reads and 8,008,492 control reads with unique match to the genome. Redundant reads were removed for identification of candidate peak regions using QuEST-2.4 (19). After removal of artifactual peaks, we obtained a final list of 84 BACH1-bound genomic regions.
Analysis of Binding Motifs, Conservation Scores, and Nearby Genes
The 300-bp genomic sequences surrounding each peak were extracted for MEME motif over-representation analysis (20) with default parameters. The strongest log-odds matrix from the MEME output was compared with known motifs in the Transfac data base using TOMTOM (21) and was used for MAST motif search (22) in the peak sequences. Average conservation scores for peaks and DNA-binding motifs were determined using Galaxy (23) with phastCons on 17-species multiZ alignment of UCSC hg18. Conservation plots were obtained from the cis-regulatory element annotation system (CEAS) (24). Nearby RefSeq genes within 15 kb of the peak regions were annotated using CisGenome version 1.2 (25).
Inhibition of BACH1 Expression by RNA Interference
RNA interference experiments for BACH1 were performed using three independent types of silencing molecules against BACH1, one unmodified synthetic small interfering RNA (siRNA1, Qiagen SI00309876), one chemically modified synthetic small interfering RNA (siRNA2) (HSS100910, Invitrogen), and one independent high complexity pool of 20–30-bp siRNA-like molecules (esiRNA). HEK 293 cells were seeded in 12-well plates together with esiRNA-HiPerFect complexes or siRNA-HiPerFect complexes according to the HiPerFect fast forward protocol (Qiagen). For the mock transfections, cells were treated with HiPerFect reagent only. Knockdown transfections were performed in triplicates and mock transfections and nontransfections in quadruplicate.
RNA Extraction, Reverse Transcription, and Hybridization on Microarrays
Total RNA was extracted from cultured cells at 24 and 72 h post-transfection using the RNeasy mini kit (Qiagen) following the manufacturer's instructions. All RNA samples were DNase-treated, purified, quantified, and inspected for integrity. For quantitative real time PCR analysis, complementary DNA was prepared from total RNA. Reverse transcription reactions were performed with random hexamer primers and SuperScript II reverse transcriptase (Invitrogen). For hybridizations on microarrays, biotinylated cRNA was synthesized using the GeneChip expression 3′ amplification one-cycle target labeling and control reagents (Affymetrix). Following integrity control, the cRNA was hybridized to the Affymetrix GeneChip HG-U133Plus2. The arrays were washed, stained, and scanned following recommended protocols from Affymetrix.
Analysis of Gene Expression Changes after BACH1 Knockdown
The knockdown efficiency of each silencing molecule was measured 24 and 72 h after transfection, both at the BACH1 mRNA and protein level. The genome-wide expression changes from the BACH1 knockdowns were analyzed in triplicate, together with four controls (mock transfections). The array probe intensities from the knockdown samples were normalized together with those from the control samples using GCRMA from the R/Bioconductor package (26). Probes with significant expression changes were defined by a p value<0.05 from t test and expression ratio >1.3 or <0.75. Genes with significant expression changes were defined by significant changes of the same probes among at least two of three different RNAi experiments. Gene ontology enrichment analysis among up-regulated genes was performed using the DAVID Bioinformatics Resources 6.7 (27).
Quantitative Real Time PCR Analysis
The ChIP enrichment as compared with input DNA was calculated from quantitative real time PCR results as fold-enrichment according to the ΔΔCt method (28). PCR was performed in triplicate using SYBR Green (Applied Biosystems) with 20 pg/μl ChIP or input DNA. The primer sequences can be found in the supplemental material. The knockdown efficiency for the BACH1 mRNA and expression changes of the vault RNAs VTRNA1-1 and VTRNA1-2 was calculated from quantitative real time PCR results as fold-changes compared with β-actin (ACTB) according to the ΔΔCt method. PCR was performed in triplicate using SYBR Green (Applied Biosystems) with a cDNA concentration of 12.5 ng/μl RNA equivalent from reverse transcription. The primer sequences can be found in the supplemental material.
Correlation between ChIP-seq Reads and Expression Ratios
ChIP-seq reads were quantified in reads/kb and million aligned reads values (RPKM) as described before (29). Mean expression ratios from the different RNAi experiments were calculated as arithmetic mean of the individual ratios. The S.E. was calculated as the root of the mean squared S.E. of the individual RNAi experiments. To assess the correlation between RPKM values and RNAi expression ratios, we calculated Pearson's correlation coefficients and Spearman's rank correlation coefficients.
Transcription Factor Affinity Prediction in Promoter Sequences
Promoter sequences of 2 kb length, stretching 1.8 kb upstream to 200 bp downstream of the respective TSS, were used for transcription factor affinity prediction as described before (30) using 554 transcription factor binding matrices in TransFac (version 12.1) and a human promoter-based background model. p values for the individual sequences were combined by Fisher's method and multiple test-corrected according to Benjamini-Hochberg. The top-ranking factors described under “Results” met the additional thresholds of a combined and corrected p <10−29 and a mean affinity score across the whole sequence sets of at least 0.01 (for up-regulated genes) or 0.005 (for down-regulated genes).
RESULTS
Genome-wide Characterization of BACH1-binding Sites in HEK 293 Cells
We identified the genome-wide gene targets of BACH1 by ChIP-seq experiments in HEK 293 cells, taking advantage of the endogenous expression of BACH1 in this cell line (31). We obtained a 137-fold enrichment of the known BACH1-binding enhancer region upstream of the HMOX1 gene (supplemental Fig. 1). We compared the sequencing output (Illumina Genome Analyzer GA1) of the enriched ChIP DNA with that of the control (input) DNA. From 8.15 million reads with unique alignment to the human genome, we identified 84 significantly enriched regions defining sharp peaks of ChIP reads (see supplemental Table 1) using the QuEST-2.4 package (19). For validation, we performed quantitative real time PCR analysis of 10 BACH1 target regions ranging from weak to strong binding based on the read density, using the DNAs from two independent ChIP experiments. The ChIP-seq and the ChIP-quantitative real time PCR results showed a strong correlation (R2 = 0.878; supplemental Fig. 2), demonstrating experimental reproducibility among biological replicates using different experimental techniques.
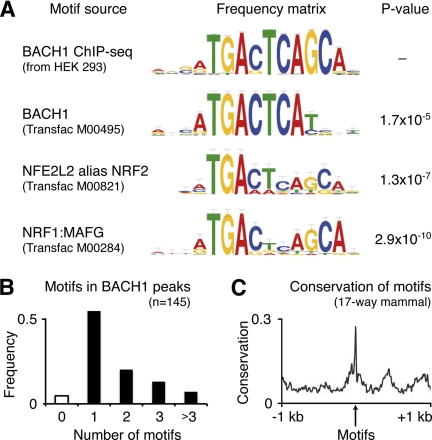
We proceeded to the discovery of transcription factor binding motifs in the 300-bp sequences around all BACH1 peaks using the MEME algorithm (20). We identified a highly significant sequence motif (E = 10−203, Fig. 1A) reminiscent of three previously known motifs, namely that of BACH1 deduced from in vitro studies (32) and those of its competitors NFE2L2 (1) and NRF1:MAFG (33). However, the newly identified consensus sequence captured the in vivo binding motif more accurately and differed by three nucleotide positions from the previously reported BACH1 motif (Fig. 1A). This binding motif was found present in 80 of the 84 identified peaks. About half of these peaks (55%) contained only one binding motif, whereas the other peaks harbored two, three, or more motifs (Fig. 1B). The highest numbers of BACH1-binding motifs were seen in the promoter of HMOX1 (seven motifs) and of the noncoding gene AFG3L1 (36 motifs).
FIGURE 1.
BACH1-binding motif comparison, occurrence, and conservation. A, top motif identified by MEME analysis of the ChIP-seq-enriched genomic regions extended the known BACH1 motif (Transfac M00495) by the three bases GCA. The comparison with all known vertebrate motifs using TOMTOM analysis revealed highly significant similarity with the known NFE2L2 motif (Transfac M00821) and the NRF1:MAFG motif (Transfac M00284). B, all binding motif occurrences in the ChIP-seq-enriched regions (n = 145) were determined by MAST analysis using the newly identified BACH1 motif. More than half of the binding peaks contained only one motif, although smaller numbers of peaks harbored two, three, or more binding motifs. C, sequence conservation analysis of the 145 motifs among 17 mammal species revealed a significantly higher mean conservation score for the motifs compared with the surrounding 2 kb of genomic DNA.
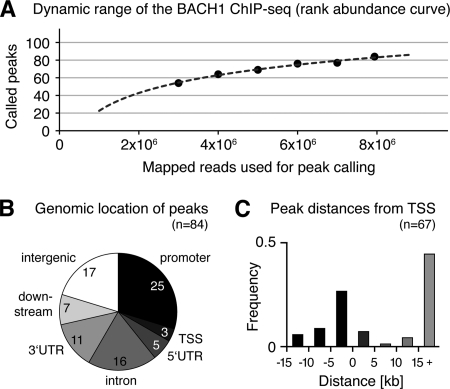
The resolution of the ChIP-seq approach was high, given that the average distance between the identified motifs and the centers of the BACH1 binding peaks was only 12 bp, with 74% of the motifs residing within 50 bp around the peak centers. The genomic binding motif sequences showed a strong conservation among mammals (phastCons on 17-species multiZ alignment), with significantly higher conservation of the motifs than the surrounding 2 kb of genomic sequence (Fig. 1C). The distance between adjacent motifs ranged between 1 and 65 bp, with over-representation of distances of 7 bp (10 motif pairs in AFG3L1 and 1 in SLC25A10) as well as 21 bp (11 motif pairs in AFG3L1), pointing to BACH1 homo- or heterodimerization (see supplemental Fig. 3 for an example region in the AFG3L1 gene). To assess whether the sequencing coverage was high enough to detect the majority of the BACH1-bound regions, we calculated the dynamic range of our BACH1 ChIP-seq approach. After sub-sampling of smaller read populations, we repeated the peak calling with the same parameters and calculated the rank abundance curve for the called peaks (Fig. 2A). This saturation analysis showed that eight million aligned reads were sufficient to call the majority of binding peaks. From extrapolation of the curve, it could be deduced that doubling the number of sequencing reads would have increased the number of called peaks by about 20%. Fig. 2A shows that the vast majority of BACH1-bound genomic regions in HEK 293 cells has been detected here.
FIGURE 2.
Dynamic range of the BACH1 ChIP-seq peak calling, enrichment of peaks at genes and transcription start sites. A, rank abundance curve of the BACH1 peak calling was simulated by repeated peak calling using subpopulations of all mapped reads. The dynamic range of the curve demonstrated that most of the peaks occurring in vivo were detected by the experimental set up used here. B, BACH1-bound peaks were found significantly enriched nearby and within annotated RefSeq genes. C, BACH1 binding peaks near genes were located mainly in the 5-kb promoter region upstream of the TSSs and within intronic regions more than 15 kb downstream of the TSSs.
Annotation of Target Genes Associated with the BACH1-bound Regions
To identify the BACH1 target genes, we investigated the location of the 84 BACH1-bound regions relative to nearby genes. A total of 67 peaks were located within 15 kb of a gene (Fig. 2B), of which 28 peaks were in promoter regions or at the TSS, 32 were intragenic, and 7 were found downstream of genes. The remaining 17 peaks were located more than 15 kb away from any annotated genes. Nevertheless, 12 of those peaks showed conservation among placental mammals, suggesting that they could be functional. Indeed, in seven cases, the conserved region was associated with features evoking transcriptional activity, from either neighboring CpG islands (34), TSS sequencing (35), or expressed sequence tags. Four of these conserved peaks were located within 5 kb of RNA polymerase IIA-bound regions in HEK 293 cells, and two of these peaks showed putative exons defined by mRNA sequencing read clusters within 15 kb (36). Regarding the five intergenic peaks without mammalian conservation, we found expressed sequence tags defining nonannotated potential target genes for four of these, and nearby mRNA sequencing read clusters for two of these. Altogether, 67 BACH1 peaks were found associated with nearby genes, and 16 of 17 intergenic peaks were located in regions displaying features suggesting transcriptional activity.
We assigned the potential target genes to the 67 peaks close to genes and identified a total of 59 genes representing the primary targets of BACH1 in HEK 293 cells according to our ChIP-seq analysis (see supplemental Table 2). We found all of the previously known BACH1 target genes, including heme oxygenase 1 (HMOX1), the ferritin heavy and light chains (FTH1 and FTL), the NAD(P)H menadione oxidoreductase (NQO2), and the glutamate-cysteine ligase catalytic and modifier subunits (GCLC and GCLM), except for thioredoxin reductase 1, which was not bound by BACH1 in this cell line. Newly identified target genes involved in oxidation-reduction processes were the malic enzyme ME1, the aldolase ALDOA, and the transketolase TKT. We identified additional target genes involved in cellular transport processes (including the cationic amino acid transporter SLC7A11, the mitochondrial dicarboxylate transporter SLC25A10, the heme transporter SLC48A1, prosaposin, and the microtubule-associated proteins Tau and calsyntenin 1). Interestingly, we also identified a significant number of target genes involved in signal transduction (calmodulin 1 and COPS6), cell cycle regulation (CDK6, MAFG, EWSR1, and LRRC8D), and apoptosis (BCL2L11, sequestosome 1, RHBDD3, and the tumor necrosis factor receptor TNFRSF1A), indicating an important function of BACH1 at the crossroads of cellular redox control and cell cycle progression.
Three genes were found associated with more than one BACH1 binding peak (AFG3L1 with nine peaks and HMOX1 and SQSTM1 with two peaks each), whereas two peaks were located in the bidirectional promoters of ZNF3/COPS6 and RHBDD3/EWSR1. In addition to 55 protein-coding genes, we could annotate two noncoding genes (AFG3L1 and ANXA2P2) and two noncoding vault RNA genes (VTRNA1-1 and VTRNA1-2) as BACH1 targets. Except for two cases (KIAA1751 and MXRA8), all targets were associated with BACH1-binding motifs. We investigated the distance of the 67 BACH1 peaks near genes relative to the closest associated TSS (Fig. 2C) and found a preference for the peaks to reside within 5 kb upstream of a TSS. Another fraction of the gene-associated peaks (45%) were found far downstream (>15 kb) of the respective TSSs, showing that BACH1 binding is not only found in the promoter/TSS regions, but also inside genes.
We compared the target genes of BACH1 in HEK 293 with the recently described list of functional Nfe2l2 target genes in mouse embryonic fibroblasts (37). Of the 59 BACH1 target genes in the human cells, 15 genes were also functional Nrf2 targets in the mouse cells. Among those were genes involved in iron ion homeostasis (Hmox1, Fth1, and Slc48a1), redox regulation (Gclm, Gclc, Tkt, and Slc7a11), cell cycle/apoptosis regulation (Sqstm1, Itpr2, Mafg, Sema3e, and Tnfrsf1a), and intracellular transport (Clstn1 and Synj2), as well as Srcrb4d, a soluble member of the scavenger receptor family. The considerable overlap of BACH1 and Nrf2 target genes between human and mouse further validates the present data and shows the strong evolutionary conservation of the core BACH1-NFE2L2 regulatory network.
Gene Expression Changes after Knockdown of BACH1 by RNA Interference
We performed knockdown experiments of BACH1 in HEK 293 cells by means of RNA interference (RNAi) and interrogated the transcriptome response by hybridization on microarrays (Affymetrix U133Plus2) at 24 and 72 h post-transfection. We used three different types of silencing molecules as follows: one high complexity pool of 20–30 bp of siRNA-like molecules (esiRNA) and two independent synthetic small interfering RNA molecules without and with chemical modification (siRNA1 and siRNA2, respectively; see supplemental Methods for details). Three replicate RNAi experiments were performed for each silencing molecule, and four replicates were performed for the controls (mock transfections, see supplemental Methods). We minimized artifactual off-target effects by considering only common targets of at least two independent silencing molecules. Consequently, genes with significant expression changes were defined by changes of the same probes among at least two of three different RNAi experiments with an expression ratio >1.3 or ratio <0.75 and a p value <0.05 from t test. Besides, we ruled out the activation of the interferon response at both time points, based on the expression analysis of six canonical genes of this pathways (IFNB1, IRF9, IFITM1, MX1, OAS1, and OAS2), ensuring the integrity of the dataset.
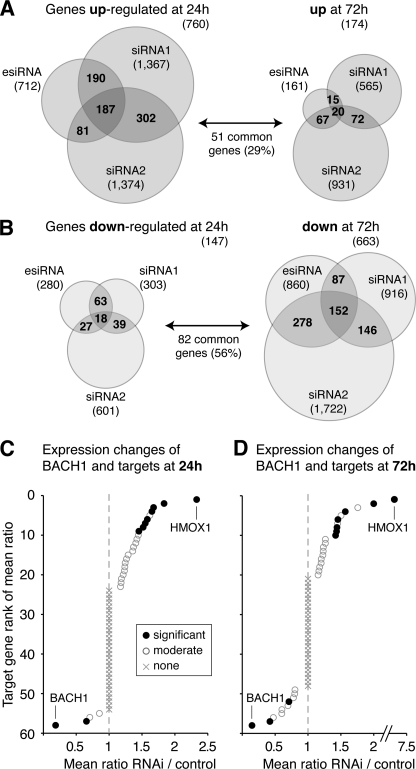
As expected, we observed a significant reduction of the BACH1 mRNA (down by 74–78%) with all three silencing molecules at both time points and a virtually complete knockdown of the BACH1 protein at 72 h (supplemental Fig. 4). Consistent with the view that BACH1 is mainly a transcriptional repressor, 760 genes were consistently up-regulated by at least two independent siRNA molecules at 24 h post-transfection (Fig. 3A and supplemental Table 3). Later on at 72 h post-transfection, a smaller number of 174 genes were significantly increased in expression (supplemental Table 4), of which 51 genes were common to those up-regulated earlier. Regarding significantly down-regulated genes, we identified 147 genes down at 24 h and 663 genes down at 72h post-transfection (Fig. 3B and supplemental Tables 5 and 6), with 82 genes common to both time points. We analyzed the genes with expression changes for enrichment in specific functional categories (Gene Ontology “biological process”) and found no significantly enriched categories for the sets of down-regulated genes after 24 and 72 h and for the up-regulated genes after 72 h. However, we observed significant enrichment of four categories of biological processes among the 760 genes with increased expression after 24 h, namely for genes involved in regulation of gene expression (167 genes, Benjamini-corrected p = 0.0016), protein catabolic processes (50 genes, p = 0.0039), negative regulation of cellular biosynthetic processes (47 genes, p = 0.0043), and regulation of macromolecule biosynthetic processes (160 genes, p = 0.0046). Additional information can be found in supplemental Table 7.
FIGURE 3.
Genome-wide expression changes after knockdown of BACH1 for 24 and 72 h. The BACH1 mRNA was knocked down by RNA interference in independent experiments using three different silencing molecules. Transcriptome changes after knockdown were monitored by hybridizations on Affymetrix GeneChip Human Genome U133 Plus 2.0 arrays, which were performed in triplicate for each siRNA together with mock transfections. A, at 24 h post-transfection, 760 genes were found significantly up-regulated with p < 0.05 and fold-change >1.3 (mean ratio RNAi/mock) for at least two of three siRNAs, depicted as overlap regions in the Venn diagram. This number decreased down to 174 genes found up-regulated after 72 h, of which 29% were common to those identified at the earlier time point. B, at 24 h post-transfection, 147 genes were found significantly down-regulated with p < 0.05 and fold-change <0.75 for at least two of three siRNAs, depicted as overlap regions in the Venn diagram. Of these genes, 56% were also identified among the larger set of 663 genes found down-regulated after 72 h. C, 59 target genes of BACH1 were sorted by descending mean expression ratio of RNAi/mock after BACH1 knockdown for 24 h. Eight target genes were found significantly up-regulated (see Table 1), among these the main BACH1 target HMOX1. The BACH1 and SLC7A11 mRNAs were found significantly down-regulated at this time point. D, 59 target genes of BACH1 were sorted by descending mean expression ratio of RNAi/mock after BACH1 knockdown for 72 h. Seven target genes showed a significant increase in expression (see Table 1). At this later time point, also the EWSR1 gene exhibited a decrease in expression, in addition to the down-regulated BACH1 and SLC7A11 genes.
A previous siRNA-based knockdown analysis of BACH1 in human skin keratinocyte-derived HaCaT cells identified only nine up-regulated genes (17), most probably due to the use of mouse oligonucleotides for microarray printing, which was sub-optimal for analyzing human RNAs. However, three genes were found in common with the knockdown performed here, namely HMOX1, STEAP3, and the interleukin 6 signal transducer (IL6ST). STEAP3 is the endosomal ferrireductase required for efficient iron uptake in erythroid cells (38). IL6ST (alias gp130) has a redox-regulatory role, as it participates in the cardioprotective IL-6 signaling via the JAK/STAT pathway following hypoxia/ischemia in the heart (39). The STEAP3 and IL6ST genes were not bound by BACH1 in HEK 293 cells.
To analyze the influence of BACH1 on the cell cycle, we inspected the gene expression changes after BACH1 knockdown for a panel of 87 genes with direct involvement in cell cycle processes, including cyclins, cyclin-dependent kinases, cyclin-dependent kinase inhibitors, retinoblastoma, and others (supplemental Table 8). We identified expression changes for 13 cell cycle genes. Intriguingly, six of these were cyclin genes involved in various phases of the cell cycle (namely the cyclins E2 and J at 24 h and the cyclins D2, G2, J, L1, and T2 at 72 h after BACH1 knockdown). All of these cyclins were found clearly down-regulated with fold-changes of 0.38–0.61. Other down-regulated cell cycle genes were the S-phase kinase-associated protein 2 (0.54-fold), BRCA2 (0.57-fold), and retinoblastoma (also 0.57-fold), whereas only four cell cycle genes were found moderately up-regulated (BIRC5, CDK2, CDK5R1, and CUL3). Taken together, these observations point toward an influence of BACH1 on the cell cycle.
Integration of the ChIP-seq and RNAi Data Sets
We inspected whether the direct BACH1-bound target genes showed expression changes after BACH1 knockdown, pointing out a direct transcriptional control of gene expression by BACH1 (Fig. 3, C and D). At 24 h post-transfection, eight BACH1-bound genes showed significantly increased expression, including the known target genes HMOX1, GCLM, and FTH1 (see Table 1 for the complete list). The strongest induction was seen for HMOX1, the major target of BACH1 (mean fold-change 2.33 ± 0.33), confirming previous observations that BACH1 acts as strong repressor of HMOX1. At this time point, only one target gene was significantly decreased in expression, namely the cystine/glutamate transporter SLC7A11, providing one example of a putative transcriptional activator role of BACH1. At 72 h post-transfection, we observed seven genes with increased expression (see Table 1), among them HMOX1 with much stronger induction (fold-change 7.19 ± 1.17). At this later time point, two BACH1 target genes were significantly decreased in expression, namely SLC7A11 and the multifunctional Ewing sarcoma breakpoint region 1 gene (EWSR1) involved in cell cycle regulation. Altogether, the microarray-based transcriptome analysis of BACH1 knockdown showed significant expression changes for 13 of 59 direct target genes. This is highly significant; the probability to find this or a higher number of genes with expression changes by random sampling is p = 0.0005.
TABLE 1.
List of the functional target genes of BACH1 in HEK 293 cells and the correlation of BACH1 binding with associated gene expression changes
| Gene symbol | Gene name | Biological process | ChIP-seq normalized reads (RPKM)a | RNAi/control mean ratio at 24 h ± S.E.b | RNAi/control mean ratio at 72 h ± S.E.b |
|---|---|---|---|---|---|
| Genes with lowered expression after BACH1 knockdown | |||||
| SLC7A11 | Solute carrier family 7, member 11 | Oxidation reduction | 7.8 | 0.66 ± 0.06 | 0.42 ± 0.03 |
| VTRNA1–1 | Vault RNA 1–1 | Unknown | 14.7 | NA | 0.49 ± 0.05 |
| EWSR1 | Ewing sarcoma breakpoint region 1 | Cell cycle regulation | 8.6 | NS | 0.71 ± 0.05 |
| Genes with elevated expression after BACH1 knockdown | |||||
| HMOX1 | Heme oxygenase (decycling) 1 | Iron ion homeostasis | 103.8 | 2.33 ± 0.33 | 7.19 ± 1.17 |
| GCLM | Glutamate-cysteine ligase, modifier subunit | Oxidation reduction | 31.1 | 1.65 ± 0.06 | 2.00 ± 0.23 |
| TTC23 | Tetratricopeptide repeat domain 23 | Unknown | 7.8 | 1.84 ± 0.41 | NS |
| FTH1 | Ferritin, heavy polypeptide 1 | Iron ion homeostasis | 21.7 | 1.68 ± 0.13 | NS |
| SQSTM1 | Sequestosome 1 | Apoptosis | 10.4 | 1.58 ± 0.10 | 1.42 ± 0.08 |
| TPR2 | Inositol 1,4,5-triphosphate receptor, type 2 | Response to hypoxia | 14.3 | NS | 1.57 ± 0.16 |
| CALM1 | Calmodulin 1 (phosphorylase kinase, δ) | Signal transduction | 9.4 | 1.55 ± 0.11 | 1.43 ± 0.09 |
| TFE3 | Transcription factor binding to IGHM enhancer 3 | Cell differentiation | 9.0 | 1.52 ± 0.07 | NS |
| ME1 | Malic enzyme 1, NADP(+)-dependent, cytosolic | Oxidation reduction | 6.9 | NS | 1.45 ± 0.11 |
| AFG3L1 | AFG3 ATPase family gene 3-like 1 (Saccharomyces cerevisiae) | Unknown | 16.0 | 1.45 ± 0.11 | NS |
| ANXA2P2 | Annexin A2 pseudogene 2 | Unknown | 9.0 | NS | 1.44 ± 0.13 |
| r (Pearson correlation coefficient for correlation of RPKM and RNAi ratio)c | 0.884 | 0.990 | |||
| RS (Spearman's rank correlation coefficient for correlation of RPKM and RNAi ratio)d | 0.333 | 0.643 | |||
a Reads were quantified in RPKM values as described previously (29).
b The S.E. was calculated as the root of the mean squared S.E. of the individual RNAi experiments (NA means not assessed; NS means not significant).
c The Pearson product-moment correlation coefficient was used as parametric measure of statistical dependence to assess how well the relationship can be described using a linear function. The expression ratios for genes with lowered expression were not included in the tests.
d Spearman's rank correlation coefficient was used as a nonparametric measure of statistical dependence to assess how well the relationship can be described using a monotonic function. The expression ratios for genes with lowered expression were not included in the tests.
We also assessed the expression changes of two noncoding vault RNA genes (VTRNA1-1 and VTRNA1-2), which were identified as BACH1 target genes. Those were not represented on the Affymetrix U133Plus2 arrays and were tested by quantitative RT-PCR. We could show that VTRNA1-2 expression was reduced in one siRNA experiment, whereas VTRNA1-1 expression was significantly reduced by two different siRNA molecules (Table 1 and supplemental Fig. 5). We observed another 23 BACH1 target genes with weaker changes in expression (1.15 to 1.3-fold for up-regulated genes and 0.87 to 0.75-fold for down-regulated genes) or only one siRNA molecule resulting in significant fold-change of <0.75 or >1.3 (see supplemental Table 2), which represent an interesting set for future analyses of an extended BACH1 regulatory network.
Quantitative Impact of BACH1 Binding on Gene Expression
To quantify the impact of BACH1 binding on target gene expression, we investigated the gene expression changes and their possible relationships with the location of BACH1-binding sites, the number of BACH1-binding motifs, and the strength of BACH1 binding. No significant enrichment of expression changes was found for any specific location of binding sites upstream, inside, or downstream of the target genes (results not shown). Regarding the number of BACH1-binding motifs, we observed that the two genes with exceptionally high numbers of motifs (AFG3L1 with 36 motifs and HMOX1 with seven motifs) were both significantly up-regulated. Apart from this, no further correlation was present between the number of binding motifs and gene expression changes. However, we identified a putative weak correlation between BACH1 binding strength (ChIP-seq reads normalized to reads per kilobase and million mapped reads, RPKM) and the expression changes of the target genes with significant increase in expression after knockdown, as shown in Table 1. The Pearson correlation coefficients between RPKM values and expression ratios observed for the up-regulated target genes were r = 0.884 and r = 0.990 for the 24- and 72-h time points, respectively. These coefficients are skewed due to the large RPKM and expression ratios for the strongest target gene, HMOX1. As a more robust statistical measure, we used Spearman's rank correlation coefficients, which were calculated as RS = 0.333 and RS = 0.643 for 24 h and 72 h, respectively, indicating a putative correlation between BACH1 binding and expression changes of the direct target genes (see supplemental Fig. 6). Because these coefficients were derived from relatively few data points, we do not regard this as a sound correlation.
Secondary Effects Mediated by Co-regulatory Transcription Factors
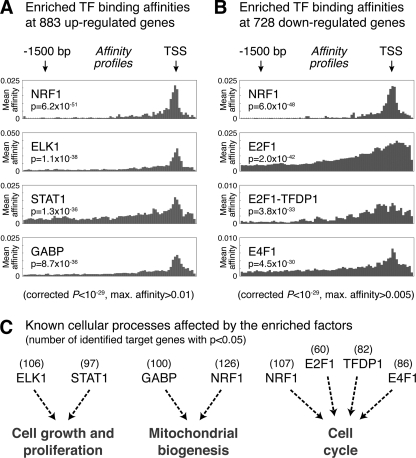
Despite the measurable impact of BACH1 on target gene expression, only a relatively small fraction of genome-wide expression changes after BACH1 knockdown can be explained by disruption of the BACH1 repressive function at direct target genes. To assess the other indirect effects on genes expression following knockdown, we analyzed the promoter regions of the up- and down-regulated genes for enrichment of specific transcription factor binding sites. Up-regulation was observed for 883 genes (760 genes after 24 h and 174 genes after 72 h, with 51 genes common to both time points), and down-regulation was observed for 728 genes (147 genes after 24 h and 663 genes after 72 h, with 82 genes common to both time points). We retrieved the RefSeq identifiers and transcription start sites for the associated transcripts and used the obtained promoter sequences for transcription factor affinity prediction. For this, we applied a physical binding model (30) to predict the relative binding affinities of the 554 vertebrate transcription factor binding matrices in the TransFac data base (version 12.1) to 2-kb promoters spanning the regions −1,800 bp upstream to 200 bp downstream of each TSS (see supplemental Methods for details). Fig. 4 shows the promoter affinity profiles for the top-scoring transcription factors with significantly enriched affinity (p value <10−29) to the promoters of the two gene sets, as compared with a human promoter background model.
FIGURE 4.
Top predicted transcription factor binding affinities in the promoters of genes with expression changes after BACH1 knockdown. We used two sets of promoter sequences for transcription factor motif analysis as follows: one set from all 883 genes with increased expression after BACH1 knockdown (24 and 72 h combined) and the other set from all 728 genes with decreased expression (also at both time points). The prediction of transcription factor binding was done using 554 binding matrices in TransFac (version 12.1) and a human promoter-based background model. The individual sequences stretched from −1800 to +200 bp respective to each TSS. p values for the individual sequences were combined by Fisher's method and multiple test-corrected according to Benjamini-Hochberg, giving a natural ranking of the transcription factors that have the most enriched binding within the whole sequence sets. A, promoters of 883 up-regulated genes were significantly enriched for binding motifs of NRF1, ELK1, STAT1, and GABP. B, promoters of 728 down-regulated genes were significantly enriched for binding motifs of NRF1, E2F1, the E2F1-TFDP1 complex, and E4F1. C, enriched transcription factors among up-regulated genes are known to be involved in the regulation of cell growth and proliferation (ELK1 and STAT1) and in mitochondrial biogenesis (GABP and NRF1). The enriched transcription factors among down-regulated genes (NRF1, E2F1, TFDP1, and E4F1) are all known to be involved in the regulation of the cell cycle.
The promoters of the 883 genes with increased expression after knockdown showed enriched binding affinities for NRF1, ELK1, STAT1, and GABP (Fig. 4A). The identified transcription factors, except for STAT1, exhibited a strong binding preference for the regions around the TSS. The NRF1 protein was also identified as the top factor with enriched binding affinity for the promoters of the 728 down-regulated genes (Fig. 4B), together with the E2F factors E2F1 and E4F1 as well as the E2F1-TFDP1 complex. The latter transcription factors generally exhibited broader affinity profiles than those identified for the up-regulated genes, with less preference for binding at the TSS. The identification of the MARE-binding factor NRF1 as top-ranking factor in both data sets shows the enrichment of MARE-containing promoters among all genes with expression changes, confirming the usefulness of the transcription factor affinity prediction.
Interestingly, the cellular processes known to be affected by the enriched transcription factors support a biological role of BACH1 in redox regulation and cell cycle control, as shown in Fig. 4C. Two factors found enriched at up-regulated genes are ELK1, a part of the MAPK signaling pathway, and STAT1, a main player in JAK-STAT signaling. Intriguingly, ELK1 has been shown during cellular stress to induce expression of the heme-regulated inhibitor (HRI), leading to inhibition of protein synthesis (40). This interconnection of cellular responses to heme and cell growth is further supported by the identification of STAT1, which is involved in the induction of heme oxygenase 1 expression following transduction of extracellular signals controlling cell growth and proliferation (41). Two other factors with enriched affinity for the promoters of up-regulated genes are GABP and NRF1, which both are well known master regulators of mitochondrial biogenesis (42, 43), thus supporting the BACH1 function in cellular responses to oxidative stress.
As shown in Fig. 4C, the transcription factors found significantly enriched in the set of 728 down-regulated genes have all been previously involved in regulatory networks controlling cell cycle progression. NRF1 and E2F factors are known to co-regulate sets of gene regulatory networks involved not only in mitochondrial biogenesis and metabolism but also cell cycle control (44). E2F1, as well as the other members of the E2F family, bind to TFDP1 and regulate the expression of genes that are required for passage through the cell cycle (45). The finding that the promoters of the down-regulated genes are enriched for binding of these factors further suggests an influence of BACH1 on the cell cycle. Our expression analysis data showed that cyclin E2 is down-regulated after BACH1 knockdown, suggesting a possible mechanism of E2F induction, because E2F activity can be directly regulated by cyclin E-CDK2 (46). Moreover, we also found among the enriched factors the atypical ubiquitin E3 ligase E4F1, a key post-translational regulator of p53 inducing cell cycle arrest but not apoptosis (47). This finding is in line with a report that BACH1 can be recruited to a subset of p53 target genes and contribute there to inhibit p53-dependent senescence (9). Taken together, the secondary effects of BACH1 knockdown are involving a number of transcription factors that can trigger concerted cellular responses influencing metabolism, mitochondrial biogenesis, cell growth, and cell cycle progression.
DISCUSSION
In this study, the genome-wide characterization of BACH1 target regions in HEK 293 cells resulted in the sensitive and specific detection of 84 BACH1-bound genomic regions. Our data suggests that BACH1 acts as a relatively specific transcription factor, as judged by the small number of target regions compared with other bZIP transcription factors for which up to several thousands of peaks have been observed (18, 37, 48).
Selectivity and Cooperativity of Genomic BACH1 Binding
The BACH1 DNA-binding motif identified here extends the previously known TGACTCA motif by the three bases GCA. The known motifs of the competitors of BACH1, namely NRF1 and NFE2L2, also contain these GCA bases, but with a smaller weight than the BACH1 motif. We suggest that the previously observed dominance of BACH1 repressor activity over NFE2L2 activator activity (1) is due to a stronger binding affinity of BACH1 at its target genes, effectively keeping target gene expression at low levels under normal conditions. Nine BACH1-bound regions harbored two or more BACH1 motifs located 7 or 21 bp away from each other, supporting the functionality of a recently discovered BACH1 homodimer interaction surface (49). In line with this observation, the two BACH1 target genes with the highest number of ChIP-seq reads were both found associated with several adjacent BACH1 motifs, namely HMOX1 (seven motifs) and AFG3L1 (36 motifs). HMOX1 also showed the strongest observed expression changes after BACH1 knockdown, whereas AFG3L1 was only moderately dysregulated.
Location and Impact of BACH1 Binding to Target Genes
Our analysis of the confirmed functional BACH1 targets, defined by the combination of BACH1 binding and expression changes after BACH1 knockdown, indicated that most active targets harbor the binding sites in their promoter region. Nevertheless, we observed several cases where functional binding sites were either intragenic or downstream of the nearest gene. An interesting subset of 17 BACH1 target regions was found located in genomic regions devoid of any annotated gene. Nevertheless, several of these sites showed sequence conservation among mammals, CpG islands, or expressed sequence tags suggesting potential transcriptional activity. Additional BACH1 functions may emerge from in-depth analysis of these anonymous binding sites.
Our approach identified all previously known BACH1 target genes except for the thioredoxin reductase 1 (7) and the NAD(P)H dehydrogenase quinone 1 (6), which exhibited no BACH1 binding in HEK 293 cells. In addition, we could annotate 50 novel BACH1 target genes. A highly significant fraction of the BACH1 target genes (14/59) showed expression changes after BACH1 depletion. Two of the functional BACH1 target genes (SLC7A11 and EWSR1) showed reduced expression after BACH1 knockdown, suggesting a potential activating role of BACH1. We observed no significant up- or down-regulation for the remaining 45 direct targets, although several of these genes showed a strong BACH1 binding (supplemental Fig. 7). Here, the repressive effect of BACH1 may not be completely alleviated by the knockdown in our experiments, leaving the possibility that the remaining BACH1 molecules could still exert repressive effects. Also, BACH1 binding may be present only in a subpopulation of the cells, so that associated gene expression changes are diluted among the entire population, where microarrays are not sensitive enough to detect subtle changes. In addition, tissue- or cell type-specific effects might render these targets nonfunctional in HEK 293 cells, although they might be active in a more complex structure such as the tissue in an organism in response to additional stimuli.
Genome-wide Effects of BACH1 on Transcription
As expected, the majority of genes with expression changes after 24 h of BACH1 knockdown showed increased expression, confirming previous observations that BACH1 acts mainly as a transcriptional repressor (8, 17). Interestingly, only 1% of these increases in expression could be attributed to direct BACH1 binding. The repressive effects of BACH1 appear to be multiplied through one or several of its direct target genes encoding transcription factors (for example TFE3) or signaling proteins (such as SQSTM1, ITPR2, and CALM1) mediating downstream effects. Although we are still missing conclusive hints how this repression could proceed in detail, we were able to pinpoint a number of potential secondary transcription factors with impact on mitochondrial biogenesis (GABP and NRF1) as well as cell growth and proliferation (ELK1 and STAT1) that could mediate the observed transcriptome changes at 24 h after knockdown.
Somewhat unexpectedly, the transcriptome changes at a later time point, 72 h after BACH1 knockdown, showed mainly a reduction in gene expression. The transcription factors that may convey these secondary effects, determined by binding site enrichment analysis, are known players in regulation of the cell cycle, including the BACH1-related transcription factor NRF1 as well as E2F1 and E4F1. Also, five cyclins were found down-regulated at this later time point (cyclin D2, G2, J, L1, and T2), with potential effects on the transcription of other genes. For example, the cyclins T1/T2 play a role in transcriptional elongation as part of the elongation factor P-TEFb (50), so that down-regulation of cyclin T2 can result in reduced expression of other genes. In addition, changes in the cellular redox state consecutive to the BACH1 knockdown can be interpreted as input for molecules sensing the redox state and conveying signals to the gene expression machinery.
Impact of BACH1 Binding on Cellular Regulatory Networks
The genes both bound by BACH1 and showing expression changes after BACH1 knockdown represent the functional direct BACH1 targets in HEK 293 cells. Fig. 5 shows an overview of different but interconnected cellular functions under the control of BACH1 repression, including cellular balance of heme, oxidative stress response, regulation of cell cycle progression, apoptosis, and cellular transport processes. Heme binding abrogates BACH1 binding to MAREs (Fig. 5A), inducing nuclear export of BACH1 (3), sequestration at microtubules (4), and polyubiquitination followed by proteasome-mediated degradation (5). Consequently, NFE2L2 can occupy the MAREs, resulting in an increase of target gene expression.
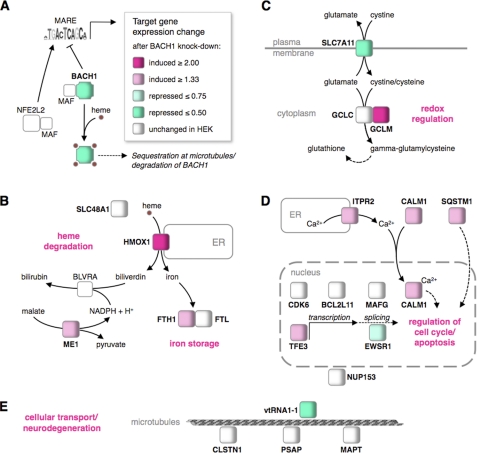
FIGURE 5.
Cellular metabolic and signaling networks affected by BACH1 target genes. A, upon heme binding, BACH1 is exported from the nucleus, sequestered at microtubules, and finally degraded. Subsequently, NFE2L2:MAF can bind and induce gene expression changes at BACH1 target genes. Knockdown of BACH1 recapitulates this process. B, increased expression of HMOX1 and ferritin heavy chain (FTH1) enhance heme degradation and iron storage, whereas the up-regulation of malic enzyme 1 (ME1) may provide reduction equivalents for biliverdin reductase. The endosomal heme transporter SLC48A1 and the ferritin light chain (FTL) remained unchanged in HEK cells. C, down-regulation of the glutamate-cystine antiporter SLC7A11 and up-regulation of the glutamate-cysteine ligase modifier subunit (GCLM) can change the cellular redox state. The glutamate-cysteine ligase catalytic subunit (GCLC) remained unchanged in HEK cells. D, up-regulation of the inositol 1,4,5-trisphosphate receptor (ITPR2) can elevate cytoplasmic calcium concentration, facilitating nuclear import of calmodulin (CALM1), with potential impact on cell cycle regulation and apoptosis. The up-regulated sequestosome 1 (SQSTM1) and the down-regulated Ewing sarcoma breakpoint region 1 (EWSR1) encode multifunctional signaling and adapter proteins that can also influence the cell cycle, as can the up-regulated transcription factor TFE3. The cyclin-dependent kinase CDK6, the apoptosis facilitator BCL2L11, and the oncogene MAFG remained unchanged in HEK cells. E, down-regulated vault RNA VTRNA1-1 is involved in cellular transport along microtubules, as are calsyntenin 1 (CLSTN1), prosaposin (PSAP), and the microtubule-associated protein Tau (MAPT), which remained unchanged in HEK cells. Also unchanged was another BACH1 target, the nucleoporin NUP153.
Control of Heme Degradation by BACH1
Regarding genes involved in the heme degradation pathway, we found five direct BACH1 targets (Fig. 5B), of which three showed increased expression after BACH1 knockdown. The induction of heme oxygenase 1 (HMOX1) expression leads to increased heme degradation forming biliverdin, iron, and carbon monoxide. The iron that is set free by this reaction can be stored by ferritin. Both the ferritin heavy chain (FTH1) and light chain (FTL) genes were found to be BACH1 targets, with elevated FTH1 expression after BACH1 knockdown. The subsequent conversion of biliverdin to bilirubin by the biliverdin reductase A (BLVRA) requires NADPH, as does the degradation of heme to biliverdin. A major source of these reduction equivalents in the cell is the BACH1 target gene malic enzyme 1 (51), which was up-regulated after BACH1 knockdown. The fifth BACH1 target gene involved in heme metabolism is the heme transporter SLC48A1, which showed no significant expression changes after BACH1 knockdown in HEK 293.
BACH1 and the Oxidative Stress Response
Regarding oxidative stress response pathways, we identified three direct BACH1 target genes (Fig. 5C), of which two showed marked expression changes after BACH1 knockdown. The decreased expression of the cystine/glutamate antiporter SLC7A11 can lower the cellular uptake of oxidized cystine, whereas reduced cysteine can still be taken up by the cells via the ubiquitous ASC transport system (52). Thus, the intracellular level of cysteine, which is important for glutathione synthesis, can be elevated. Accordingly, SLC7A11 is involved in glutathione-dependent neuroprotection from oxidative stress (53). Two more BACH1 targets involved in the oxidative stress response are the GCLC and GCLM subunits, which together catalyze the first rate-limiting step during glutathione synthesis. We found GCLM strongly up-regulated after BACH1 knockdown, leading to increased synthesis of glutathione, the most abundant antioxidant in the cell and a crucial factor in the response to oxidative stress and aging. Two additional redox-regulatory BACH1 target genes, for which we observed no expression changes after BACH1 knockdown (the NAD(P)H quinone oxidoreductase NQO2 and the mitochondrial dicarboxylate transporter SLC25A10), are not shown in Fig. 5C.
Potential Impact of BACH1 on Cell Cycle Progression and Apoptosis
Our analysis revealed that eight BACH1 target genes are involved in control of cell cycle progression and apoptosis (Fig. 5D), with five of these genes showing expression changes after BACH1 knockdown. Increased expression of the inositol 1,4,5-triphosphate receptor type 2 (ITPR2), one of the main regulators of intracellular calcium concentration with an important role in apoptosis, can lead to an elevated calcium efflux from the endoplasmic reticulum. The calcium-binding protein calmodulin (CALM1), also up-regulated after knockdown, is activated by calcium binding and can subsequently promote cell cycle progression for example via calcium/calmodulin-dependent protein kinases (54) or via calmodulin binding to cyclin E1/CDK2 (55). As mentioned above, we observed a marked decrease of cyclin E2 transcript levels following BACH1 knockdown, indicating that BACH1 influences the expression of members of the cyclin E family. The transcription factor TFE3, another BACH1 target with increased expression after knockdown, could mediate this BACH1 action in cell growth and proliferation through its ability to directly regulate cyclin E expression (56). Moreover, the BACH1 target Ewing sarcoma breakpoint region 1 (EWSR1), a multifunctional RNA-binding protein, can regulate the ratio of cyclin D1a and D1b transcripts by increasing the transcription elongation rate in EwSa cells (57), and we found decreased EWSR1 expression after BACH1 knockdown.
Finally, we observed increased expression of the BACH1 target sequestosome 1 (SQSTM1), a multifunctional ubiquitin-binding protein that serves as a storage place for ubiquitinated proteins (58) and can regulate activation of the NFκB signaling pathway inducing apoptosis (59). Other direct targets of BACH1 involved in cell cycling and apoptosis, but showing no significant expression changes in HEK 293, were the cyclin-dependent kinase 6 (CDK6), the BCL2-like 11 apoptosis facilitator (BCL2L11), and the v-maf MAFG.
BACH1 Targets Involved in Cellular Transport Processes and Neurodegeneration
After heme binding and nuclear export, BACH1 has been reported to dynamically bind to the hyaluronan-mediated motility receptor (4), suggesting an influence of BACH1 on cellular transport processes. Interestingly, we identified four BACH1 target genes involved in subcellular transport (CLSTN1, PSAP, MAPT, and NUP153, see Fig. 5E), none of which showed expression changes after BACH1 knockdown in HEK 293 cells. Calsyntenin 1 (CLSTN1), a calcium-binding protein present in vesicles transiting to neuronal growth cones, can interact with kinesin-1 to regulate transport of cellular vesicles along microtubule tracks (60). Interestingly, we also identified the microtubule-associated protein Tau (MAPT) as a direct BACH1 target, which has been intensely studied for its involvement in the stabilization of microtubules and Tau-mediated neuro-degeneration in Alzheimer disease (61). The other BACH1 target genes involved in cellular transport processes are PSAP, especially abundant in the nervous system, and nucleoporin 153 kDa (NUP153), a key component of the nuclear pore complex mediating the regulated movement of macromolecules between the nucleus and cytoplasm. NUP153 was shown to be mislocated following oxidative stress (62).
Noncoding RNAs as Targets of BACH1
In addition to 55 protein-coding BACH1 target genes, we found four noncoding genes with BACH1-binding sites, namely two vault RNA genes (VTRNA1-1 and VTRNA1-2) and two noncoding RNA genes (ANXA2P2 and AFG3L1). The vault VTRNA1-1 identified as direct BACH1 target showed significantly reduced expression after BACH1 knockdown (Table 1 and supplemental Fig. 5). Because the early identification of vaults as particles mediating multidrug resistance in cancer cells (63), these highly conserved ribonucleoprotein particles with a hollow barrel-like structure have been under investigation for their cellular functions. As shown in Fig. 5E, vault particles have been reported to be associated with nucleopores and to travel along microtubules (64, 65), suggesting that vaults function as specific cargo transporters. Vault particles form complexes with hypoxia-induced factor-1 (HIF-1) and can promote the ubiquitination and degradation of HIF-1 induced by hypoxia (66), a finding that links the largely unknown function of vault particles with the redox regulatory network controlled by BACH1. Together with the recently evolved notion that vaults promote cell survival through resistance to apoptosis (67, 68), the binding and regulation of vault RNAs by BACH1 open an interesting area for further studies.
The two other BACH1 targets annotated as noncoding genes were the annexin A2 pseudogene 2 (ANXA2P2) and the AFG3 ATPase family gene 3-like 1 (AFG3L1). Our RNAi experiments showed significantly increased expression of both AFG3L1 and ANXA2P2 transcripts after BACH1 knockdown, demonstrating that these genes are transcribed in HEK 293 and repressed by BACH1. Although the function of ANXA2P2 remains elusive, the ATP-dependent mouse ortholog Afg3l1 has been identified as a subunit of the mitochondrial m-AAA protease that is involved in the balance between maintenance or fragmentation and removal of damaged mitochondria under stress conditions (69). However, the human transcript of AFG3L1 does not seem to be translated into a protein (70), but it may have acquired other post-transcriptional regulatory functions, which are partly under control of BACH1.
Potential Involvement of BACH1 in Pathogenic Mechanisms
From a disease-related point of view, our study revealed PSAP and MAPT as direct BACH1 target genes, both of which have been identified as potential biomarkers for the detection of Alzheimer disease (10, 71). Also, the CLSTN1 gene was identified as BACH1 target, which encodes the Alzheimer-related cadherin-like protein calsyntenin 1. Interactions of CLSTN1 with kinesin-1 can block the transport of amyloid precursor protein-containing vesicles and increase β-amyloid generation, which may promote aberrant amyloid precursor protein metabolism in Alzheimer disease (72). We observed no significant expression changes for CLSTN1, PSAP, and MAPT after BACH1 knockdown in HEK 293 cells, pointing to context-dependent mechanisms for BACH1 repression of these genes, for example in neuronal cells. Here, further studies may unveil age-related changes in BACH1-dependent cellular responses to oxidative stress, potentially resulting in pathological overactivation and aberrant function of these target genes in neurodegenerative diseases.
Other aspects of BACH1-mediated gene repression are currently emerging in the field of cancer research. Several recent reports have linked BACH1 to viral infection-associated cancerogenesis via microRNA-mediated down-regulation of BACH1 expression. For example, the Kaposi sarcoma-associated herpesvirus encodes miR-K12-11, a microRNA suppressing BACH1 in infected macrophages and endothelial cells, protecting these cells from apoptotic death in oxidative stress environments (12). Somewhat similarly, BACH1 is down-regulated in lymphocytes infected by Epstein-Barr virus by the virus-induced miR-155 (13). Overexpression of miR-155 has been observed in several types of B-cell lymphomas and induces B-cell malignancies in transgenic mice (73). Down-regulation of BACH1 in human acute myeloid leukemia cells decreases the cytotoxic effect of the anticancer drug cytosine arabinoside, rendering functional up-regulation of BACH1 a potential strategy for antileukemic therapy (11). In this respect, we have identified here two noncoding vault RNAs as BACH1 targets whose expression has been linked to Mitoxantrone resistance in human malignant cells (74). Further studies have to be carried out on these and other disease-relevant BACH1 targets.
The identification of the functional BACH1 target genes in HEK 293 cells provides a strong basis for future dissection of the roles of BACH1 in gene regulation. The combination of genome-wide BACH1 binding data with RNAi-mediated knockdown allowed for the identification of the global transcriptional regulatory networks targeted by BACH1. Further studies will be needed to elucidate all aspects of BACH1-mediated gene repression, focusing also on the currently emerging involvement of BACH1 in the fields of neurodegeneration and cancerogenesis.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of an Escherichia coli RNase III clone from Dr. Frank Buchholz (Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstrasse 108, 01307 Dresden, Germany). We also thank Dr. Ralf Herwig for advice during microarray analysis.
Note Added in Proof
Please note that the transcription factor NRF1 (nuclear respiratory factor 1) identified by motif enrichment analysis in promoters is not identical with the ARE-binding transcription factor NFE2L1 (which is also often called NRF1).
This work was supported by the Max Planck Society (Munich, Germany) and the European Commission under its Sixth Framework Programme Grant AnEUploidy LSHG-CT-2006-037627.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Figs. 1–7, Tables 1–8, and additional references.
The genome-wide binding data and gene expression data reported in this paper can be accessed through the NCBI Gene Expression Omnibus (GEO) database under accession number GSE28053.
- MARE
- MAF recognition element
- ChIP-seq
- chromatin immunoprecipitation sequencing
- esiRNA
- endoribonuclease-prepared siRNA
- RPKM
- reads per kilobase and million reads
- TSS
- transcription start site
- GCLC
- glutamate-cysteine ligase catalytic
- GCLM
- glutamate-cysteine ligase modifier
- MAFG
- musculoaponeurotic fibrosarcoma oncogene homolog G
- MAPT
- microtubule-associated protein Tau
- PSAP
- prosaposin.
REFERENCES
- 1. Sun J., Hoshino H., Takaku K., Nakajima O., Muto A., Suzuki H., Tashiro S., Takahashi S., Shibahara S., Alam J., Taketo M. M., Yamamoto M., Igarashi K. (2002) EMBO J. 21, 5216–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hira S., Tomita T., Matsui T., Igarashi K., Ikeda-Saito M. (2007) IUBMB Life 59, 542–551 [DOI] [PubMed] [Google Scholar]
- 3. Suzuki H., Tashiro S., Hira S., Sun J., Yamazaki C., Zenke Y., Ikeda-Saito M., Yoshida M., Igarashi K. (2004) EMBO J. 23, 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamasaki C., Tashiro S., Nishito Y., Sueda T., Igarashi K. (2005) J. Biochem. 137, 287–296 [DOI] [PubMed] [Google Scholar]
- 5. Zenke-Kawasaki Y., Dohi Y., Katoh Y., Ikura T., Ikura M., Asahara T., Tokunaga F., Iwai K., Igarashi K. (2007) Mol. Cell. Biol. 27, 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dhakshinamoorthy S., Jain A. K., Bloom D. A., Jaiswal A. K. (2005) J. Biol. Chem. 280, 16891–16900 [DOI] [PubMed] [Google Scholar]
- 7. Hintze K. J., Katoh Y., Igarashi K., Theil E. C. (2007) J. Biol. Chem. 282, 34365–34371 [DOI] [PubMed] [Google Scholar]
- 8. MacLeod A. K., McMahon M., Plummer S. M., Higgins L. G., Penning T. M., Igarashi K., Hayes J. D. (2009) Carcinogenesis 30, 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dohi Y., Ikura T., Hoshikawa Y., Katoh Y., Ota K., Nakanome A., Muto A., Omura S., Ohta T., Ito A., Yoshida M., Noda T., Igarashi K. (2008) Nat. Struct. Mol. Biol. 15, 1246–1254 [DOI] [PubMed] [Google Scholar]
- 10. Mueller C., Zhou W., Vanmeter A., Heiby M., Magaki S., Ross M. M., Espina V., Schrag M., Dickson C., Liotta L. A., Kirsch W. M. (2010) J. Alzheimers Dis. 19, 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyazaki T., Kirino Y., Takeno M., Samukawa S., Hama M., Tanaka M., Yamaji S., Ueda A., Tomita N., Fujita H., Ishigatsubo Y. (2010) Cancer Sci. 101, 1409–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qin Z., Freitas E., Sullivan R., Mohan S., Bacelieri R., Branch D., Romano M., Kearney P., Oates J., Plaisance K., Renne R., Kaleeba J., Parsons C. (2010) PLoS Pathog. 6, e1000742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yin Q., McBride J., Fewell C., Lacey M., Wang X., Lin Z., Cameron J., Flemington E. K. (2008) J. Virol. 82, 5295–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun J., Brand M., Zenke Y., Tashiro S., Groudine M., Igarashi K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Copple I. M., Lister A., Obeng A. D., Kitteringham N. R., Jenkins R. E., Layfield R., Foster B. J., Goldring C. E., Park B. K. (2010) J. Biol. Chem. 285, 16782–16788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okada S., Muto A., Ogawa E., Nakanome A., Katoh Y., Ikawa S., Aiba S., Igarashi K., Okuyama R. (2010) J. Biol. Chem. 285, 23581–23589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reichard J. F., Sartor M. A., Puga A. (2008) J. Biol. Chem. 283, 22363–22370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt D., Wilson M. D., Spyrou C., Brown G. D., Hadfield J., Odom D. T. (2009) Methods 48, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Valouev A., Johnson D. S., Sundquist A., Medina C., Anton E., Batzoglou S., Myers R. M., Sidow A. (2008) Nat. Methods 5, 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bailey T. L., Elkan C. (1994) Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 [PubMed] [Google Scholar]
- 21. Gupta S., Stamatoyannopoulos J. A., Bailey T. L., Noble W. S. (2007) Genome Biol. 8, R24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bailey T. L., Gribskov M. (1998) Bioinformatics 14, 48–54 [DOI] [PubMed] [Google Scholar]
- 23. Taylor J., Schenck I., Blankenberg D., Nekrutenko A. (2007) Curr. Protoc. Bioinformatics, Chapter 10, Unit 10.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ji X., Li W., Song J., Wei L., Liu X. S. (2006) Nucleic Acids Res. 34, W551–W554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ji H., Jiang H., Ma W., Johnson D. S., Myers R. M., Wong W. H. (2008) Nat. Biotechnol. 26, 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A. J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J. Y., Zhang J. (2004) Genome Biol. 5, R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3 [PubMed] [Google Scholar]
- 28. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 29. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 30. Roider H. G., Kanhere A., Manke T., Vingron M. (2007) Bioinformatics 23, 134–141 [DOI] [PubMed] [Google Scholar]
- 31. Warnatz H. J., Querfurth R., Guerasimova A., Cheng X., Haas S. A., Hufton A. L., Manke T., Vanhecke D., Nietfeld W., Vingron M., Janitz M., Lehrach H., Yaspo M. L. (2010) Nucleic Acids Res. 38, 6112–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kanezaki R., Toki T., Yokoyama M., Yomogida K., Sugiyama K., Yamamoto M., Igarashi K., Ito E. (2001) J. Biol. Chem. 276, 7278–7284 [DOI] [PubMed] [Google Scholar]
- 33. Igarashi K., Kataoka K., Itoh K., Hayashi N., Nishizawa M., Yamamoto M. (1994) Nature 367, 568–572 [DOI] [PubMed] [Google Scholar]
- 34. Bock C., Walter J., Paulsen M., Lengauer T. (2007) PLoS Comput. Biol. 3, e110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamashita R., Wakaguri H., Sugano S., Suzuki Y., Nakai K. (2010) Nucleic Acids Res. 38, D98–D104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sultan M., Schulz M. H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D., Schmidt D., O'Keeffe S., Haas S., Vingron M., Lehrach H., Yaspo M. L. (2008) Science 321, 956–960 [DOI] [PubMed] [Google Scholar]
- 37. Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T. W., Wasserman W. W., Biswal S. (2010) Nucleic Acids Res. 38, 5718–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ohgami R. S., Campagna D. R., Greer E. L., Antiochos B., McDonald A., Chen J., Sharp J. J., Fujiwara Y., Barker J. E., Fleming M. D. (2005) Nat. Genet. 37, 1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fischer P., Hilfiker-Kleiner D. (2007) Basic Res. Cardiol. 102, 279–297 [DOI] [PubMed] [Google Scholar]
- 40. Singh V. B., Pavithra L., Chattopadhyay S., Pal J. K. (2009) Biochem. Biophys. Res. Commun. 379, 710–715 [DOI] [PubMed] [Google Scholar]
- 41. Weis N., Weigert A., von Knethen A., Brüne B. (2009) Mol. Biol. Cell 20, 1280–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Evans M. J., Scarpulla R. C. (1990) Genes Dev. 4, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 43. Virbasius J. V., Virbasius C. A., Scarpulla R. C. (1993) Genes Dev. 7, 380–392 [DOI] [PubMed] [Google Scholar]
- 44. Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R. C., Young R., Kluger Y., Dynlacht B. D. (2004) Mol. Cell 16, 399–411 [DOI] [PubMed] [Google Scholar]
- 45. Wu C. L., Zukerberg L. R., Ngwu C., Harlow E., Lees J. A. (1995) Mol. Cell. Biol. 15, 2536–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris L., Allen K. E., La Thangue N. B. (2000) Nat. Cell Biol. 2, 232–239 [DOI] [PubMed] [Google Scholar]
- 47. Le Cam L., Linares L. K., Paul C., Julien E., Lacroix M., Hatchi E., Triboulet R., Bossis G., Shmueli A., Rodriguez M. S., Coux O., Sardet C. (2006) Cell 127, 775–788 [DOI] [PubMed] [Google Scholar]
- 48. Raha D., Wang Z., Moqtaderi Z., Wu L., Zhong G., Gerstein M., Struhl K., Snyder M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ito N., Watanabe-Matsui M., Igarashi K., Murayama K. (2009) Genes Cells 14, 167–178 [DOI] [PubMed] [Google Scholar]
- 50. Taube R., Lin X., Irwin D., Fujinaga K., Peterlin B. M. (2002) Mol. Cell. Biol. 22, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sanz N., Díez-Fernández C., Valverde A. M., Lorenzo M., Benito M., Cascales M. (1997) Br. J. Cancer 75, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bussolati O., Laris P. C., Rotoli B. M., Dall'Asta V., Gazzola G. C. (1992) J. Biol. Chem. 267, 8330–8335 [PubMed] [Google Scholar]
- 53. Shih A. Y., Erb H., Sun X., Toda S., Kalivas P. W., Murphy T. H. (2006) J. Neurosci. 26, 10514–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li N., Wang C., Wu Y., Liu X., Cao X. (2009) J. Biol. Chem. 284, 3021–3027 [DOI] [PubMed] [Google Scholar]
- 55. Choi J., Husain M. (2006) Cell Cycle 5, 2183–2186 [DOI] [PubMed] [Google Scholar]
- 56. Nijman S. M., Hijmans E. M., El Messaoudi S., van Dongen M. M., Sardet C., Bernards R. (2006) J. Biol. Chem. 281, 21582–21587 [DOI] [PubMed] [Google Scholar]
- 57. Sanchez G., Bittencourt D., Laud K., Barbier J., Delattre O., Auboeuf D., Dutertre M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6004–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shin J. (1998) Arch. Pharm. Res. 21, 629–633 [DOI] [PubMed] [Google Scholar]
- 59. Geetha T., Wooten M. W. (2002) FEBS Lett. 512, 19–24 [DOI] [PubMed] [Google Scholar]
- 60. Konecna A., Frischknecht R., Kinter J., Ludwig A., Steuble M., Meskenaite V., Indermühle M., Engel M., Cen C., Mateos J. M., Streit P., Sonderegger P. (2006) Mol. Biol. Cell 17, 3651–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ballatore C., Lee V. M., Trojanowski J. Q. (2007) Nat. Rev. Neurosci. 8, 663–672 [DOI] [PubMed] [Google Scholar]
- 62. Kodiha M., Tran D., Qian C., Morogan A., Presley J. F., Brown C. M., Stochaj U. (2008) Biochim. Biophys. Acta 1783, 405–418 [DOI] [PubMed] [Google Scholar]
- 63. Scheffer G. L., Wijngaard P. L., Flens M. J., Izquierdo M. A., Slovak M. L., Pinedo H. M., Meijer C. J., Clevers H. C., Scheper R. J. (1995) Nat. Med. 1, 578–582 [DOI] [PubMed] [Google Scholar]
- 64. Paspalas C. D., Perley C. C., Venkitaramani D. V., Goebel-Goody S. M., Zhang Y., Kurup P., Mattis J. H., Lombroso P. J. (2009) Cereb. Cortex 19, 1666–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Slesina M., Inman E. M., Moore A. E., Goldhaber J. I., Rome L. H., Volknandt W. (2006) Cell Tissue Res. 324, 403–410 [DOI] [PubMed] [Google Scholar]
- 66. Iwashita K., Ikeda R., Takeda Y., Sumizawa T., Furukawa T., Yamaguchi T., Akiyama S., Yamada K. (2010) Cancer Sci. 101, 920–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Berger W., Steiner E., Grusch M., Elbling L., Micksche M. (2009) Cell. Mol. Life Sci. 66, 43–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ryu S. J., Park S. C. (2009) Expert Opin. Ther. Targets 13, 479–484 [DOI] [PubMed] [Google Scholar]
- 69. Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J. C., Westermann B., Rugarli E. I., Langer T. (2009) J. Cell Biol. 187, 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kremmidiotis G., Gardner A. E., Settasatian C., Savoia A., Sutherland G. R., Callen D. F. (2001) Genomics 76, 58–65 [DOI] [PubMed] [Google Scholar]
- 71. Snider B. J., Fagan A. M., Roe C., Shah A. R., Grant E. A., Xiong C., Morris J. C., Holtzman D. M. (2009) Arch. Neurol. 66, 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Araki Y., Kawano T., Taru H., Saito Y., Wada S., Miyamoto K., Kobayashi H., Ishikawa H. O., Ohsugi Y., Yamamoto T., Matsuno K., Kinjo M., Suzuki T. (2007) EMBO J. 26, 1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gopinath S. C., Wadhwa R., Kumar P. K. (2010) Mol. Cancer Res. 8, 1536–1546 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.