Abstract
TSG-6 (TNF-α-stimulated gene/protein 6), a hyaluronan (HA)-binding protein, has been implicated in the negative regulation of inflammatory tissue destruction. However, little is known about the tissue/cell-specific expression of TSG-6 in inflammatory processes, due to the lack of appropriate reagents for the detection of this protein in vivo. Here, we report on the development of a highly sensitive detection system and its use in cartilage proteoglycan (aggrecan)-induced arthritis, an autoimmune murine model of rheumatoid arthritis. We found significant correlation between serum concentrations of TSG-6 and arthritis severity throughout the disease process, making TSG-6 a better biomarker of inflammation than any of the other arthritis-related cytokines measured in this study. TSG-6 was present in arthritic joint tissue extracts together with the heavy chains of inter-α-inhibitor (IαI). Whereas TSG-6 was broadly detectable in arthritic synovial tissue, the highest level of TSG-6 was co-localized with tryptases in the heparin-containing secretory granules of mast cells. In vitro, TSG-6 formed complexes with the tryptases murine mast cell protease-6 and -7 via either heparin or HA. In vivo TSG-6-tryptase association could also be detected in arthritic joint extracts by co-immunoprecipitation. TSG-6 has been reported to suppress inflammatory tissue destruction by enhancing the serine protease-inhibitory activity of IαI against plasmin. TSG-6 achieves this by transferring heavy chains from IαI to HA, thus liberating the active bikunin subunit of IαI. Because bikunin is also present in mast cell granules, we propose that TSG-6 can promote inhibition of tryptase activity via a mechanism similar to inhibition of plasmin.
Keywords: Antigen, Cytokine, Extracellular Matrix, Heparin, Hyaluronate, Inflammation, Mast Cell, Mouse, PGIA (PG-induced Arthritis), TSG-6
Introduction
TSG-6 (the product of tumor necrosis factor α-stimulated gene-6; also called Tnfaip6 or Tnfip6) is a hyaluronan (HA)3-binding protein, secreted by a variety of cells in response to proinflammatory stimuli (1). TSG-6 protein has been detected in large quantities in the synovial fluids and synovial tissues of inflamed joints of patients with rheumatoid arthritis (2–4). Recombinant mouse TSG-6 (rmTSG-6) has demonstrated anti-inflammatory and chondroprotective effects in mouse models of rheumatoid arthritis (5). TSG-6 forms a stable complex with a heavy chain (HC) of inter-α-trypsin inhibitor (IαI), a major serine protease inhibitor in serum (3). Because IαI exhibits increased inhibitory activity against plasmin, a key activator of matrix metalloproteinases after encountering TSG-6, it has been postulated that TSG-6 exerts its anti-inflammatory and chondroprotective effects primarily through inhibition of the protease network (6, 7).
TSG-6 consists of a “Link” module and a “CUB” (complement C1s/C1r-, uEGF-, BMP-1-like) domain. The positively charged Link module binds various glycosaminoglycans (GAGs), including HA, chondroitin sulfate, heparin, and heparan sulfate (1, 8–10). The CUB domain of TSG-6 is similar to the CUB module found in several developmentally regulated proteins (11) that are thought to be involved in protein-protein interactions (12–14). However, to date, only fibronectin has been shown to bind to the CUB domain of TSG-6 (15). TSG-6 is not expressed constitutively but can be induced by proinflammatory cytokines or LPS (2, 16, 17). In contrast, anti-inflammatory cytokines, such as IL-4 or IL-10, suppress TSG-6 expression either directly or via inhibition of LPS/Toll-like receptor-induced cell activation (17).
TSG-6 can modulate the binding of HA to the cell surface HA receptor, CD44 (18). Treatment of leukocytes with soluble HA·TSG-6 complex has been shown to inhibit the CD44-mediated adhesion of these cells to immobilized HA in vitro (19). Because the CD44-supported adhesion (rolling) of leukocytes on HA-covered surfaces of inflamed vascular endothelium is required for the emigration of these cells from the bloodstream into inflamed tissue (20), inhibition of this adhesion step by the HA·TSG-6 complex could have a negative impact on the extravasation of inflammatory cells. The in vitro observations are consistent with in vivo studies reporting reduced leukocyte influx into the arthritic joints of TSG-6-treated mice (5, 6) and enhanced leukocyte extravasation in the joints of TSG-6-deficient mice (21). Collectively, the in vivo observations lend support to the concept that TSG-6 has a critical role in the resolution of inflammation, but this function of TSG-6 may rely on more than one mechanism.
One of the initial goals of the present study was to develop a sensitive detection method for measuring the concentrations of TSG-6 in serum and synovial fluid samples of mice with arthritis. Using cartilage proteoglycan (PG)-induced arthritis (PGIA) in BALB/c mice, we monitored serum levels of TSG-6 in correlation with the onset and progression of arthritis and identified TSG-6-positive cells in the joints. Although many connective tissue cells were TSG-6-positive in the arthritic joints, unexpectedly, the strongest immunostaining of TSG-6 was detected in the granules of mast cells that accumulated in inflamed paws of mice. In vitro, rmTSG-6 bound to both heparin and the mast cell-restricted tryptases, murine mast cell protease-6 (mMCP-6) and mMCP-7, two major serine proteases present in mast cell secretory granules (22). Further, TSG-6 could be co-immunoprecipitated with both mMCP-6 and mMCP-7 from tissue extracts of arthritic paws, in which the heavy chains (HC1 and HC2) of IαI were also present. These data suggest that TSG-6 may modulate mast cell function via its interactions with key components of secretory granules.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
Chemicals were obtained from Sigma-Aldrich or Fisher, and molecular biology grade reagents were from Invitrogen. Recombinant human TSG-6 and rmTSG-6, rmMCP-6 and rmMCP-7 were obtained from R&D Systems (Minneapolis, MN), and cytokine ELISAs were from either BD Biosciences or R&D Systems. Horseradish peroxidase (HRP)-conjugated polyclonal antibodies (Abs) against murine IgG and IgG2a and HRP-labeled rabbit anti-goat IgG were purchased from BD Biosciences. Goat polyclonal Ab to murine HC1 and HC2 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and goat Abs to mMCP-6 and mMCP-7 were from R&D Systems. Chinese hamster ovary (CHO-K1) cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA), and murine CD44-transfected CTLL-2 cells (CTLL-2/CD44+) were kindly donated by Robert Hyman (18). Unless noted otherwise, the standard cell culture medium was DMEM, containing 4.5 g/liter glucose and supplemented with 1% non-essential amino acid solution, 1 mm sodium pyruvate, 1% l-glutamine, 100 mg/liter gentamicin sulfate, 0.5 μm β-2-mercaptoethanol, and 10% FBS (Hyclone, Logan, UT). All cell cultures were performed in a humidified atmosphere of 5% CO2 in air at 37 °C.
Production and Purification of rmTSG-6 Fusion Protein
A 753-bp-long cDNA fragment of mouse IgG2a heavy chain was obtained by reverse transcription of RNA purified from a mAb-producing murine B-cell hybridoma. The cDNA fragment was amplified by PCR using primers with linkers for restriction enzyme (EcoRI and BclI) cleavage sites. The 5′-end of the cDNA included the hinge region of the mouse IgG2a heavy chain, and it was inserted into a Lonza pEE14.1 mammalian expression vector (Lonza Biologics Ltd., Slough, UK) (Fig. 1A). The stop codon of the full-length (828-bp) mouse TSG-6 cDNA (23) in a pBlueScript S/K vector (Stratagene, La Jolla, CA) was replaced with a sequence coding for the cleavage site of the endopeptidase factor Xa (Ile-Glu-Gly-Arg) followed with a 9-bp spacer before the EcoRI cleavage site (ATAGAAGGTCGT/GACTCGAGG/GAATTC) at the 3′-end of the TSG-6 cDNA. Purified mTSG-6 cDNA was inserted into the EcoRI site between the Lonza vector and the 5′-end of the mouse IgG2a heavy chain (Fig. 1A). Insert orientation was determined by PCR, and the construct was sequenced. Semiconfluent CHO-K1 cells were transfected with the mTSG-6-Xa-mFc2a Lonza construct using CaCl2 precipitation according to a standard protocol (24).
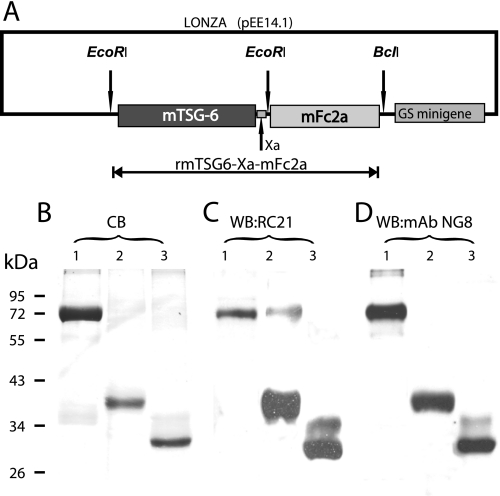
FIGURE 1.
Schematics of the mammalian expression vector containing the rmTSG-6 fusion protein and analysis of purified rmTSG-6. A, structure of the mammalian expression vector (Lonza pEE14.1-rmTSG-6-Xa-mFc2a) incorporating the fusion protein. Shown is detection of rmTSG-6 by SDS-PAGE and Coomassie Blue (CB) staining (B) and Western blotting (WB) with affinity-purified and biotinylated RC21 polyclonal rabbit anti-TSG-6 antibody (C) and monoclonal mouse anti-TSG-6 antibody (clone NG8) (D). The lanes were loaded with rmTSG-6 in the same order. Lane 1, the 72-kDa rmTSG-6-Xa-mFc2a fusion protein purified from CHO serum-free medium (a CHO line transfected with pEE14.1-rmTSG-6-Xa-mFc2a and cloned as described); lane 2, purified rmTSG-6 (∼39 kDa without the mFc2a-tail) after factor-Xa cleavage and repurification on protein G-Sepharose; lane 3, the same rmTSG-6 as shown in lane 2 but after deglycosylation (∼30 kDa) using a kit containing peptide:N-glycosidase F, O-glycosidase, sialidase, β-galactosidase, and glucosaminidase. B, the TSG-6 proteins were loaded on 12% SDS-PAGE (20 μg protein/lane), run under reducing conditions and stained with Coomassie Blue (CB). C and D, the separated proteins (1 μg/lane) were transferred to nitrocellulose membrane and probed with biotinylated RC21 and NG8 antibodies, respectively, followed by HRP-streptavidin. Molecular mass is indicated in kDa. WB, Western blot.
The Lonza expression vector contains a minigene encoding glutamine synthase, an enzyme responsible for the biosynthesis of glutamine (using glutamate and ammonia as substrates). The transfected glutamine synthase gene (a part of the Lonza vector) can act as a selection tool in the presence of methionine sulfoximine, and CHO cells containing the Lonza vector with the glutamine synthase gene can survive in the absence of glutamine and in the presence of 25–50 μm methionine sulfoximine. Approximately 2 weeks after the transfection, individual CHO colonies were transferred into 96-well cell culture plates. Glutamine-free DMEM was replaced with CHO serum-free medium (Lonza Inc., Mapleton, IL), and 48 h later, 100 μl of supernatant from each well was transferred to 96-well Maxisorp ELISA plates (Nunc International, Hanover Park, IL) and incubated overnight. Free binding sites of the wells were blocked with 1% BSA, and the clones expressing the fusion protein (TSG-6-mFc2a) were identified with HRP-conjugated goat anti-mouse IgG2a. Positive colonies were retested using affinity-purified and biotinylated RC21 rabbit Ab raised against mouse TSG-6 (5, 7, 25) followed by incubation with HRP-labeled streptavidin and tetramethylbenzidine substrate (TMB; BD Biosciences OptEiA TMB substrate set). Cell lines secreting the highest amounts of fusion protein were cloned using the limiting dilution (0.5 cell/well) method and cultured in the presence of irradiated (70 grays) mouse embryonic fibroblast feeder cell layers. Positive colonies (retested by ELISA as described above) were recloned by limiting dilution, and stable clones, secreting high amounts of fusion protein, were subjected to further testing.
The rmTSG-6-Xa-mFc2a fusion protein from the serum-free supernatant of the CHO transfectant (clone 514) was purified on Protein G-Sepharose 4 Fast Flow (GE Healthcare) according to the manufacturer's instructions. The eluted product was dialyzed against ultrapure H2O, lyophilized, and stored at −20 °C until further use. The purity of the fusion protein was determined using SDS-PAGE with Coomassie Blue staining. Western blots were performed with HRP-labeled goat anti-mouse IgG2a or with affinity-purified and biotinylated rabbit RC21 anti-TSG-6 antibody (7). Purified rmTSG-6-Xa-mFc2a fusion protein was cleaved with factor Xa (200 units/mg protein) in 2.5 ml of factor Xa cleavage/capture buffer (Novagen, Madison, WI) (100 mm NaCl, 50 mm Tris-HCl, 5 mm CaCl2, pH 8.0) overnight at room temperature. The enzyme was removed by Xarrest Sepharose (Novagen), and the IgG2a-Fc fragment was absorbed onto Protein G-Sepharose.
After optimization of factor Xa cleavage, we tested the glycosylation level of purified rmTSG-6 (clone 514). Twenty μg of purified protein was digested using a deglycosylation kit (Enzymatic DeglycoMx kit; QA-Bio LLC, Palm Desert, CA) containing a mixture of peptide:N-glycosidase F, O-glycosidase, sialidase, β-galacosidase, and glucosaminidase, which removed all N-linked and most of the O-linked oligosaccharides. The digested product was loaded on 12% SDS-PAGE and transferred onto nitrocellulose membrane for immunostaining as described above.
Testing of HA Binding and Enhancement of HA-CD44 Interaction by rmTSG-6
Binding of rmTSG-6 to immobilized HA or heparin was tested using a microplate titration assay (supplemental Fig. S1, A and B). In brief, rooster comb HA (Sigma) was serially diluted in PBS (from 200 μg/ml to 3.1 μg/ml). One hundred μl of each HA solution was dispensed into duplicate wells of 96-well Maxisorp plates and incubated overnight at room temperature. Free binding sites were blocked with 1% BSA. rmTSG-6 (from clone 514, free of IgG-Fc tail) was diluted in PBS (concentration range, 0.031–2.0 μg/100 μl per well) and incubated with the HA-coated wells for 2 h at 37 °C. HA-bound rmTSG-6 was detected with biotinylated RC21 antibody, followed by HRP-streptavidin and TMB substrate. rmTSG-6-mediated enhancement of HA binding to cell surface CD44 was tested using murine CTLL-2/CD44+ cells (18). Relative amounts of cell surface-bound fluorescein-conjugated HA (FL-HA) in the presence or absence of rmTSG-6 were measured by flow cytometry as described before (19). Briefly, HA-rmTSG-6 complex was preformed by incubating FL-HA with rmTSG-6 at a 5:1 ratio (w/w) in PBS (pH 7.2) for 30 min at room temperature. The solution containing this complex was then added to CTLL-2/CD44+ cells and incubated for 1 h at 4 °C. Base-line HA binding was determined by incubating the cells with the same amount of FL-HA alone, and PBS alone served as the background control. Cell surface fluorescence was detected and analyzed using a FACSCalibur flow cytometer and CellQuest software (BD Biosciences) (19) (supplemental Fig. S1C).
TSG-6-specific Monoclonal Antibody (mAb) Production and Development of Capture ELISA
To generate murine B cells for hybridoma fusion and subsequent TSG-6-specific mAb production, we used rmTSG-6 to immunize TSG-6-deficient BALB/c mice (21) (now available from The Jackson Laboratory; stock number 012903). All animal experiments were approved by the Institutional Animal Care and Use Committee of Rush University Medical Center (Chicago, IL). The TSG-6-deficient BALB/c mice were immunized intraperitoneally with 50 μg of purified rmTSG-6 emulsified with 2 mg of dimethyldioctadecyl-ammonium bromide (DDA) adjuvant in a total volume of 100 μl of PBS. Peripheral blood samples were taken after the third injection, and anti-TSG-6 serum titers were determined by ELISA (using immobilized rmTSG-6 as the antigen and HRP-conjugated anti-mouse IgG for detection). Spleen cells from positive mice were fused with the Sp2/0-Ag14 myeloma cell line (ATCC) employing a standard protocol (26, 27), and mAb-producing hybridomas were selected by ELISA. Positive hybridomas were repeatedly cloned by limiting dilution, and ascites fluid was produced in BALB/c mice after hybridoma injection. The isotypes and light chains of each mAb were identified using a mouse IgG isotype determination kit (Santa Cruz). Immunoglobulin fractions of five hybridoma clones (designated NG2, NG3, NG4, NG5, and NG8) were purified on protein G columns, and a portion of each was biotinylated and tested for cross-reactivity using a standard inhibition ELISA (27). In brief, 0.1 μg of rmTSG-6 was coated to each well, and serial dilutions of purified anti-TSG-6 mAbs were mixed with optimally diluted aliquots of each biotinylated anti-mTSG-6 mAb. Recombinant mouse and human TSG-6 proteins (R&D Systems) were used as additional positive controls and for the in-house development of new murine and human TSG-6 capture ELISA systems employing non-cross-reactive pairs of anti-TSG-6 mAbs. Purified mAb NG3 (IgG2b κ) was used for the capture of either murine or human TSG-6. Biotinylated NG8 mAb (IgG1 κ) was employed for the detection of murine TSG-6, and NG4 (IgG1 κ) was used for the detection of human TSG-6 (an example of the standard curve of murine TSG-6 ELISA is shown in supplemental Fig. S1D). The sensitivity of capture ELISAs ranged from 500 pg/ml to 200 ng/ml TSG-6.
Immunization with Cartilage PG Aggrecan and Assessment of Arthritis
Human osteoarthritic cartilage was obtained from patients undergoing knee joint replacement surgery. The collection of cartilage from consenting patients was approved by the Institutional Review Board of Rush University Medical Center. PG isolation from osteoarthritic cartilage and deglycosylation of PG have been described in detail (28, 29). Female BALB/c mice of 16–24 weeks of age were purchased from the NCI-Frederick, National Institutes of Health. A standard procedure was applied for induction of PGIA in BALB/c mice (29). In brief, 100 μg of core protein of PG was emulsified with 2 mg of DDA in PBS (pH 7.4) and injected intraperitoneally at 3-week intervals (days 0, 21, and 42). Control animals received PBS emulsified with DDA. Arthritis severity was determined using a visual scoring system based on the degree of swelling and redness of the front and hind paws (29, 30). Animals were examined at least three times a week; the degree of inflammation was scored from 0 to 4 for each paw, resulting in a cumulative arthritis score ranging from 0 to 16 for each animal (29, 30). All mice were scored by two different investigators in a blinded manner. The incidence of arthritis was expressed as the percentage of immunized mice that were arthritic. Upon sacrifice, limbs were removed, fixed in 10% formalin, acid-decalcified, embedded in paraffin, and processed according to standard histological procedures (29–31). Serum cytokines IL-1β, IL-6, IL-17, and TNF-α were quantified using commercially available ELISA kits.
Preparation of Synovial Fluid and Tissue Extracts, Histology, and Immunohistochemistry
Blood and tissue samples were collected from the animals at different time points of immunization, prior to and after the onset of arthritis (5, 30, 32, 33). Synovial fluid from arthritic ankle joints (with the highest arthritis score of 4) was harvested immediately after euthanasia (34) in 20 μl of PBS/joint and centrifuged at 5,000 rpm for 10 min, and the supernatant was stored at −70 °C until use. For preparation of tissue extracts, the skin was removed from arthritic or normal paws, and the tissues of ankle and tarso-metatarsal joints (including bone) were dissected and placed in cold radioimmune precipitation assay lysis buffer containing a Halt protease inhibitor mixture (both from Pierce). Homogenates were prepared by ultrasonication (Virsonic Digital 50, VirTis, Gardiner, NY) for 90 s on ice and centrifuged at 5,000 rpm. Protein contents of the supernatants of tissue extracts were measured using the bichinconic acid assay (Pierce).
For histology, macroscopically similar paws were dissected, fixed in 10% formalin, decalcified, and embedded in paraffin. Deparaffinized sections were stained with hematoxylin and eosin, toluidine blue, or safranin O. For immunohistochemistry, paws with similar degrees of arthritis were embedded in optimal cutting temperature medium, frozen, and sectioned with a tungsten knife at −32 °C in a MICROM HM 550 cryostat (MICROM International, Walldorf, Germany). Frozen sections were immunostained with mAbs to TSG-6 or goat Abs to the mouse tryptases mMCP-6 and mMCP-7. Non-immune goat or mouse IgG served as a background control.
In Vitro Detection of Interactions of TSG-6 with HA or Heparin and Mast Cell Tryptases
Using ELISA-based systems for the detection of HA- or heparin-bound proteins (TSG-6 and the mast cell tryptases mMCP-6 and mMCP-7), optimal concentrations of rooster comb HA or heparin (both from Sigma); rmTSG-6, rmMCP-6, and rmMCP-7; and Abs against TSG-6 and mMCP-6 and mMCP-7 were determined in preliminary experiments. To detect a tripartite interaction of HA or heparin with TSG-6 and tryptases, we developed a triple layer sandwich microplate assay. In brief, 2 μg of HA in 100 μl of PBS (first layer) was coated onto the wells of Maxisorp (Nunc) plates overnight at room temperature. Heparin (10 μg) in 1% N-(3-dimethylaminopropyl)-N-ethyl-carbodiimide hydrochloride was dissolved in H2O and coated onto the wells of CovaLink (Nunc) plates overnight at 37 °C (35). After blocking the free binding sites with 5% BSA, the wells were incubated with 0.1 μg of rmTSG-6 (second layer) for 1 h at 37 °C, followed by incubation with rmMCP-6 or rmMCP-7 (third layer; concentration range, 0.025–0.2 μg/well). The third layer of bound tryptases was detected with anti-mMCP-6 and anti-mMCP-7 Abs. Reciprocally, rmMCP-6 or rmMCP-7 was used as a second layer, and rmTSG-6 was used as the third layer (at the concentrations described above), followed by detection of bound rmTSG-6 with biotinylated NG8 mAb. In each assay, reference wells contained only a single protein (TSG-6 or tryptase), and immobilized proteins detected with the irrelevant Ab (rmTSG-6 with anti-tryptase Abs and vice versa) served as background controls. Reactions were developed with HRP-conjugated secondary reagents and TMB substrate, and values were expressed as the absorbance measured at 450 nm using a Synergy 2 ELISA reader (BioTek Instruments, Winooski, VT).
Identification of Mast Cell Tryptases by Western Blotting and Co-immunoprecipitation of Tryptases and HCs of IαI with TSG-6
Tissue extracts were prepared from arthritic and non-arthritic paws as described above, and 50 μg of each was loaded onto 12% SDS-PAGE, along with 0.2 μg of rmTSG-6, 0.1 μg of rmMCP-6, and 0.05 μg of rmMCP-7. The proteins were resolved under reducing conditions, transferred to a nitrocellulose membrane, and probed with different antibodies: affinity-purified biotinylated rabbit RC21 anti-TSG-6 Ab, anti-MCP-6, or anti-MCP-7. In most cases, membranes were stripped and reprobed with another antibody to detect comigration of the two proteins. The supernatants of paw extracts were subjected to Western blotting with or without immunoprecipitation as described (18, 36).
For co-immunoprecipitation, supernatants of paw extracts (200 μg of protein of each) were preabsorbed with Protein G-Sepharose, washed, and then incubated with 10 μg of TSG-6 mAb mixture (NG3, NG4, NG5, and NG8; 2.5 μg of each) overnight at 4 °C on a rotary shaker. Immune complexes were absorbed to Protein G-Sepharose for 30 min at room temperature, washed, and boiled in reducing buffer. One-third of each sample was loaded on 12% SDS-PAGE, and proteins were separated under reducing conditions. The proteins were transferred onto nitrocellulose membrane and stained with biotinylated Abs to TSG-6 (RC21 or NG8) or with goat antibodies to HC1 or HC2 (both at 1:500 dilution) or goat antibodies to mMCP-6 or mMCP-7 (both at 1:2,000 dilution) followed by HRP-conjugated rabbit antibody to goat IgG (Invitrogen; 1:5,000 dilution). HRP-labeled mouse mAb to β-actin (Invitrogen; 1:5,000 dilution) was used to ensure that equal amounts of cell extracts were loaded. All incubations were performed at room temperature for 1–2 h. Positive protein bands were detected by ECL (Amersham Biosciences).
Statistical Analysis
Data were analyzed using Student's t-tests to compare two groups or analysis of variance with the least significant difference post hoc test for comparison of multiple groups. Spearman's ρ test was used to determine correlation between two sets of data. p values of <0.05 were considered statistically significant. All analyses were performed using the SPSS (version 16.0) statistical software package (SPSS, Chicago, IL).
RESULTS
Production, Purification, and Cleavage of Recombinant Mouse TSG-6 (rmTSG-6) Fusion Protein
The first goal of this study was to develop a simple expression system for high yield production of functionally active rmTSG-6. The schematics of the construct used for CHO cell transfection are shown in Fig. 1A, and the stepwise cloning method is described under “Experimental Procedures.” Positive selection of transfectants, followed by a limiting dilution cloning procedure and a direct ELISA system using either a tag-specific (mouse IgG2a-Fc) or a protein-specific (TSG-6) Ab, allowed us to select clones with the highest yield of the fusion protein. Real-time PCR confirmed the presence of >80 copy numbers using the template of genomic DNA of the CHO transfectant, and ∼0.8–1.2 mg of recombinant fusion protein could be purified from 100 ml of conditioned medium of clone 514.
The purity of the rmTSG-6-Xa-mFc2a fusion protein (∼72 kDa) and rmTSG-6 (∼39 kDa after cleavage with factor Xa and repurification on Protein G-Sepharose), were over 95%, although some degradation occurred during the enzymatic cleavage and repeated purification (Fig. 1, B–D). Because rmTSG-6 was synthesized by CHO cells in serum-free medium, serum immunoglobulins and IαI could not affect the purity (first and second lanes in Fig. 1, B–D) or the functionality of secreted rmTSG-6. The molecular mass of purified rmTSG-6 after Xa cleavage was ∼39 kDa (instead of ∼30 kDa), indicating that the CHO cell-secreted rmTSG-6 was glycosylated (Fig. 1, B–D, second lanes). Indeed, enzymatic removal of all N-linked and most O-linked oligosaccharides reduced the mass of rmTSG-6 to the expected ∼30 kDa size (third lanes, Fig. 1, B–D). The Fc-free rmTSG-6 was largely water-insoluble but could be solubilized by the addition of 5% BSA and dialysis against serum-free CHO medium. Purified rmTSG-6 bound HA in a concentration-dependent manner, and, similar to recombinant human TSG-6 (19), it enhanced the binding of fluorescence-labeled HA to cell surface CD44 (supplemental Fig. S1C). rmTSG-6 was used for immunization of TSG-6-deficient mice and subsequent generation of TSG-6-specific mAbs (clones NG2, NG3, NG4, NG5, and NG8). rmTSG-6 also served as a reference standard (supplemental Fig. S1D) for the development and use of mouse (and human) TSG-6 ELISA systems.
Serum TSG-6 Concentrations Correlate with Arthritis Severity and Serum Levels of Proinflammatory Cytokines but Not with Immune Responses in PGIA
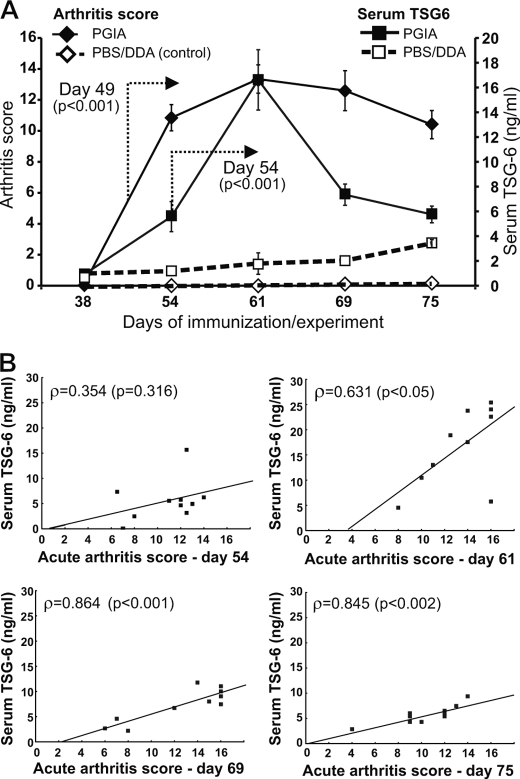
Purified rmTSG-6 and optimally paired mAbs were used to measure TSG-6 concentrations in serum and synovial fluid samples and in tissue extracts of inflamed joints of BALB/c mice immunized with cartilage PG on days 0, 21, and 42 to induce PGIA. Serum TSG-6 reached the maximum levels by day 61 of immunization, around the time when the acute joint inflammation (swelling, redness, and cell infiltration) also reached the maximum (Fig. 2, A and 3). However, whereas joint inflammation (arthritis score) declined slowly (Fig. 2A, solid diamonds), serum TSG-6 concentrations declined more rapidly (Fig. 2A, solid squares). Low levels of TSG-6 could also be detected in the serum of PBS/DDA-injected mice (open squares in Fig. 2A), which was probably due to the activation of innate immune cells at the injection site (peritoneal cavity) (32). Thus, the amount of TSG-6 in serum can be regarded as an indicator of preinflammatory and inflammatory processes (37) and appears to be a more reliable “biomarker” of the severity of acute disease than the levels of proinflammatory cytokines in PGIA (Table 1). The correlation between arthritis score and serum TSG-6 was not significant at the early stage of disease (day 54) but became significant at the peak of arthritis (day 61) and remained such during progression toward chronic disease (Fig. 2B and Table 1). However, TSG-6 became undetectable in serum samples harvested from mice at late stages of PGIA (120–150 days after the first immunization), when acute synovial inflammation had given way to pathologic joint remodeling, leading to deformities and loss of function (data not shown).
FIGURE 2.
Correlation between arthritis severity and serum TSG-6 levels at different time points during the development of PGIA. A, mice immunized with human cartilage PG in DDA adjuvant were scored for arthritis severity twice a week after the second immunization (week 21), and serum was collected weekly up to day 75 after the first immunization. Control groups were injected with PBS/DDA emulsion. Arthritis scores on the left Y axis, and serum TSG-6 concentrations on the right y axis are shown at five selected time points of immunization. Data shown are the mean ± S.E. (error bars) (n = 10 mice/group/time point). Statistically significant differences (p < 0.001) between the PGIA and PBS/DDA-injected groups from the indicated time point are depicted with arrows. Data were analyzed using repeated measures analysis of variance. B, correlations between serum TSG-6 levels are indicated by Spearman's ρ values and corresponding p values at four different time points after the first PG/DDA immunization.
TABLE 1.
Serum TSG-6 and cytokine concentrations and their correlation with arthritis and/or serum TSG-6 levels at selected time points in PGIA
The numbers in the first rows indicate mean ± S.E. of TSG-6 or cytokine concentrations (pg/ml) in serum at five selected time points (days 38, 54, 61, 69, and 75 after the first PG immunization) (n = 10 mice). Numbers in parentheses in italic type (Spearman's correlation coefficient/corresponding p value) in the second and third rows depict significant correlation with serum TSG-6 levels and arthritis score, respectively. NS, correlation is not significant.
| Day 38 | Day 54 | Day 61 | Day 69 | Day 75 | |
|---|---|---|---|---|---|
| TSG-6 | 0.0 | 5.59 ± 1.31 | 16.6 ± 2.45 | 7.34 ± 1.06 | 5.77 ± 0.57 |
| Arthritis | (NS) | (NS) | (0.631/0.050) | (0.864/0.001) | (0.845/0.002) |
| IL-6 | 6.68 ± 0.68 | 17.5 ± 1.57 | 19.18 ± 3.29 | 14.30 ± 0.85 | 15.65 ± 1.01 |
| TSG-6 | (NS) | (NS) | (0.760/0.017) | (NS) | (NS) |
| Arthritis | (NS) | (NS) | (0.720/0.029) | (NS) | (NS) |
| IL-17 | 0.61 ± 0.39 | 5.27 ± 0.74 | 7.54 ± 1.30 | 5.35 ± 0.47 | 4.81 ± 0.20 |
| TSG-6 | (NS) | (NS) | (0.894/0.007) | (NS) | (NS) |
| Arthritis | (NS) | (NS) | (0.935/0.002) | (NS) | (NS) |
| TNF-α | 1.38 ± 0.51 | 9.27 ± 0.77 | 9.28 ± 0.39 | 8.34 ± 1.14 | 8.25 ± 0.31 |
| TSG-6 | (NS) | (NS) | (NS) | (0.841/0.018) | (NS) |
| Arthritis | (NS) | (NS) | (NS) | (0.741/0.057) | (NS) |
| IL-1β | 7.40 ± 0.92 | 9.50 ± 1.23 | 9.20 ± 1.20 | 9.31 ± 0.96 | 7.85 ± 0.89 |
| TSG-6 | (NS) | (NS) | (NS) | (NS) | (NS) |
| Arthritis | (NS) | (NS) | (NS) | (NS) | (NS) |
Whereas serum TSG-6 concentrations showed a strong positive correlation with arthritis severity from day 61 to 75 after the first immunization (Fig. 2A and Table 1), serum levels of other “arthritis signature” proinflammatory cytokines (such as IL-6, IL-17, and TNF-α) correlated with the arthritis scores and serum TSG-6 at the acute or subacute phase (day 61 or 69) of PGIA, whereas serum IL-1β concentrations increased in response to immunization and subsequent arthritis onset but did not seem to correlate with disease severity or serum TSG-6 (Table 1). We could not detect any correlation between serum TSG-6 levels and the concentrations of anti-PG Abs in serum or the magnitude of PG-specific T-cell responses (data not shown).
TSG-6 in Synovial Fluid and Tissue Extracts of Inflamed Joints of Mice with PGIA
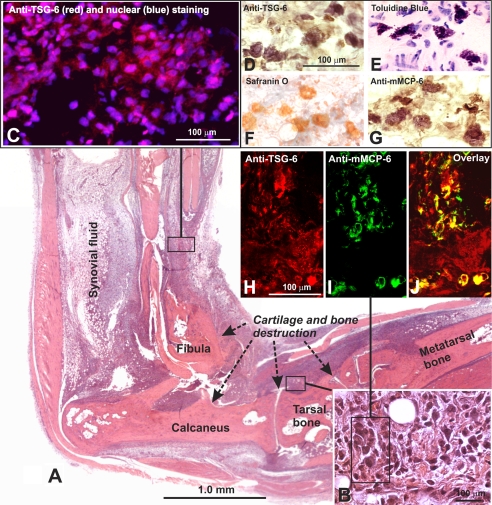
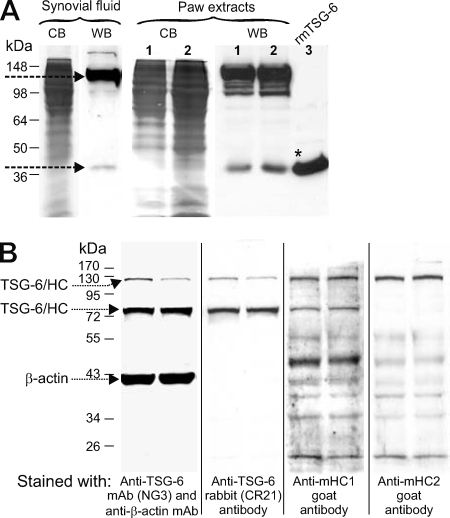
As illustrated in the histology panel in Fig. 4A, at an advanced stage of acute arthritis (61 days after the first immunization), the involved paws were heavily infiltrated by inflammatory cells and also showed histological evidence of cartilage and bone destruction. At this stage of PGIA, we harvested synovial fluid from arthritic ankle joints and prepared tissue extracts from the inflamed paws for measurement of TSG-6 content by ELISA and Western blotting. The amount of TSG-6 in pooled samples of synovial fluid was 110 ± 7 pg/mg protein, and it ranged from 20 to 550 pg/mg protein in the extracts of inflamed paws and from 0 to 60 pg/mg protein in non-arthritic control paws, as determined by ELISA. Western blotting demonstrated the presence of TSG-6 in both synovial fluid and tissue samples (Fig. 3A). In both cases, TSG-6 was found mostly in the form of high molecular weight complexes (most likely composed of TSG-6 and a heavy chain (HC1 and/or HC2) of IαI (3, 25)); only small amounts of the free form could be detected (Fig. 3A). Interestingly, synovial fluid contained a single TSG-6-positive band at ∼120–125 kDa, whereas the extracts of inflamed paws showed multiple bands below that size (Fig. 3A, lanes 1 and 2). A Western blot of paw extracts revealed two TSG-6 bands at ∼125 and ∼80 kDa, respectively (Fig. 3B). Both of these bands were also recognized by an anti-mouse HC1 Ab, but only the upper band was recognized by anti-HC2 Ab (Fig. 3B).
FIGURE 4.
Histology of the inflamed paw on day 61 of PGIA and localization of TSG-6 in inflamed tissue and mast cells. A, the entire ankle-paw region of an arthritic hind limb is shown in a low magnification montage picture. The section was stained with H&E. The arrows depict areas of cartilage and bone destruction by inflammatory synovium, and the area from which synovial fluid was harvested is also indicated. B, a high magnification insert at the bottom right corner represents inflamed periarticular tissue where TSG-6-positive cells (including mast cells) were found in frozen sections of hind limbs with similar arthritis scores. C, immunostaining of inflamed tissue with anti-TSG-6 mAbs (red fluorescence) identified several TSG-6-expressing cells (nuclei were stained blue). D, mast cells, which showed strong and distinct immunostaining for TSG-6 in their secretory granules, were identified by toluidine blue (E), safranin O (F), and anti-mouse mast cell protease (G) (mMCP-6; mast cell tryptase) staining. In areas similar to that framed in B TSG-6 (red) (H) and mMCP-6 (green) (I) co-localized in mast cells as shown in the overlaid image (yellow) (J).
FIGURE 3.
Western blot analysis of TSG-6 in synovial fluid and in tissue extracts of inflamed paws of mice with PGIA. A, the Coomassie Blue (CB)-stained strips represent total protein from synovial fluid (first lane) or tissue extracts of arthritic paws (first and second lanes in the left panel under Paw extracts), harvested on the peak of arthritis (day 61). Pooled synovial fluids (50 μl) after digestion with Streptomyces hyaluronidase or from 2 mg (lane 1) and 4 mg (lane 2) of protein of paw extracts from arthritic mice were separated by 12% SDS-PAGE. Lane 3 contains 1 μg of purified rmTSG-6 protein (*) (without Fc2a tail). TSG-6 was detected with biotinylated RC21 antibody. The arrows point to the high molecular mass (∼120-kDa) TSG-6 complex and free (∼39-kDa) TSG-6 (B) Western blot analysis of TSG-6 and the heavy chains (HC1 and HC2) of IαI in tissue extracts of inflamed paws. Tissue extracts from two arthritic paws were loaded on 12% SDS-PAGE (20 μg of protein/lane). The proteins were transferred onto a nitrocellulose membrane, probed with mAbs to TSG-6 and β-actin, and then stripped and sequentially reprobed with RC21 anti-TSG-6 antibody, anti-HC1, and anti-HC2 antibody, respectively. The arrows point to the TSG-6-containing complexes (no free TSG-6 was detected in this case) and to β-actin (loading control). WB, Western blot.
Cellular Localization of TSG-6 in the Inflamed Paws of Mice with PGIA
Using immunohistochemistry on frozen sections of arthritic hind limbs (similar to that shown in Fig. 4, A and B), we could detect TSG-6 in most of the cells of the inflamed tissue (Fig. 4, C and H). Unexpectedly, the strongest immunostaining appeared to be localized within large, granulated cells (Fig. 4D). Such cells also showed metachromatic staining with toluidine blue (Fig. 4E) and red staining with safranin O (Fig. 4F), indicating that they could be connective tissue mast cells. To further identify this cell type, we used an Ab against the mast cell-restricted tryptase mMCP-6 for immunostaining of tissue sections. Indeed, anti-TSG-6 (Fig. 4H) and anti-mMCP-6 (Fig. 4, G and I) Abs both stained the same type of cell. Co-localization of TSG-6 (Fig. 4H, red) and mMCP-6 (Fig. 4I, green) was further confirmed by the overlay of images (Fig. 4J, yellow) of sections simultaneously stained with fluorescent Abs against the respective molecules.
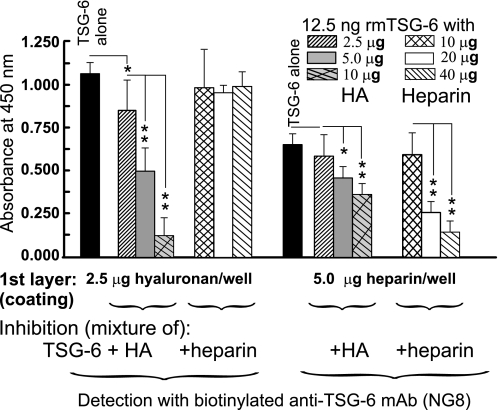
HA Is a Stronger Competitor Than Heparin for TSG-6 Binding
rmTSG-6 bound to either HA- or heparin-coated plates, although the amount of TSG-6 bound to heparin appeared to be less than the amount that bound to HA. This difference might be due to the different molecular mass of HA versus heparin or different affinity of TSG-6 with HA or heparin (10). Although TSG-6 binding sites to HA and heparin are different (10, 38), and an octasaccharide of HA is sufficient to bind TSG-6, these two GAGs may compete for and/or interfere with TSG-6 binding either in vivo or in vitro. In a competitive inhibition assay, we compared how HA or heparin could compete for TSG-6 binding (Fig. 5). Indeed, whereas the in vitro preformed TSG-6·heparin complex could not inhibit TSG-6 binding to HA, the in vitro preformed TSG-6·HA complex inhibited TSG-6 binding to heparin. The controls showed that the TSG-6·HA complex inhibited binding to HA, and the TSG-6·heparin complex inhibited the TSG-6 binding to heparin (Fig. 5).
FIGURE 5.
Competitive inhibition assay of TSG-6 binding to HA and heparin. The optimal range of TSG-6 binding to HA- or heparin-coated plates was determined and then a competitive inhibition assay was performed. Three different concentrations were used for coating (2.5, 5, and 10 μg of HA or 5.0, 10, and 20 μg of heparin), and four different concentrations of TSG-6 in duplicate wells were assayed simultaneously in two plates. TSG-6 alone (50 ng in 100 μl of 0.5% BSA/PBS, closed columns), or the same amounts of TSG-6 were preincubated with HA or heparin (amounts of these GAGs are indicated) for 2 h at 37 °C prior to transferring to either HA- or heparin-coated wells. TSG-6-binding was detected with biotinylated mAb NG8, and the reaction was developed with TMB substrate. Heparin only slightly inhibited or did not inhibit the binding of TSG-6 to HA, whereas HA could significantly inhibit TSG-6 binding to heparin (in a concentration-dependent manner). Significant differences are indicated with asterisks (*, p < 0.05; **, p < 0.01). Error bars, S.E.
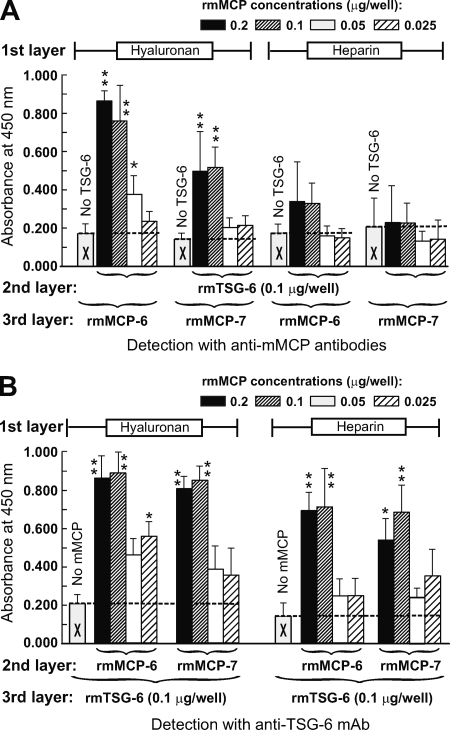
TSG-6 Binds Mast Cell Tryptases in the Presence of Heparin or HA
Co-localization of TSG-6 and mMCP-6 in mast cells in inflamed joint tissue suggested that TSG-6 might be stored in mast cell granules, which are known to contain tryptases (mMCP-6 (22) and mMCP-7 (39)) and heparin (40). Among other constituents, TSG-6 has been shown to bind either HA (1, 8) or heparin (10). In addition, TSG-6 can form a ternary complex with IαI in the presence of HA (3) or heparin (10). Therefore, it was of interest to determine whether TSG-6 could interact with mast cell tryptases in a similar manner in vitro. rmTSG-6, prebound to HA, could bind rmMCP-6 or (to a lesser extent) rmMCP-7, but this was not the case with TSG-6 prebound to heparin (Fig. 6A). In a reciprocal system, rmMCP-6 and rmMCP-7, bound to either HA or heparin, could bind TSG-6 (Fig. 6B).
FIGURE 6.
Measurement of the association of rmTSG-6 with mast cell tryptases (rmMCP-6 or rmMCP-7) and HA or heparin in vitro. The graphs represent the relative amounts of bound rmMCP-6 or rmMCP-7 after incubation of 0.1 μg of rmTSG-6/well with immobilized HA or heparin and the subsequent addition of various amounts of rmMCPs (concentrations are indicated at the top) (A) and the relative amounts of bound rmTSG-6 after incubation of rmMCP-6 or rmMCP-7 (at concentrations indicated at the top) with HA or heparin and the subsequent addition of rmTSG-6 (0.1 μg/well) (B). Immobilization of HA and heparin and subsequent binding and detection of complexes formed between rmTSG-6 and rmMCP-6 or between rmTSG-6 and rmMCP-7 are described in detail under “Experimental Procedures.” Base-line values (absorbance values resulting from direct binding of either TSG-6 or mMCPs to either HA or heparin) are indicated with empty columns with an X, and the base-line levels are indicated with horizontal broken lines. Mean ± S.E. values (error bars) of duplicate samples of three independent experiments are shown. Significant differences are indicated with asterisks (*, p < 0.05; **, p < 0.01).
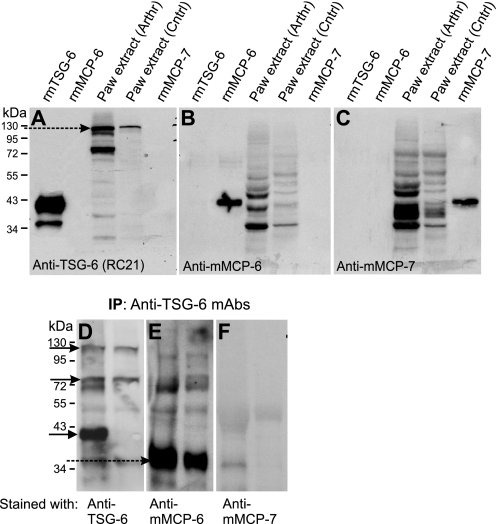
TSG-6 Can Be Co-immunoprecipitated with Mast Cell Tryptases from Tissue Extracts of Arthritic Joints
TSG-6, mMCP-6, and mMCP-7 were present in relatively high amounts in tissue extracts of inflamed paws but were detected in lower amounts in tissue extracts of non-inflamed paws (Fig. 7, A–C). TSG-6 could be co-immunoprecipitated with both tryptases (especially with mMCP-6) (Fig. 7, D–F), suggesting that complex formation between TSG-6 and mast cell tryptases (most likely via HA or heparin binding in secretory granules) did occur in vivo under inflammatory conditions.
FIGURE 7.
Western blots of TSG-6 and mast cell tryptases in arthritic and normal paw extracts and co-immunoprecipitation of in vivo formed TSG-6-tryptase complexes. A–C, Western blots show the same membrane, which was stained with an antibody to TSG-6 (A) and then stripped and restained with antibodies to mMCP-6 (B) and mMCP-7 (C), respectively. The lanes were loaded with samples as follows: 0.2 μg of purified rmTSG-6, 0.1 μg of recombinant mMCP-6, tissue extract (50 μg of protein) from arthritic mouse paw (Arthr), extract (50 μg of protein) from normal control paw (Cntrl), and 0.05 μg of mMCP-7. D–F, extracts of arthritic (left lanes) or normal (right lanes) mouse paws were immunoprecipitated (IP) with a mixture of anti-TSG-6 mAbs (as described under “Experimental Procedures”) and then blotted (WB) with anti-TSG-6 (RC21) (D), anti-mMCP-6 (E), and anti-mMCP-7 antibodies (F). The arrows point to the highest and lowest molecular weight TSG-6 species in D. The broken arrow indicates the TSG-6 coimmunoprecipitated mMCPs.
DISCUSSION
The beneficial effects of TSG-6 treatment have been described in different murine models of rheumatoid arthritis (5–7, 41), and the protective function of this protein is consistent with the development of more severe arthritis in TSG-6-deficient mice than in wild type mice upon immunization with cartilage PG (21). In adoptively transferred PGIA, TSG-6 gene expression was detected in synovial tissue of recipient mice before the clinical symptoms of arthritis developed (37), and TSG-6 was also described as a signature gene of early inflammatory events in a number of other diseases (for more details, see reviews (42, 43)). However, no systemic studies have been carried out to investigate the kinetics of TSG-6 production during the development and progression of any inflammatory joint disease, including rheumatoid arthritis.
As a first step of generating a highly sensitive assay, we constructed an expression vector that contained both TSG-6 and mouse IgG2a-Fc fusion partner cDNA, separated by an endoprotease cleavage site (Ile-Glu-Gly-Arg) specific for factor Xa (which converts prothrombin to thrombin). We also developed a stepwise cloning procedure for rapid selection of CHO transfectants with the highest yield of rmTSG-6 fusion protein released into serum-free medium. rmTSG-6 protein was then used to generate B cell hybridomas from TSG-6-deficient mice (21) and to select pairs of mAbs for capture ELISAs, where rmTSG-6 could also be employed as a reference standard. Some of the anti-TSG-6 mAbs were also suitable for immunoprecipitation, Western blotting, and immunohistochemistry. With this repertoire of detection tools, we immunized BALB/c mice with cartilage PG to induce PGIA, with the goals to determine the kinetics of serum TSG-6 levels in correlation with arthritis severity and other disease markers and to detect the accumulation of this protein at the site of inflammation.
Serum TSG-6 concentrations showed significant positive correlation with the progression of arthritis (Fig. 2). In this regard, TSG-6 appeared to be a better biomarker of joint inflammation than the arthritis-related proinflammatory cytokines IL-6, IL-17, TNF-α, and IL-1β, which showed correlation with the disease development only at certain time points or not at all (Table 1). Serum TSG-6 levels declined when arthritis progressed toward the chronic phase (Fig. 2), during which massive tissue destruction (Fig. 4) was followed by synovial fibrosis and bone remodeling (21, 31). We could accurately measure TSG-6 levels in synovial fluid samples and tissue extracts of severely inflamed paws of mice. Western blots carried out on these samples revealed that the majority of TSG-6 was present as a high molecular weight species (Fig. 3), indicating that at these inflammatory sites, TSG-6 was probably complexed with HCs from IαI, as described before (3, 7). The inflammatory paw extracts consistently contained both a ∼125-kDa species (probably a TSG-6·HC1 and a TSG-6·HC2 complex) (Figs. 3 and 7) and an ∼80-kDa species (Fig. 3B). The latter band could be a degradation product of the TSG-6·HC1 complex and was not detected in synovial fluids of arthritic joints.
For the first time, we were able to immunolocalize TSG-6-producing cells in acutely inflamed joint tissues of mice using the newly generated mAbs. Whereas most cells in the arthritic joints stained positively for TSG-6 at low to moderate levels, very high amounts of TSG-6 protein were detected in the secretory granules of mast cells, most of which cells showed evidence of activation (degranulation) in the inflamed tissue (Fig. 4). Beyond metachromatic and positive safranin O staining, we sought to identify these cells more precisely by employing Abs to mouse mast cell-specific proteins. An Ab against the mast cell-specific tryptase mMCP-6 (also a component of secretory granules) gave a signal that overlapped with TSG-6-specific staining, indicating colocalization of these two proteins in mast cell granules (Fig. 4).
The fact that both mMCP-6 (22, 39) and TSG-6 (10) can bind heparin prompted us to investigate whether TSG-6 could associate with mMCP-6 via heparin. Indeed, our in vitro studies revealed a tripartite association of rmTSG-6 with rmMCP-6 (and, to a lesser extent, with rmMCP-7) through heparin as well as through HA (Fig. 6). mMCP-6 and, to a lesser extent, mMCP-7 could be pulled down by TSG-6 immunoprecipitates of arthritic paw extracts (Fig. 7), suggesting that these proteins were associated (or formed a complex) in vivo under the conditions of joint inflammation.
Mast cells constitute the first line of defense in various IgE-mediated allergic reactions and helminth infections, and they are strategically positioned near blood vessels, which enables them to capture allergen-specific IgE, IgG, or other substances from both blood and tissue (44). Mast cell secretory granules contain proteases, GAGs, and proinflammatory cytokines that are released into extracellular matrix upon degranulation (22). Tryptases, such as mMCP-6 and mMCP-7 (murine orthologs of human β-tryptases), are the predominant serine proteases stored in mast cell secretory granules (45). These enzymes are tightly packaged in complexes with serglycin PGs through electrostatic interactions between their positively charged amino acid residues and the negatively charged GAG (heparin or chondroitin sulfate E) side chains of serglycin (39, 45). In addition to packaging, heparin may play a role in the maintenance of tryptases in an active conformation at low pH inside the mast cell granules, but proteolytic activity increases at neutral pH after release of the tryptase-heparin complexes into the extracellular environment (39, 45).
The presence of TSG-6 in mast cells has not been reported. Here we have found that the tryptases mMCP-6 and mMCP-7, upon binding to either HA or heparin, associate with TSG-6. As noted above, HA-bound TSG-6 has a stronger association with both tryptases than heparin-bound TSG-6 (Fig. 6). Some of the differences between HA- and heparin-bound complexes might be explained with the better adsorption of high molecular mass HA (1,000–2,000 kDa) than low molecular mass heparin (12–15 kDa) to plastic surfaces. However, in a competitive inhibition assay (Fig. 5), we found that HA inhibited the binding of rmTSG-6 to heparin, but heparin did not inhibit the binding of rmTSG-6 to HA. Nonetheless, enhancement of TSG-6-tryptase interactions on the GAG-coated surfaces is obvious under the conditions of our in vitro assays (Fig. 6). Moreover, co-immunoprecipitation of TSG-6 and tryptases (Fig. 7) suggests that complex formation may also occur in vivo. However, more extensive studies are required to specify the type of molecular interactions among HA, heparin, TSG-6, and tryptases.
What could be the relevance of these tripartite interactions to pathophysiological events involving mast cells? As mentioned before, TSG-6 readily forms a covalent complex with the HC1 or HC2 subunit of the serine protease inhibitor IαI (composed of two HCs bound to the chondroitin sulfate chain on bikunin, a Kunitz type protease inhibitor) and augments the inhibitory capacity of this molecule (3). Recent studies indicate that potentiation of IαI activity is achieved by the TSG-6-mediated transfer of a HC from the single chondroitin sulfate side chain of bikunin onto HA (46, 47). This is followed by the release of bikunin with a single HC (i.e. pre-IαI) or without HC (although release of free bikunin has not been observed in vitro). Bikunin, which harbors the protease-inhibitory activity of IαI, is more active in the absence than in the presence of the HCs on its GAG chain (10). Intriguingly, the tryptase inhibitor (designated “trypstatin”) present in mast cells has been identified as bikunin (48, 49). Because only bikunin protein but not mRNA could be detected in mast cells, the authors concluded that bikunin was trapped by these cells from serum of circulating blood (48, 49). It is tempting to speculate that TSG-6 has a role in liberating bikunin from IαI and trapping it inside the mast cell granules or on the surface of granule membranes. Upon mast cell exocytosis (i.e. the inflammation-induced release of granule contents into the extracellular space), the tryptases would meet bikunin and thus be inactivated. Whether or not TSG-6 plays a regulatory role in these tryptase-bikunin interactions in mast cell granules remains to be determined.
The role of mast cells in non-IgE-mediated diseases, such as arthritis, is being increasingly recognized. Mast cells are abundant in inflamed synovial tissues of both humans and mice (50, 51). Mast cell-restricted tryptases can convert matrix metalloproteinase-3 (stromelysin) from latent to active form, which cleaves cartilage aggrecan and also activates synoviocyte procollagenase (52). Mice lacking mast cells (W/Wv mutant) are less susceptible to inflammatory arthritis than the wild type counterparts (53). Moreover, mice deficient in both mMCP-6 and mMCP-7 or lacking the enzyme responsible for heparin biosynthesis are remarkably protected from cartilage degradation after induction of destructive arthritis (54). In our earlier studies, intra-articularly injected rmTSG-6 (5) and cartilage-specific expression of a TSG-6 transgene (7) were both found to exhibit substantial chondroprotective effects in murine models of arthritis. These observations are consistent with the involvement of TSG-6 in the negative regulation of the activity of matrix-degrading proteases, probably including mast cell-restricted tryptases.
Mast cell granule components, specifically mMCP-6·heparin complexes, also stimulate neutrophil extravasation via induction of chemokines (55). Influx of neutrophils into the joints is a prominent feature of arthritis in both humans and experimental animals (56, 57). Systemic administration of rmTSG-6 to mice with PGIA significantly suppressed leukocyte infiltration into the joints (5). TSG-6 (or its isolated Link module) also inhibited neutrophil influx into zymosan-stimulated air pouches, apparently due to reduced amounts of neutrophil-attracting chemokines at the site of inflammation (58). Conversely, the most distinctive phenotype of TSG-6-deficient mice is the exaggerated recruitment of neutrophils in the joints after the development of PGIA (21) and in the peritoneal cavity after peritonitis induction (21). Collectively, these data argue for a role of TSG-6 in the resolution of inflammation, in part due to a negative influence on mast cell protease-induced chemokine production and subsequent leukocyte extravasation. These novel functions of TSG-6, proposed here in inflammatory joint destruction, warrant further investigation.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants P01 AR45652, R01 AR051163, R01 AR40310, and R01 HL081064. This work was also supported by a grant from the Grainger Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HA
- hyaluronan
- GAG
- glycosaminoglycan
- HC
- heavy chain
- IαI
- inter-α-inhibitor
- IP
- immunoprecipitation
- mMCP-6 and -7
- murine mast cell protease-6 and -7, respectively
- rmTSG-6
- recombinant mouse TSG-6
- rmMCP-6 and -7
- recombinant mouse MCP-6 and -7, respectively
- PG
- proteoglycan
- PGIA
- proteoglycan-induced arthritis
- Ab
- antibody
- TMB
- tetramethylbenzidine substrate
- FL-HA
- fluorescein-conjugated HA
- DDA
- dimethyldioctadecyl-ammonium bromide.
REFERENCES
- 1. Lee T. H., Wisniewski H. G., Vilcek J. (1992) J. Cell Biol. 116, 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wisniewski H. G., Maier R., Lotz M., Lee S., Klampfer L., Lee T. H., Vilcek J. (1993) J. Immunol. 151, 6593–6601 [PubMed] [Google Scholar]
- 3. Wisniewski H. G., Burgess W. H., Oppenheim J. D., Vilcek J. (1994) Biochemistry 33, 7423–7429 [DOI] [PubMed] [Google Scholar]
- 4. Stanker L. H., Vanderlaan M., Juarez-Salinas H. (1985) J. Immunol. Methods 76, 157–169 [DOI] [PubMed] [Google Scholar]
- 5. Bárdos T., Kamath R. V., Mikecz K., Glant T. T. (2001) Am. J. Pathol. 159, 1711–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mindrescu C., Thorbecke G. J., Klein M. J., Vilcek J., Wisniewski H. G. (2000) Arthritis Rheum. 43, 2668–2677 [DOI] [PubMed] [Google Scholar]
- 7. Glant T. T., Kamath R. V., Bárdos T., Gál I., Szántó S., Murad Y. M., Sandy J. D., Mort J. S., Roughley P. J., Mikecz K. (2002) Arthritis Rheum. 46, 2207–2218 [DOI] [PubMed] [Google Scholar]
- 8. Kohda D., Morton C. J., Parkar A. A., Hatanaka H., Inagaki F. M., Campbell I. D., Day A. J. (1996) Cell 86, 767–775 [DOI] [PubMed] [Google Scholar]
- 9. Parkar A. A., Day A. J. (1997) FEBS Lett. 410, 413–417 [DOI] [PubMed] [Google Scholar]
- 10. Mahoney D. J., Mulloy B., Forster M. J., Blundell C. D., Fries E., Milner C. M., Day A. J. (2005) J. Biol. Chem. 280, 27044–27055 [DOI] [PubMed] [Google Scholar]
- 11. Bork P., Beckmann G. (1993) J. Mol. Biol. 231, 539–545 [DOI] [PubMed] [Google Scholar]
- 12. Zhang P., Pan W., Rux A. H., Sachais B. S., Zheng X. L. (2007) Blood 110, 1887–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ng D., Pitcher G. M., Szilard R. K., Sertié A., Kanisek M., Clapcote S. J., Lipina T., Kalia L. V., Joo D., McKerlie C., Cortez M., Roder J. C., Salter M. W., McInnes R. R. (2009) PLoS Biol. 7, e41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Major B., Kardos J., Kékesi K. A., Lorincz Z., Závodszky P., Gál P. (2010) J. Biol. Chem. 285, 11863–11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuznetsova S. A., Mahoney D. J., Martin-Manso G., Ali T., Nentwich H. A., Sipes J. M., Zeng B., Vogel T., Day A. J., Roberts D. D. (2008) Matrix Biol. 27, 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wisniewski H. G., Hua J. C., Poppers D. M., Naime D., Vilcek J., Cronstein B. N. (1996) J. Immunol. 156, 1609–1615 [PubMed] [Google Scholar]
- 17. Maina V., Cotena A., Doni A., Nebuloni M., Pasqualini F., Milner C. M., Day A. J., Mantovani A., Garlanda C. (2009) J. Leukoc. Biol. 86, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lesley J., English N. M., Gál I., Mikecz K., Day A. J., Hyman R. (2002) J. Biol. Chem. 277, 26600–26608 [DOI] [PubMed] [Google Scholar]
- 19. Lesley J., Gál I., Mahoney D. J., Cordell M. R., Rugg M. S., Hyman R., Day A. J., Mikecz K. (2004) J. Biol. Chem. 279, 25745–25754 [DOI] [PubMed] [Google Scholar]
- 20. Hutás G., Bajnok E., Gál I., Finnegan A., Glant T. T., Mikecz K. (2008) Blood 112, 4999–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szántó S., Bárdos T., Gál I., Glant T. T., Mikecz K. (2004) Arthritis Rheum. 50, 3012–3022 [DOI] [PubMed] [Google Scholar]
- 22. Pejler G., Rönnberg E., Waern I., Wernersson S. (2010) Blood 115, 4981–4990 [DOI] [PubMed] [Google Scholar]
- 23. Fülöp C., Kamath R. V., Li Y., Otto J. M., Salustri A., Olsen B. R., Glant T. T., Hascall V. C. (1997) Gene 202, 95–102 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 25. Fülöp C., Szántó S., Mukhopadhyay D., Bárdos T., Kamath R. V., Rugg M. S., Day A. J., Salustri A., Hascall V. C., Glant T. T., Mikecz K. (2003) Development 130, 2253–2261 [DOI] [PubMed] [Google Scholar]
- 26. Köhler G., Milstein C. (1975) Nature 256, 495–497 [DOI] [PubMed] [Google Scholar]
- 27. Glant T. T., Mikecz K., Poole A. R. (1986) Biochem. J. 234, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Glant T. T., Cs-Szabó G., Nagase H., Jacobs J. J., Mikecz K. (1998) Arthritis Rheum. 41, 1007–1018 [DOI] [PubMed] [Google Scholar]
- 29. Glant T. T., Mikecz K. (2004) Methods Mol. Med. 102, 313–338 [DOI] [PubMed] [Google Scholar]
- 30. Glant T. T., Mikecz K., Arzoumanian A., Poole A. R. (1987) Arthritis Rheum. 30, 201–212 [DOI] [PubMed] [Google Scholar]
- 31. Glant T. T., Finnegan A., Mikecz K. (2003) Crit. Rev. Immunol. 23, 199–250 [DOI] [PubMed] [Google Scholar]
- 32. Hanyecz A., Berlo S. E., Szántó S., Broeren C. P., Mikecz K., Glant T. T. (2004) Arthritis Rheum. 50, 1665–1676 [DOI] [PubMed] [Google Scholar]
- 33. Adarichev V. A., Valdez J. C., Bárdos T., Finnegan A., Mikecz K., Glant T. T. (2003) J. Immunol. 170, 2283–2292 [DOI] [PubMed] [Google Scholar]
- 34. Jaen O., Rullé S., Bessis N., Zago A., Boissier M. C., Falgarone G. (2009) Immunology 126, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zielen S., Bröker M., Strnad N., Schwenen L., Schön P., Gottwald G., Hofmann D. (1996) J. Immunol. Methods 193, 1–7 [DOI] [PubMed] [Google Scholar]
- 36. Shi M., Dennis K., Peschon J. J., Chandrasekaran R., Mikecz K. (2001) J. Immunol. 167, 123–131 [DOI] [PubMed] [Google Scholar]
- 37. Adarichev V. A., Vermes C., Hanyecz A., Mikecz K., Bremer E. G., Glant T. T. (2005) Arthritis Res. Ther. 7, R196–R207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Higman V. A., Blundell C. D., Mahoney D. J., Redfield C., Noble M. E., Day A. J. (2007) J. Mol. Biol. 371, 669–684 [DOI] [PubMed] [Google Scholar]
- 39. Kolset S. O., Prydz K., Pejler G. (2004) Biochem. J. 379, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eggli P. S., Graber W. (1993) J. Invest. Dermatol. 100, 121–125 [DOI] [PubMed] [Google Scholar]
- 41. Mindrescu C., Dias A. A., Olszewski R. J., Klein M. J., Reis L. F., Wisniewski H. G. (2002) Arthritis Rheum. 46, 2453–2464 [DOI] [PubMed] [Google Scholar]
- 42. Milner C. M., Day A. J. (2003) J. Cell Sci. 116, 1863–1873 [DOI] [PubMed] [Google Scholar]
- 43. Wisniewski H. G., Vilcek J. (2004) Cytokine Growth Factor Rev. 15, 129–146 [DOI] [PubMed] [Google Scholar]
- 44. Galli S. J., Grimbaldeston M., Tsai M. (2008) Nat. Rev. Immunol. 8, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McNeil H. P., Adachi R., Stevens R. L. (2007) J. Biol. Chem. 282, 20785–20789 [DOI] [PubMed] [Google Scholar]
- 46. Mukhopadhyay D., Asari A., Rugg M. S., Day A. J., Fülöp C. (2004) J. Biol. Chem. 279, 11119–11128 [DOI] [PubMed] [Google Scholar]
- 47. Sanggaard K. W., Hansen L., Scavenius C., Wisniewski H. G., Kristensen T., Thøgersen I. B., Enghild J. J. (2010) Biochim. Biophys. Acta 1804, 1011–1019 [DOI] [PubMed] [Google Scholar]
- 48. Itoh H., Ide H., Ishikawa N., Nawa Y. (1994) J. Biol. Chem. 269, 3818–3822 [PubMed] [Google Scholar]
- 49. Ide H., Itoh H., Yoshida E., Kobayashi T., Tomita M., Maruyama H., Osada Y., Nakahata T., Nawa Y. (1999) Cell Tissue Res. 297, 149–154 [DOI] [PubMed] [Google Scholar]
- 50. Crisp A. J., Chapman C. M., Kirkham S. E., Schiller A. L., Krane S. M. (1984) Arthritis Rheum. 27, 845–851 [DOI] [PubMed] [Google Scholar]
- 51. Shin K., Gurish M. F., Friend D. S., Pemberton A. D., Thornton E. M., Miller H. R., Lee D. M. (2006) Arthritis Rheum. 54, 2863–2871 [DOI] [PubMed] [Google Scholar]
- 52. Gruber B. L., Marchese M. J., Suzuki K., Schwartz L. B., Okada Y., Nagase H., Ramamurthy N. S. (1989) J. Clin. Invest. 84, 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee D. M., Friend D. S., Gurish M. F., Benoist C., Mathis D., Brenner M. B. (2002) Science 297, 1689–1692 [DOI] [PubMed] [Google Scholar]
- 54. McNeil H. P., Shin K., Campbell I. K., Wicks I. P., Adachi R., Lee D. M., Stevens R. L. (2008) Arthritis Rheum. 58, 2338–2346 [DOI] [PubMed] [Google Scholar]
- 55. Shin K., Nigrovic P. A., Crish J., Boilard E., McNeil H. P., Larabee K. S., Adachi R., Gurish M. F., Gobezie R., Stevens R. L., Lee D. M. (2009) J. Immunol. 182, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bjelle A., Norberg B., Sjögren G. (1982) Scand. J. Rheumatol. 11, 124–128 [DOI] [PubMed] [Google Scholar]
- 57. Angyal A., Egelston C., Kobezda T., Olasz K., László A., Glant T. T., Mikecz K. (2010) Arthritis Res. Ther. 12, R44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Getting S. J., Mahoney D. J., Cao T., Rugg M. S., Fries E., Milner C. M., Perretti M., Day A. J. (2002) J. Biol. Chem. 277, 51068–51076 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.