Abstract
p70 ribosomal protein S6 kinase 1 (S6K1) is regulated by multiple phosphorylation events. Three of these sites are highly conserved among AGC kinases (cAMP dependent Protein Kinase, cGMP dependent Protein Kinase, and Protein Kinase C subfamily): the activation loop in the kinase domain, and two C-terminal sites, the turn motif and the hydrophobic motif. The common dogma has been that phosphorylation of the hydrophobic motif primes S6K1 for the phosphorylation at the activation loop by phosphoinositide-dependent protein kinase 1 (PDK1). Here, we show that the turn motif is, in fact, phosphorylated first, the activation loop second, and the hydrophobic motif is third. Specifically, biochemical analyses of a construct of S6K1 lacking the C-terminal autoinhibitory domain as well as full-length S6K1, reveals that S6K1 is constitutively phosphorylated at the turn motif when expressed in insect cells and becomes phosphorylated in vitro by purified PDK1 at the activation loop. Only the species phosphorylated at the activation loop by PDK1 gets phosphorylated at the hydrophobic motif by mammalian target of rapamycin (mTOR) in vitro. These data are consistent with a previous model in which constitutive phosphorylation of the turn motif provides the key priming step in the phosphorylation of S6K1. The data provide evidence for regulation of S6K1, where hydrophobic motif phosphorylation is not required for PDK1 to phosphorylate S6K1 at the activation loop, but instead activation loop phosphorylation of S6K1 is required for mTOR to phosphorylate the hydrophobic motif of S6K1.
Keywords: ATP, Cyclic Nucleotides, Enzyme Structure, Protein Kinases, Protein Kinase A (PKA), Protein Purification, Protein Structure
Introduction
p70 ribosomal protein S6 kinase 1 (S6K1)2 is a member of the AGC subfamily of serine/threonine protein kinases, which are key mediators of growth factor and insulin signaling pathways. They regulate various cellular processes including growth, division, differentiation, and metabolism, and their dysregulation has been implicated in various diseases and disorders (1, 2). The AGC subfamily includes cAMP-dependent protein kinase (PKA), cGMP-dependent protein kinase (PKG), protein kinase C (PKC), protein kinase B (PKB; also known as Akt), phosphoinositide-dependent protein kinase 1 (PDK1), p70 S6 kinase 1 (S6K1), p90 S6 kinase 1 (RSK1) and mitogen and stress-activated protein kinase (3) as key representative members. All AGC kinases have a catalytic kinase domain and a conserved C-terminal tail after the kinase domain (4). The kinase domain has a conserved phosphorylation site on the activation loop whereas the C-terminal tail usually harbors two other conserved phosphorylation sites, namely the turn motif and the hydrophobic motif (HM) site (Fig. 1a) (4–6). Activation of AGC kinases typically requires phosphorylation of these three sites by distinct mechanisms (5, 7). In the case of PKA and PKC, the active fully phosphorylated kinase is stored in an inactive state that is then activated by second messengers. For others such as PKB (or Akt), phosphorylation at two of these sites, the activation loop and HM site are agonist-evoked (6). Phosphorylation also critically controls S6K1.
FIGURE 1.
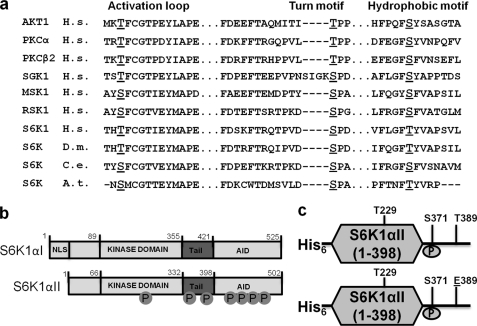
Domain organization, S6K1 constructs used in study, and purification. a, sequence alignment of various representative AGC kinases showing three conserved phosphorylation sites (underlined) in three separate sites, namely activation loop, turn motif, and hydrophobic motif. Also shown is conservation of these sites in S6K1 across various orthologs. b, schematic representation of S6K1αI and S6K1αII isoforms showing seven phosphorylation sites. c, the two baculovirus S6K1αII isoform constructs with truncation of AID (1–398) used in the study. When expressed in insect cells, both have phosphorylation at turn motif site Ser-371. The lower panel shows construct with HM residue Thr mutated to a phosphomimetic Glu.
S6K1 is activated by multiple independent inputs, which are tightly regulated by growth factor signaling and nutrient availability. Two S6K1 isoforms (accession no. NM003161, αI and αII isoforms) are transcribed from a single gene by alternative mRNA splicing, using alternative translational start site (8). The αI isoform (525 residues) contains an N-terminal 23-residue segment that encodes a nuclear localization motif, whereas the αII isoform (502 residues) is cytoplasmic and starts at a Met residue equivalent to Met-24 in the αI isoform, and the sequences of both isoforms are identical thereafter (here onward S6K1 means S6K1αII isoform). S6K1 has an N-terminal kinase domain and a 104-amino acid C-terminal autoinhibitory domain (AID) separated by the conserved C-terminal tail region (Fig. 1b). The AID blocks phosphorylation of the kinase domain by upstream kinases, and this inhibition is relieved by phosphorylation at four specific Ser/Thr sites in the AID, namely Ser-411, Ser-418, Thr-421, and Ser-424, that are each followed by Pro residues (9). Mutation of these four sites in AID with phosphomimetic residues (abbreviated S6K1D3E) relieves the inhibition and generates an enzyme that is as active as the wild-type construct and is activated by mitogen in cultured cells (10). However, the S6K1D3E activity was sensitive to rapamycin and phosphatase treatment, suggesting that there are additional phosphorylation sites regulating the activity. Additionally, truncation of the complete AID resulted in a construct with the same basal activity as that of endogenous full-length S6K1, suggesting that eliminating AID does not affect S6K1 activity toward downstream substrates, but instead prevents the activation of S6K1 by upstream kinases (11).
Rapamycin-sensitive phosphorylation sites in S6K1 were identified by two-dimensional thin layer electrophoresis/chromatography followed by mass spectrometry to be Thr-229 and Thr-389 (11). Thr-389 was the primary target of rapamycin. Mutations of Thr-229 to Ala or Glu led to kinase inactivation, suggesting that Thr-229 is a critical residue for S6K1 activity (11). The Thr-389 residue when mutated to Glu had 3-fold higher basal activity than the wild-type enzyme, and the activity was 50–70% rapamycin-resistant. This feature was not seen in the T389A mutant construct. Interestingly, substituting all four sites in the AID region along with HM site with phosphomimetic residues and generating mutant S6K1E389D3E showed 2-fold higher basal activity than either S6K1D3E or S6K1T389E (12). One additional phosphorylation site, Ser-371, was subsequently identified in S6K1 (13). Mutational studies showed Ser-371 mutation to Ala or Asp abolished kinase activity. Moreover, Ser-371 is highly conserved in S6K1 homologs from Drosophila to yeast (Fig. 1a). The study also showed that the S371D mutation led to complete absence of Thr-389 phosphorylation but did not affect phosphorylation of Thr-229 as well as the four sites in AID (12). Overall, S6K1 requires as many as seven distinct phosphorylations (Fig. 1b) for activation, four of which are in the AID, whereas the remaining three sites, Thr-229, Ser-371, and Thr-389 are equivalent to the conserved signature phosphorylation sites for the AGC subfamily members (Fig. 1a). Thr-229 is the activation loop site (also known as T loop), Ser-371 is the turn motif site, which is usually followed by a Pro, and Thr-389 is the HM site (5, 7, 14).
The four sites in the AID region are phosphorylated under mitogen stimulus, and mitogen-activated protein kinases which have proline-directed kinase activity have been shown to phosphorylate AID sites in vitro (14). The activation loop site (Thr-229) was found to be phosphorylated in vivo and in vitro by PDK1 (15, 16). The kinase responsible for the HM site (Thr-389) phosphorylation was found to be mammalian target of rapamycin (mTOR) (17, 18). It was also found that the turn motif site (Ser-371) was phosphorylated by a rampamycin-resistant upstream kinase which remains to be identified (13). Although it was reported that the turn motif phosphorylation is required for substantial phosphorylation of the HM site of S6K1 in vivo and is independent of activation loop (Thr-229) phosphorylation, S6K1 activity was largely understood to be dependent on strong constructive interplay between the activation loop (Thr-229) and the HM site (Thr-389) phosphorylations (15), whereas the role of the turn motif site was mostly ignored until recently when it was found to be once again important for the S6K1 activity and also as a regulator of HM phosphorylation (7). Moreover, two models were proposed for stepwise order of phosphorylation events leading to activation of S6K1 by Avruch and co-workers (19, 20). By mutating the phosphorylation sites to alanine, the earlier model supports the idea that PDK1 phosphorylation of Thr-229 is followed by HM phosphorylation, although their data showed that mutation of the HM site to alanine incorporated 0.75 mol of phosphate and was a better substrate for PDK1 than the wild-type S6K1 (19). The latter model using phospho-specific antibody suggested that the activation loop phosphorylation is followed by HM site phosphorylation (20). The latter model was also supported by another study wherein PDK1−/− knock-out mouse embryonic fibroblasts were shown to have no phosphorylation of the HM site (21). Both models suggested the turn motif site to be phosphorylated first, followed by the AID sites. The bias toward support of the earlier model increased with modeling studies of the PDK1 kinase domain (the activation loop kinase) which identified a pocket that bound the hydrophobic PIF (PDK1-interacting fragment) peptide (22, 23) and also the crystal structure of the PDK1 kinase domain that had a sulfate residue next to the PIF pocket (22, 24) resulting in a very attractive model, suggesting that the HM site of the substrate kinase (S6K1) needs to be phosphorylated thereby allowing PDK1 to dock onto the phospho-HM site to phosphorylate Thr-229 in the activation loop (25). Moreover, mutation and truncation data of RSK2 (p90 ribosomal S6 kinase 2), a member of AGC subfamily, showed that PDK1 is recruited and activated by a phosphoserine-regulated docking site (26). Although mutational, phospho-specific antibodies and knock-out data argued against the earlier model (19–21, 27), it has been generally accepted. Another reason for persistence of this model was the unavailability of a PDK1-S6K1 co-crystal or in vitro biochemical data with purified S6K1 and PDK1 rigorously examining the order of the phosphorylation events that lead to activation of S6K1.
In this paper, we use purified S6K1 kinase domain constructs as well as full-length S6K1 to elucidate the order of phosphorylation and the role of the negative charge at the HM site. Specifically, we examine the phosphorylation of constructs lacking the AID (S6K1ΔAID, residues 1–398) as well as an HM T389E mutant (S6K1ΔAIDT389E) expressed in Sf9 insect cells (Fig. 1c) and full-length S6K1 immunopurified from HEK293 cells, by incubating in vitro with both upstream kinases PDK1ΔPH (expressed in insect cells) and mTOR (expressed in HEK293 cells). Our phosphorylation data reveal that the turn motif (Ser-371) gets phosphorylated first, followed by the activation loop (Thr-229), and the HM (Thr-389) is the third site to be phosphorylated. HM site phosphorylation is thus not required for PDK1 phosphorylation of S6K1. Instead, PDK1 phosphorylation of the activation loop is required for mTOR to phosphorylate the HM site.
EXPERIMENTAL PROCEDURES
Materials
The baculovirus for S6K1ΔAID, S6K1ΔAIDT389E, as well as PDK1ΔPH were as described in Refs. 28, 29 (Figs. 1c and 2a). HA-S6K1, myc-mTOR vector, and anti-myc antibody were generous gifts from Dr. Kun-Liang Guan, University of California at San Diego. Antibody against Thr(P)-229 was purchased from Abcam. Ser(P)-371, Thr(P)-389, Ser(P)-421/Ser(P)-424 (S6K1), and Ser(P)-241 (PDK1) were from Cell Signaling. Transfection reagent FuGENE was from Roche. Dulbecco's modified Eagle's medium (DMEM) and insect cell Sf9 medium were from Invitrogen. All other chemicals, salt, and buffers were from Sigma.
FIGURE 2.
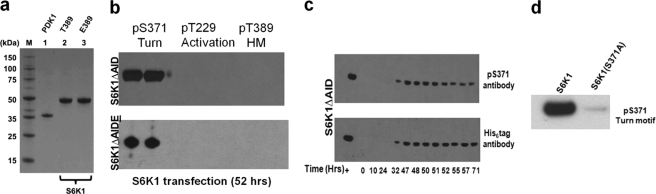
Phosphorylation status of S6K1 overexpression in insect cells. a, Coomassie gel shows purification of S6K1 constructs as well as PDK1ΔPH, all generated from insect cells. b, S6K1ΔAID and S6K1ΔAIDE, when overexpressed solely in insect cells for 52 h, get phosphorylated only at turn motif Ser-371(upper and lower panel, respectively) as probed by Ser(P)-371 antibody (in duplicate). c, transfection time course (72 h) of S6K1ΔAID expression using His6 tag antibody in parallel with Ser(P)-371 antibody shows that phosphorylation of S6K1 at turn motif is constitutive. d, specificity check for Ser(P)-371 antibody shows that the S371A mutant was not detected by the antibody.
Protein Expression and Purification
The S6K1ΔAID, S6K1ΔAIDT389E, and PDK1ΔPH were generated as reported in Keshwani et al. (29). Briefly, insect cells at 2 × 106 cells per ml were transfected with individual virus for each of the above constructs with a multiplicity of infection of 2 for 58–60 h. The cells were then pelleted and resuspended in lysis buffer (50 mm Tris, pH 7.5, 300 mm NaCl, 20 mm imidazole, 1 mm DTT, and protease inhibitor mixture). The cell suspension was frozen at −80 °C overnight and thawed at room temperature to lyse the cells. The cell lysate was then spun at 14,000 rpm for 1 h and the supernatant loaded onto a metal affinity column using Profinia (Bio-Rad) and eluted with elution buffer (50 mm Tris, pH 7.5, 300 mm NaCl, 300 mm imidazole, 1 mm DTT). To obtain S6K1ΔAID completely phosphorylated at all three sites, equal volumes of S6K1ΔAID and PDK1ΔPH virus were used to co-transfect insect cells. S6K1ΔAID was purified first using metal affinity chromatography followed by anion exchange chromatography as described in Ref. 29. The homogeneity was verified using SDS-PAGE and the concentration estimated by Bradford assay (30).
Cell Culture
HEK293 cells were maintained in DMEM with 10% FBS. The cells were transfected with HA-S6K1 for 24 h using FuGENE followed by serum starvation of 16–24 h. After serum starvation, the cells were treated with PI3K inhibitor (gift from Dr. Newton's laboratory, University of California at San Diego) for 15 min, followed by EGF stimulation (50 ng/ml) for 10 min.
Phosphorylation Assay
The S6K1ΔAID and S6K1ΔAIDT389E were resolved on 4–12% gel (Invitrogen) and transferred on a PVDF membrane (Bio-Rad) and blotted against phospho-antibodies for Thr-229, Ser-371, and Thr-389. For in vitro kinase assay, 5 μm S6K1ΔAID was incubated with 10 mm Mg2+, 500 μm ATP, and 5 × kinase buffer (50 mm MOPS, pH 7.25, 125 mm pyrophosphate, 1 mm EGTA, 5 mm sodium orthovanadate, 5 mm DTT) in a reaction volume of 100 μl. The mix was incubated in a waterbath for 1 h at 30 °C and quenched by 100 μl of 2 × SDS buffer and probed for phosphorylation at Thr-229 and Thr-389 using Western blots against specific phosphoantibodies.
Immunoprecipitation
Myc-mTOR was expressed in HEK293 cells using FuGENE as transfection reagent. Briefly, HEK cells maintained in DMEM with 10% FBS with confluence of 50–70% were transfected using 2 μg of DNA for 16 h. Cells were harvested and lysed as described by Millipore. The anti-mTOR immunocomplex was prepared using anti-myc antibody and was immunoprecipitated using protein A beads (Millipore). Immunoprecipitation was followed by in vitro kinase assay as described above. Phosphorylation of Thr-389 was probed using Western blot analysis.
RESULTS
Turn Motif Site Ser-371 Is Constitutively Phosphorylated
Both S6K1ΔAID and S6K1ΔAIDT389E constructs were expressed in insect cells in serum-free medium and purified to homogeneity (Fig. 2a). Both constructs were phosphorylated at the turn motif site Ser-371 (Fig. 2b). There was no detectable phosphorylation of the activation loop (Thr-229) and HM site (Thr-389) in S6K1ΔAID and no phosphorylation of the activation loop (Thr-229) in the S6K1ΔAIDT389E construct. Similar observations were made with full-length S6K1 overexpression in COS-7 cells (7). We also did a time course of insect cell expression of S6K1ΔAID (His6 tag) over 72 h. Using Ser(P)-371 antibody and His6 tag antibody in parallel, we found that the protein gets phosphorylated constitutively, and there is no lag in phosphorylation of Ser-371, suggesting that the Ser-371 phosphorylation may be a co-translational event (Fig. 2c). To check for phospho-specificity of the Ser-371 antibody, we overexpressed the S371A mutant. As shown in Fig. 2d, the antibody specifically binds the phosphorylated Ser-371. Overall, our data suggest that phosphorylation of the turn motif site (Ser-371) is constitutive and the first phosphorylation event in S6K1 constructs lacking the AID.
Activation Loop Site Thr-229 Gets Phosphorylated Next
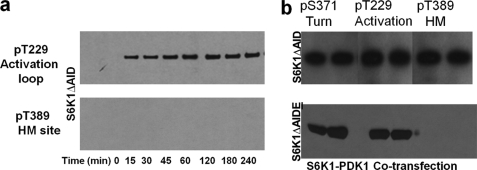
To address the question of whether the activation loop or the HM site gets phosphorylated after the turn motif, we carried out an extensive in vitro kinase assay using purified PDK1ΔPH (the upstream activation loop kinase) (Fig. 2a, lane 1). S6K1ΔAID on incubation with PDK1 kinase domain in vitro with Mg-ATP gets phosphorylated only at Thr-229 (Fig. 3a, upper panel). PDK1 does not phosphorylate HM Thr-389 in vitro (Fig. 3a, lower panel). In a similar experiment with S6K1ΔAIDT389E, we found that PDK1ΔPH phosphorylated only at the activation loop (Thr-229) of S6K1ΔAIDT389E (data not shown). Interestingly, both S6K1 constructs were equally good substrates for PDK1, strongly suggesting that the HM site phosphorylation is not required for PDK1 to dock and phosphorylate at the activation loop site Thr-229. This was also suggested in our previous study involving kinetics of PDK1 phosphorylation of the activation loop site (Thr-229) of S6K1 (28). In another independent study, the S6K1 HM T389A mutant was shown to be phosphorylated by PDK1, 2–3-fold better than the wild-type S6K1 construct (19). Recently, Sumani et al., reported the structure of the S6K1 (1–377, PDB Id 3A60, 3A61, 3A62) construct where they used PDK1 to in vitro phosphorylate the Thr-229 site (31). The construct of S6K1 used in the study did not have the HM segment but was phosphorylated by PDK1 in vitro and subsequently crystallized. This further suggests that phosphorylated HM is not required for PDK1 phosphorylation of S6K1. Phosphorylation of the turn motif site (Ser-371) was not investigated. Surprisingly, when we co-transfected S6K1ΔAID and PDK1ΔPH in insect cells, we found all three sites completely phosphorylated (Fig. 3b). A similar finding was earlier reported by Romanelli et al., who showed that co-expression of S6K1 with PDK1, PKCζ, or PKB resulted in increased phosphorylation of the HM site Thr-389 (27). Our finding that pure PDK1 phosphorylates the turn motif phosphorylated S6K1ΔAID suggests that this site is the second site to be phosphorylated and does not require the HM site (Thr-389) phosphorylation.
FIGURE 3.
In vitro and in vivo phosphorylation of S6K1ΔAID with PDK1. a, 4-h time course of PDK1 phosphorylation of S6K1ΔAID using Mg-ATP. As shown in the figure only activation loop site (Thr-229) gets phosphorylated (upper panel), but the HM site (Thr-389) is never phosphorylated in vitro (lower panel). b, co-transfection of S6K1ΔAID with PDK1ΔPH (upper panel) resulting in phosphorylation of all three sites, namely activation loop (Thr-229), turn motif (Ser-371), and the HM (Thr-389). Co-transfection of S6K1ΔAIDE with PDK1ΔPH (lower panel) results in phosphorylation of two sites, namely activation loop (Thr-229) and turn motif (Ser-371), and the HM (Thr-389) site is mutated to Glu. All blots are representative of three different experiments.
HM Site Thr-389 Gets Phosphorylated Only If Thr-229 Is Phosphorylated in S6K1ΔAID
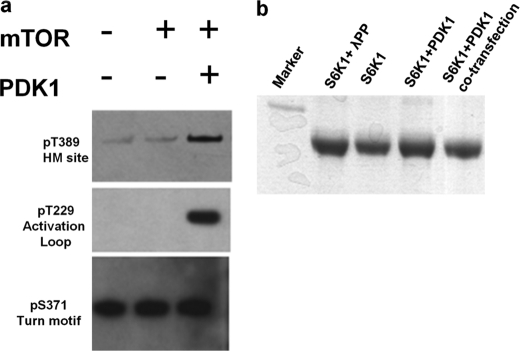
We expressed myc-mTOR in HEK293 cells and immunoprecipitated mTOR using myc monoclonal antibody (generous gifts from Dr. Guan, University of California at San Diego). S6K1ΔAID was then incubated with mTOR-precipitated beads and Mg-ATP for 1 h at 30 °C and then probed for phosphorylation of Thr-389 in vitro (Fig. 4a, second lane). We did not find any detectable phosphorylation at HM site Thr-389 when S6K1ΔAID was incubated with mTOR-immunoprecipitated beads, but when S6K1ΔAID was preincubated with PDK1 and Mg-ATP for 1 h followed by incubation with mTOR-precipitated beads and Mg-ATP, Thr-389 was phosphorylated (Fig. 4a, third lane). We tried to carry out a mobility shift assay of the different phospho-isoforms of S6K1ΔAID to find out the stoichiometry of each phosphorylation, but none of the phospho-isoforms showed any mobility shift (Fig. 4b). Our results clearly suggested that PDK1 phosphorylation was a prerequisite for mTOR phosphorylation of the HM site Thr-389 and once again support the observations that the HM phosphorylation is not an essential docking site for PDK1 to phosphorylate the activation loop Thr-229 of S6K1ΔAID as suggested previously. Further evidence in support of the activation loop being phosphorylated before the HM site comes from studies with PDK1−/− embryonic stem cells which do not show any Thr-229 and Thr-389 phosphorylation (21). In another independent study, by co-immunoprecipitation experiments of PDK1 with various S6K1 mutants, it was clearly shown that phosphorylation of Thr-389 was not a prerequisite for PDK1 interaction of S6K1, and these authors alluded to the fact that the activation loop site phosphorylation may be prior to the HM site phosphorylation (27). The same study showed that S6K1 T229A mutant showed dramatic reduction in Thr-389 phosphorylation under EGF stimulation. All of these findings clearly suggest that the activation loop Thr-229 has to be the second site to be phosphorylated followed by the HM site Thr-389. We would like to emphasize that Thr-389 phosphorylation is a key regulator of S6K1 activity, but the phosphorylated HM site does not serve as the docking site for PDK1.
FIGURE 4.
In vitro phosphorylation of S6K1ΔAID with mTOR and mobility shift of phospho-isoforms of S6K1ΔAID. a, S6K1ΔAID, when incubated with mTOR (myc-mTOR expressed in HEK293 cells and immunoprecipitated), and Mg-ATP showed no phosphorylation at HM site Thr-389, but when S6K1ΔAID was preincubated with Mg-ATP and PDK1 followed by mTOR incubation, showed Thr-389 phosphorylation. Turn motif phosphorylation was used as a loading control. b, 10% SDS-PAGE is shown. Lane 1, marker; lane 2, Lambda protein phosphatase 1-treated (1 h) S6K1ΔAID (no phosphate); lane 3, S6K1ΔAID (Ser(P)-371 turn motif phosphorylated); lane 4, S6K1ΔAID incubated with PDK1 and Mg-ATP for 1 h (turn motif and activation loop phosphorylated); lane 5, S6K1ΔAID and PDK1ΔPH co-transfected (turn motif, activation loop and HM site phosphorylated).
HM Site Phosphorylation Is Not Required for Phosphorylation of Activation Loop Thr-229 in Full-length S6K1
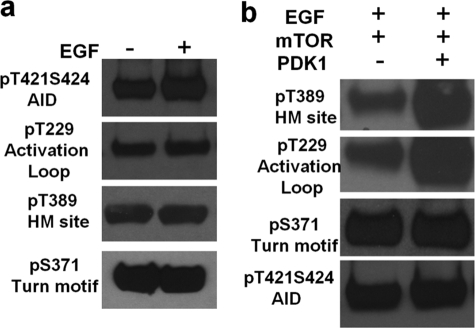
To complement our data with S6K1ΔAID and conclusively prove that HM site phosphorylation is the last step in activation of S6K1, we overexpressed HA-S6K1 in HEK293 cells and immunopurified it using anti-HA antibody as described in Ref. 27. The S6K1 was constitutively phosphorylated on Ser-371 in serum-starved conditions, and the level of phosphorylation did not change on EGF stimulation (Fig. 5a). Moreover, the AID site phosphorylation increased on EGF stimulation. To prevent effects of EGF on Thr-229 and Thr-389 phosphorylations, we treated the cells with PI3K inhibitor before EGF treatment, and as seen in Fig. 5a, the basal level of Thr-229 and Thr-389 did not change with EGF treatment in the presence of inhibitor. We then immunopurified HA-S6K1 after inhibitor and EGF stimulation and incubated them with immunopurified myc-mTOR to see whether mTOR can phosphorylate S6K1 at Thr-389 in the absence of Thr-229 phosphorylation by PDK1. As shown in Fig. 5b, only pretreatment with PDK1 leads to phosphorylation of Thr-389 by mTOR, suggesting that Thr-389 is followed by Thr-229.
FIGURE 5.
HM site phosphorylation is not required for phosphorylation of activation loop Thr-229 in full-length S6K1. a, EGF stimulation of HA-S6K1 pretreated with PI3K inhibitor, shows increase in AID site Ser-421/Ser-424 but has no effect on basal levels of Thr-229 and Thr-389. Ser-371 is constitutively phosphorylated and also serves as loading control. b, EGF stimulated and immunopurified HA-S6K1, when incubated with mTOR alone shows no increase in basal Thr-389 phosphorylation but when pretreated with PDK1 followed by mTOR, shows increase in basal Thr-389 and Thr-229 phosphorylation. Ser(P)-421/Ser(P)-424 and Ser(P)-371 show no enhancement due to mTOR and PDK1 treatment and serve as control.
Model of S6K1ΔAID Activation via Sequential Phosphorylation and Role of Each Phosphorylation
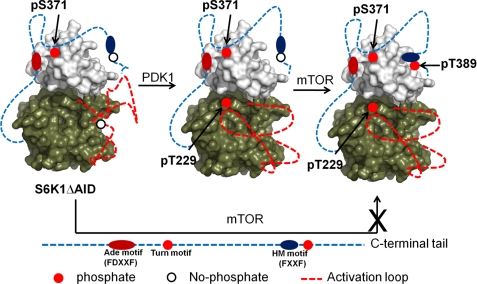
Based on our phosphorylation assays, we would like to propose a three-step sequential phosphorylation model for activation of S6K1ΔAID (Fig. 6). The turn motif site (Ser-371) phosphorylation is constitutive (7), and our data suggest that it could occur during translation (Fig. 2c). The turn motif phosphorylation and its interaction with the basic pocket on the N-lobe of the kinase core are not only critical for activity of S6K1 but may also be important for activation of, or binding to PDK1. The turn motif phosphate presumably sequesters the conserved C-terminal tail away from the active site cleft thereby making the activation loop accessible for PDK1. Additionally, the turn motif phosphate along with the Ade motif (conserved adenine binding motif, FDXXF) on the C-terminal tail (Fig. 6), stabilizes the glycine-rich loop which positions the ATP (32). The second step is phosphorylation of the activation loop by constitutively active PDK1. Phosphorylation of the HM segment is not required for PDK1 to phosphorylate the activation loop of S6K1ΔAID. The activation segments contain the conserved DFG motif (conserved Asp-Phe-Gly motif) which aligns the ATP for phosphoryl transfer. In addition, phosphorylation of the activation loop creates an ionic interaction with Arg of HRD motif that is part of catalytic loop (33). The Phe-237 residue in the DFG motif is a regulatory spine residue, and phosphorylation of the activation loop completes the regulatory spine by aligning DFG Phe and Tyr-216 of the H/YRD motif (conserved His/Tyr-Arg-Asp motif) with Leu-147 of the αC helix and Leu-158 of the β4 strand (33). Phosphorylation of the activation loop thus primes the C-lobe of the kinase core for catalysis (33). The final step is HM site phosphorylation. The turn motif phosphorylation helps to dock the HM segment to a hydrophobic pocket in the N-lobe formed by the αC helix, and phosphorylation of activation loop results in ordering of the activation segment clearing the way for the mTOR to phosphorylate the HM site of S6K1. The HM site phosphorylation is important for optimal activity and possibly for stabilization of S6K1. It is also believed to be very critical for the interaction of S6K1 to the eukaryotic initiation factor 3 preinitiation complex and downstream targets of S6K1 (34).
FIGURE 6.
Model of S6K1ΔAID activation via sequential phosphorylation. The turn motif Ser-371 phosphorylation is constitutive and results in ordering of the C-terminal tail onto the N-lobe of S6K1ΔAID (the conserved C-terminal tail with Ade motif, the HM segment, and two phosphorylation sites are pictured in detail at the bottom). This allows for phosphorylation of activation loop by PDK1 followed by the HM site by mTOR kinase. The HM site may not get phosphorylated after the turn motif because the unphosphorylated activation loop presumably causes steric hindrance for the accessibility of the HM segment by mTOR kinase. PDK1 phosphorylation of activation loop is a prerequisite for mTOR phosphorylation of HM site in S6K1ΔAID.
DISCUSSION
Eukaryotic protein kinases are distinct from their prokaryotic precursors in terms of their intricate regulation. Activation by multiple site-specific phosphorylations is a feature shared by most eukaryotic protein kinases (4). These are achieved by autophosphorylation both cis and trans, as well as by other activating kinases under a specific signaling impetus. In the AGC subfamily there are multiple upstream kinases that are involved in such activation. The catalytic kinase core of the AGC kinases has a classic bilobal structure with a small N-lobe and a larger C-lobe (35). Another conserved feature of AGC kinases is the presence of a C-terminal tail that follows the kinase core and harbors two of the three phosphorylation sites (4, 32). Each phosphorylation is distinct and important for kinase activity. The two lobes need priming events for catalysis, and this is rendered by phosphorylation. The turn motif and HM site phosphorylation prime the N-lobe for catalysis while the activation loop phosphorylation primes the C-lobe for catalysis.
The importance of multiple site phosphorylation regulated by divergent signaling pathways for AGC kinase activity has long been known. The identification of the activation loop site in PKA (35) followed by identification of the turn motif and HM in PKC (5) provided the first clues of the importance of these sites in AGC kinase regulation. For S6K1, the common understanding was that the two sites critical for activation are the activation loop and the HM site, with little attention paid to the turn motif. However, for other AGC kinases, such as PKC, it has long been known that the turn motif is critical for function (5). The importance of this latter site was highlighted in a study by Frodin and co-workers, showing that it plays an important role in stabilizing the C-terminal tail by interacting with a Ser(P)/Thr(P) binding site above the Gly-rich loop in the kinase core (7). PKB is also constitutively phosphorylated at the turn motif (36). Elegant mutational analysis by Thomas and co-workers revealed that the turn motif site in S6K1 is important for S6K1 activity and needs to be phosphorylated (13). For unknown reasons, this critical site was subsequently ignored in S6K1. We have expressed S6K1ΔAID, PKB, PKCα, and PKCβII, and all of these show constitutive and complete phosphorylation at the turn motif in serum-free medium using insect cells. Phosphorylation of the activation loop and the HM site was variable in all kinases (data not shown). Using S6K1ΔAID as model, we have comprehensively shown that the turn motif site is constitutively phosphorylated and is indeed the first site to be phosphorylated. Because the turn motif is constitutively phosphorylated and additionally, relatively resistant to phosphatase treatment, its role as a key activation site for S6K1 has been largely unappreciated.
The PDK1ΔPH crystal structure showed a phosphate binding pocket (24), and it was also proposed by molecular modeling and surface plasmon resonance analysis using PIF (26) that PDK1 gets activated if the HM residue of its substrate is phosphorylated or replaced by a phosphomimetic. It is important to note that affinity of PIF toward PDK1 is 0.3 μm (37), and no crystal structure of PIF bound to PDK1 has been solved yet. Moreover, PDK1 also has C-terminal PH domain, and the structure of full-length PDK1 has also not been solved so how the PH domain interacts with the kinase domain is also unknown. We previously reported detailed kinetic analysis of S6K1ΔAID as well as S6K1ΔAIDT389E, Thr-229 phosphorylation by PDK1ΔPH. Our data suggested that both S6K1ΔAID and S6K1ΔAIDT389E phosphorylated to an equal extent at Thr-229 by PDK1ΔPH. Also, the kinetic parameters for both constructs of S6K1 were similar. The Km for S6K1ΔAIDT389E was 2-fold lower than S6K1ΔAID, but the Vmax was similar. The 2-fold increase in Km was not significant but underscored the fact that S6K1ΔAID had 2-fold reduction in the formation of the reactive ternary complex for Thr-229 phosphorylation (28).
Based on all of the previous findings of various independent studies with S6K1 and results presented here, we propose a model for the order of phosphorylation events leading to activation of S6K1ΔAID. As shown in Fig. 6, S6K1ΔAID has a disorderd unphosphorylated activation loop (in red) and C-terminal tail (in blue) which has the turn motif site Ser-371 phosphorylated and the HM segment partially ordered. PDK1 can phosphorylate the activation loop at Thr-229 and induce structural ordering of the activation loop. Once the activation loop is phosphorylated, mTOR complex 1 can phosphorylate Thr-389 in the HM site, resulting in a fully active S6K1. Thus, our data are consistent with a model for the phosphorylation of S6K1 in which the turn motif provides the first and key step to prime S6K1 for further phosphorylations by PDK1 and mTOR, respectively. This model of S6K1ΔAID activation is suitable for other AGC kinases as most of them, like S6K1, have three conserved phosphorylation sites and can have significant impact on general understanding of signaling networks that are regulated by phosphorylation.
Acknowledgments
We thank Dr. Kun-Liang Guan, Moores Cancer Center, University of California, San Diego, for the myc-mTOR and HA-S6K1 plasmid and anti-myc antibody, and Dr. Alexandr Kornev and Jon Steichen for reading the manuscript.
The work was supported, in whole or in part, by National Institutes of Health Grants 19301 (to S. S. T.) and DK054441 through the NIDDK (to S. S. T. and A. C. N.).
- S6K1
- p70 ribosomal protein S6 kinase 1
- AGC
- cAMP dependent protein kinase, cGMP dependent protein kinase and Protein Kinase C
- AID
- autoinhibitory domain
- HM
- hydrophobic motif
- mTOR
- mammalian target of rapamycin
- PDK1
- phosphoinositide-dependent protein kinase 1
- PIF
- PDK1-interacting fragment.
REFERENCES
- 1. Montagne J., Stewart M. J., Stocker H., Hafen E., Kozma S. C., Thomas G. (1999) Science 285, 2126–2129 [DOI] [PubMed] [Google Scholar]
- 2. Martin K. A., Blenis J. (2002) Adv. Cancer Res. 86, 1–39 [DOI] [PubMed] [Google Scholar]
- 3. Hanks S. K., Hunter T. (1995) FASEB J. 9, 576–596 [PubMed] [Google Scholar]
- 4. Kannan N., Haste N., Taylor S. S., Neuwald A. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1272–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keranen L. M., Dutil E. M., Newton A. C. (1995) Curr. Biol. 5, 1394–1403 [DOI] [PubMed] [Google Scholar]
- 6. Newton A. C. (2009) J. Lipid Res. 50, S266–S271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hauge C., Antal T. L., Hirschberg D., Doehn U., Thorup K., Idrissova L., Hansen K., Jensen O. N., Jørgensen T. J., Biondi R. M., Frödin M. (2007) EMBO J. 26, 2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grove J. R., Banerjee P., Balasubramanyam A., Coffer P. J., Price D. J., Avruch J., Woodgett J. R. (1991) Mol. Cell. Biol. 11, 5541–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferrari S., Bannwarth W., Morley S. J., Totty N. F., Thomas G. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7282–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferrari S., Pearson R. B., Siegmann M., Kozma S. C., Thomas G. (1993) J. Biol. Chem. 268, 16091–16094 [PubMed] [Google Scholar]
- 11. Weng Q. P., Andrabi K., Kozlowski M. T., Grove J. R., Avruch J. (1995) Mol. Cell. Biol. 15, 2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pearson R. B., Dennis P. B., Han J. W., Williamson N. A., Kozma S. C., Wettenhall R. E., Thomas G. (1995) EMBO J. 14, 5279–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moser B. A., Dennis P. B., Pullen N., Pearson R. B., Williamson N. A., Wettenhall R. E., Kozma S. C., Thomas G. (1997) Mol. Cell. Biol. 17, 5648–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis R. J. (1993) J. Biol. Chem. 268, 14553–14556 [PubMed] [Google Scholar]
- 15. Alessi D. R., Deak M., Casamayor A., Caudwell F. B., Morrice N., Norman D. G., Gaffney P., Reese C. B., MacDougall C. N., Harbison D., Ashworth A., Bownes M. (1997) Curr. Biol. 7, 776–789 [DOI] [PubMed] [Google Scholar]
- 16. Pullen N., Dennis P. B., Andjelkovic M., Dufner A., Kozma S. C., Hemmings B. A., Thomas G. (1998) Science 279, 707–710 [DOI] [PubMed] [Google Scholar]
- 17. Burnett P. E., Barrow R. K., Cohen N. A., Snyder S. H., Sabatini D. M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isotani S., Hara K., Tokunaga C., Inoue H., Avruch J., Yonezawa K. (1999) J. Biol. Chem. 274, 34493–34498 [DOI] [PubMed] [Google Scholar]
- 19. Alessi D. R., Kozlowski M. T., Weng Q. P., Morrice N., Avruch J. (1998) Curr. Biol. 8, 69–81 [DOI] [PubMed] [Google Scholar]
- 20. Weng Q. P., Kozlowski M., Belham C., Zhang A., Comb M. J., Avruch J. (1998) J. Biol. Chem. 273, 16621–16629 [DOI] [PubMed] [Google Scholar]
- 21. Williams M. R., Arthur J. S., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D. R. (2000) Curr. Biol. 10, 439–448 [DOI] [PubMed] [Google Scholar]
- 22. Biondi R. M., Kieloch A., Currie R. A., Deak M., Alessi D. R. (2001) EMBO J. 20, 4380–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Biondi R. M., Cheung P. C., Casamayor A., Deak M., Currie R. A., Alessi D. R. (2000) EMBO J. 19, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biondi R. M., Komander D., Thomas C. C., Lizcano J. M., Deak M., Alessi D. R., van Aalten D. M. (2002) EMBO J. 21, 4219–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mora A., Komander D., van Aalten D. M., Alessi D. R. (2004) Semin. Cell Dev. Biol. 15, 161–170 [DOI] [PubMed] [Google Scholar]
- 26. Frödin M., Antal T. L., Dümmler B. A., Jensen C. J., Deak M., Gammeltoft S., Biondi R. M. (2002) EMBO J. 21, 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Romanelli A., Dreisbach V. C., Blenis J. (2002) J. Biol. Chem. 277, 40281–40289 [DOI] [PubMed] [Google Scholar]
- 28. Keshwani M. M., Gao X., Harris T. K. (2009) J. Biol. Chem. 284, 22611–22624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keshwani M. M., Ross D. B., Ragan T. J., Harris T. K. (2008) Protein Expr. Purif. 58, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 31. Sunami T., Byrne N., Diehl R. E., Funabashi K., Hall D. L., Ikuta M., Patel S. B., Shipman J. M., Smith R. F., Takahashi I., Zugay-Murphy J., Iwasawa Y., Lumb K. J., Munshi S. K., Sharma S. (2010) J. Biol. Chem. 285, 4587–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Romano R. A., Kannan N., Kornev A. P., Allison C. J., Taylor S. S. (2009) Protein Sci. 18, 1486–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kornev A. P., Haste N. M., Taylor S. S., Eyck L. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17783–17788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holz M. K., Ballif B. A., Gygi S. P., Blenis J. (2005) Cell 123, 569–580 [DOI] [PubMed] [Google Scholar]
- 35. Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M. (1991) Science 253, 407–414 [DOI] [PubMed] [Google Scholar]
- 36. Bellacosa A., Chan T. O., Ahmed N. N., Datta K., Malstrom S., Stokoe D., McCormick F., Feng J., Tsichlis P. (1998) Oncogene 17, 313–325 [DOI] [PubMed] [Google Scholar]
- 37. Balendran A., Casamayor A., Deak M., Paterson A., Gaffney P., Currie R., Downes C. P., Alessi D. R. (1999) Curr. Biol. 9, 393–404 [DOI] [PubMed] [Google Scholar]