Abstract
Deregulated IL-12 and IL-23 production from activated myeloid lineage cells is a key driver of numerous T cell-dependent autoimmune and inflammatory diseases. IL-12 and IL-23 share a common p40 subunit encoded by Il12b, which is negatively regulated at the transcriptional level by the STAT3 (signal transducer and activator of transcription 3)-activating anti-inflammatory cytokine IL-10. We found that IL-10 targets an enhancer 10 kb upstream of the Il12b transcriptional start site. Within the enhancer, a single 10-bp site is required for the inhibitory effects of IL-10 and is bound by NFIL3 (nuclear factor, interleukin 3-regulated), a B-ZIP transcription factor. Myeloid cells lacking NFIL3 produce excessive IL-12p40 and increased IL-12p70. Thus, the STAT3-dependent expression of NFIL3 is a key component of a negative feedback pathway in myeloid cells that suppresses proinflammatory responses.
Keywords: Dendritic Cell, DNA Binding Protein, Gene Regulation, Macrophage, Myeloid Cell, IL-10, IL-12, NFIL-3, STAT3, p40
Introduction
IL-12 and IL-23 are heterodimeric cytokines composed of the p40-p35 and p40-p19 subunits, respectively, and are produced primarily by macrophages and dendritic cells activated by diverse inflammatory stimuli (1, 2). IL-12 is decisive in regulating lineage commitment to Th1 cell development, whereas IL-23 is essential for the maturation and stability of IL-17-producing Th17 cells (3). The IL-23 p19 subunit also has proinflammatory and protumorigenic effects that extend beyond the effects of IL-23 on Th17 lineages and IL-17 (4–7). Polymorphisms in multiple genes involved in IL-12 and IL-23 production and signaling have been discovered by genome-wide association studies for Crohn's disease, ulcerative colitis, psoriatic arthritis, psoriasis, and Behcet's disease, including IL12B (encoding p40), IL23R (encoding the IL-23 receptor), and STAT4 and STAT3 (encoding key signaling components downstream of the IL-12 and IL-23 receptors) (8–11). Consistent with these genetic studies, excessive Th1 responses are implicated in multiple forms of intestinal ailments (12, 13) coupled to specific pathogenic effects of IL-12 and IL-23 in mouse models of inflammatory bowel disease (4, 14–18). Because of the link between IL-12p40 and inflammatory diseases, several efforts to target p40 using humanized antibodies have undergone clinical trials in Crohn's disease, whereas IL-12- and IL-23-specific neutralization strategies are in development for inflammatory disorders (19, 20), and one, ustekinumab (Stelara), is approved for psoriasis therapy.
Production of IL-12 and IL-23 is regulated at several levels. First, the common IL-12p40 subunit is transcriptionally regulated by inflammatory stimuli and often produced in excess to the mature IL-12 and IL-23 cytokines. Furthermore, p40 subunits can homodimerize, although the precise biological effects of these homodimers remain unclear (21). Second, the p35 and p19 subunits are themselves subject to transcriptional regulation. For example, p19 (encoded by Il23a) is strongly induced by yeast cell wall particles, whereas p35 is induced by Toll-like receptor 4 (TLR4)2 and TLR9 agonists. The final formation of IL-12 and IL-23 is dependent in part on the relative ability of the p40-p35 and p40-p19 heterodimers to form complexes in the endoplasmic reticulum and then be secreted.
A key step in IL-12 and IL-23 regulation is the transcriptional repression of IL-12p40 production by IL-10. By inhibiting the output of IL-12 and IL-23, IL-10 restricts the amounts of cytokine available to regulate Th1 and Th17 development and stability in immune microenvironments (22). Extensive in vivo evidence has shown that IL-10 signaling to suppress IL-12p40 is an essential mechanism to maintain inflammatory homeostasis (23–26). The underlying mechanism of IL-10-mediated p40 inhibition is thus likely to illuminate multiple components of immune regulation. IL-10 targets Il12b transcription through a mechanism that requires STAT3-dependent production of new proteins (27–29). Surprisingly, however, IL-10 does not appear to target the promoter of Il12b, where NF-κB family members, including c-Rel, bind following TLR signaling (28, 29). Here we identify a mechanism that IL-10 uses to suppress IL-12p40 transcription.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 and Il10−/− mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Nfil3−/− mice on a C57BL/6 background (30) were a gift of Tak Mak, Jillian Haight, and Thomas Look. Cebpb−/− mice (31) were on a C57BL/6:129Sv F1 background, and Cebpd−/− mice (32) were on a C57BL/6 background. All mice were used in accordance with the St. Jude Children's Research Hospital Institutional Animal Care and Use Committee.
Luciferase Reporter Constructs and Macrophage Transfections
The β-globin insulated vector containing the basal Il12b promoter driving GFP expression (33) was modified to insert luciferase and Il12b enhancer fragments as follows: The luc2 gene from pGL4.10 (Promega) was isolated with NheI and ClaI and cloned into the same sites in the Il12b-GFP reporter to replace GFP. The Il12b enhancer and promoters were isolated by PCR from C57BL/6 genomic DNA with NotI and MluI ends, or MluI and NotI ends, respectively. These two fragments were digested with the appropriate enzymes and cloned 5′ to luc2. Enhancer fragments of various lengths were cloned into the reporter vector using the same strategy. Luciferase reporter stable lines were derived by transfecting RAW264.7 macrophages as described (34). Cells were selected with G418 and expanded. To assay luciferase reporter lines, cells were plated in 12-well plates at 2 × 106 cells/well and stimulated with LPS (100 ng/ml) or LPS plus IL-10 (10 ng/ml) for 2, 4, or 8 h followed by lysis and standard luciferase assays. In some experiments, luciferase reporter constructs were used in transient assays as described (34).
Primary Macrophage and Dendritic Cell Isolation
BMDMs were isolated as described (35). PDMs were isolated following intraperitoneal injection of thioglycollate as described (36, 37). Gut and spleen CD11b+ cells were isolated by magnetic bead purification (Miltenyi Biotec). CD11c+ DCs2 were isolated by magnetic bead purification from GM-CSF cultures as described (38).
Quantitative RT-PCR
qRT-PCR was performed as described previously (39). Sequences for all oligonucleotide primers and TaqMan probes are available from P. J. M. on request.
Immunoblotting
Immunoblotting for NFIL3 was performed using polyclonal anti-NFIL3 antibodies (C-18, goat polyclonal, Santa Cruz Biotechnology, Inc., catalog no. sc-9550 or V-19, goat polyclonal Santa Cruz Biotechnology, Inc., catalog no. 9549) used at a 1:1000 dilution.
In Vitro Transcription/Translation (IVTT) and DNA Binding Assays
To produce NFIL3, we used the TnT quick coupled transcription/translation system (Promega, L1170). 1 μg of murine NFIL3 cDNA in pcDNA3.1 was used as the template in each reaction. In some experiments, cDNAs encoding C/EBPβ and arginase-1 were used under the same conditions. From the final 50 μl of IVTT product, we used 6 μl in DNA binding assays as described previously (34). Briefly, IVTT reactions were blocked with 2 μg of poly(dI-dC) on ice for 20 min, followed by binding to 32P-labeled and annealed primers at RT for 30 min.
Mass Spectrometry
We prepared nuclear extracts from 5 × 106 IL-10-deficient BMDMs stimulated with media, LPS, or LPS plus IL-10 for 2 h in two independent experiments. From the nuclear extract, 450 μg of protein was used per sample and resolved on SDS-PAGE gels followed by Sypro Ruby staining. Ten consecutive gel slices were prepared from ∼10 kDa to ∼100 kDa. Proteins were eluted from each slice and subjected to LC/MS analysis.
LPS Challenge and Toxoplasma Infections
LPS challenges were performed as described using Escherichia coli LPS (40). Mice were weighed prior to injection followed by intraperitoneal administration of LPS on a mg/kg basis. Mice were infected intraperitoneally with Toxoplasma gondii N28E2 cysts grown in human foreskin fibroblasts (40, 41). Mice were bled on day 3 post-infection and bled terminally on day 7.
Statistics
Statistics were determined by unpaired, two-tailed Student's t test and are noted by *, **, and *** for p values < 0.05, < 0.01, and < 0.001, respectively.
RESULTS
IL-10 Targets an Enhancer ∼10 kb Upstream of the Il12b Promoter
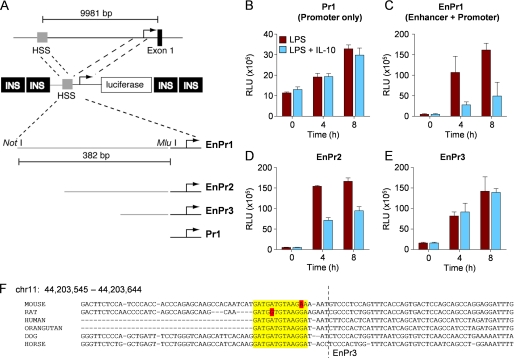
Because IL-10 potently suppresses Il12b transcription but does not target the proximal promoter (29), we suspected that IL-10 negatively controlled a distal site upstream or downstream of the Il12b promoter. We focused on a conserved enhancer ∼10 kb upstream of the Il12b start site that is targeted by TLR signaling to increase Il12b transcription in response to LPS (33). We made luciferase reporter constructs that fused the Il12b promoter to the upstream enhancer (Fig. 1A). Each reporter construct was flanked by tandem insulator elements derived from the β-globin gene to reduce the effects of surrounding chromatin in stable transfection assays (33) and to potentially allow the reporter constructs to adopt a more native chromatin structure (Fig. 1A, INS). We transfected each linearized construct into RAW264.7 cells and selected for resistance to G418. We next used each cell line in reporter assays using stimulation with LPS or LPS plus IL-10 over time. Although IL-10 did not inhibit the Il12b promoter as anticipated on the basis of experiments by Trinchieri and colleagues (29), suppression of luciferase activity was observed when the enhancer was linked to the promoter in the EnPr1 and EnPr2 constructs (Fig. 1, B–E). The enhancer was required for full inducibility to LPS as reported previously (compare Fig. 1, B and C) (33). Using deletion mutants and site-directed mutagenesis, we narrowed the IL-10 target sequence to a region bounded within 294 bp and 309 bp in the enhancer (Fig. 1, D and E, and supplemental Fig. S1). To further define the IL-10-responsive region in the enhancer, we made a series of contiguous 10-bp deletion mutants in the ∼100-bp region in the EnPr2 construct that was responsive to IL-10 inhibition. We found that both LPS-inducible enhancer activity and IL-10-mediated inhibition was ablated in two mutants (h and i) close to the boundary of EnPr2 and EnPr3 (supplemental Fig. S1). Given that the enhancer is required for full LPS inducibility of Il12b, the clearest interpretation of these data is that the 20-bp region defined by the mutants h and i is required to integrate multiple positive and negative signals that mediate normal Il12b transcriptional regulation. On the basis of the boundaries of the inhibitory effects of IL-10 on Il12b reporter activity, we concluded that the key IL-10 target site contains an evolutionarily conserved sequence (Fig. 1F).
FIGURE 1.
IL-10 restricts Il12b transcription by a mechanism that targets an enhancer. A, fusion constructs between the enhancer (depicted as hyper-sensitive site, HSS) and the Il12b promoter. Each construct was flanked by duplicated β-globin insulators (INS). Four basic constructs (EnPr1, 2, 3, and Pr1) are shown. B–E, luciferase reporter activity in stably transfected RAW264.7 macrophages are shown following stimulation with LPS or LPS plus IL-10 for the times indicated. Note that the activity of the basal Pr1 reporter is ∼5-fold less that when the enhancer is attached. Data are presented as mean relative luciferase units (RLU) plus the S.D. and are representative of 10 experiments using stable transfections and five experiments using transient transfections. F, sequence conservation between species around the region of minimal IL-10 responsiveness within the Il12b enhancer (yellow).
Analysis of IL-10-induced Transcription Factors Narrows the Field to NFIL3
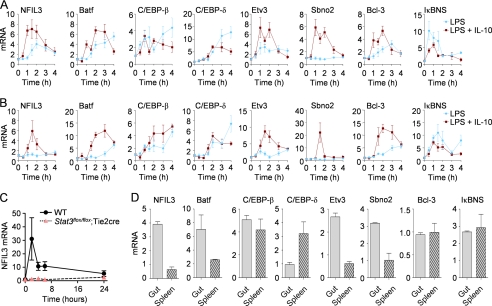
The anti-inflammatory effects of IL-10 require STAT3-mediated expression of factors that target inflammatory gene transcription (26, 27, 42, 43). To search for transcription factors that could be involved in the suppression of Il12b transcription, we used qRT-PCR to measure the expression of transcription factor mRNAs induced by IL-10 in the presence of an inflammatory costimulus. Using existing microarray screens as a starting point, we focused on C/EBPβ, C/EBPδ, NFIL3, and Batf (all B-ZIP family members); IκBNS and Bcl-3 (IκB family members); Etv3 (an ETS family repressor); and Sbno2 (a large IL-10-induced helicase) as potential candidates (39, 44). Our results from primary bone marrow-derived macrophages (BMDMs), peritoneal-derived inflammatory macrophages (PDMs) indicated that each transcription factor mRNA was induced to some extent by IL-10 in the presence of LPS with the exception of IκBNS (Fig. 2A). However, the kinetics and degree of gene expression varied widely between each candidate. Furthermore, LPS alone induced the expression of some transcription factor mRNAs with slower kinetics that LPS plus IL-10 (e.g. Fig. 2A, NFIL3 and Bcl-3). We attribute the LPS-mediated expression of each mRNA predominantly to aurocrine-paracrine IL-10 production following LPS stimulation, although other secreted factors such as IL-6 could also potentially contribute to the delayed expression kinetics of each mRNA (37, 39). To test this idea directly, we isolated BMDMs from control or Stat3flox/flox;Tie2-cre mice that have a >95% deletion of STAT3 in primary macrophages. We stimulated each population with LPS and measured NFIL3 mRNA over time. As shown in Fig. 2C, the expression of NFIL3 was ablated in the absence of STAT3. Because LPS does not signal directly through STAT3, the LPS-mediated induction of NFIL3 observed in Fig. 2, A–C is mediated by the production of autocrine-paracrine factors that signal through STAT3.
FIGURE 2.
NFIL3 mRNA and protein levels are regulated by IL-10 signaling. A and B, BMDMs (A) and PDMs (B) from C57BL/6 mice (n = 4) were stimulated with LPS or LPS plus IL-10 over time. RNA was collected and analyzed by qRT-PCR for the indicated targets, and the data are presented as the mean ± S.E. C, BMDMs from Stat3+/+;Tie2-cre (WT, n = 2) or Stat3flox/flox;Tie2-cre (n = 2) were stimulated with LPS over time, and NFIL3 mRNA amounts were detected by qRT-PCR. Data are presented as the mean NFIL3 expression relative to GAPDH expression plus the S.D. D, Gut lamina propria and splenic CD11b+ cells were collected. RNA was analyzed by qRT-PCR for the indicated targets, and the data are presented as the mean plus the S.D.
We also measured the expression of each candidate transcription factor in freshly isolated CD11b+ myeloid cells purified from the gut and spleen of wild-type mice. We observed that NFIL3, Batf, Etv3 and Sbno2 all had higher relative expression in the gut relative to the spleen (Fig. 2, B and C). Because gut macrophages are continuously exposed to IL-10, these data indicate that a subset of transcription factors are constitutively expressed in myeloid populations naturally exposed to IL-10.
To further narrow the field to transcription factors linked with the Il12b enhancer, we used a 37-bp biotinylated oligonucleotide probe spanning the IL-10 target sequence of Il12b to isolate nuclear proteins bound after 1 h of stimulation with LPS, or LPS plus IL-10, when IL-10 anti-inflammatory effects on Il12b transcription becomes measurable (27). Using qualitative mass spectrometry analysis, we compared peptide sequences between the two stimulation conditions and found that two B-ZIP proteins, NFIL3 and C/EBPδ, were enriched following LPS plus IL-10 stimulation relative to LPS stimulation alone (supplemental Table S1). NFIL3 and/or C/EBPδ may therefore contribute to the IL-10-mediated suppression of Il12b transcription.
We first, however, tested whether the related C/EBP family member C/EBPβ, previously shown to bind to the Il12b enhancer (33) and partially induced by IL-10 in PDMs (Fig. 2B), was responsible for the inhibitory effects of IL-10 on Il12b transcription. Using BMDMs from Cebpb−/− mice, we observed that the inhibitory effects of IL-10 on IL-12p40 production was intact, ruling out a direct role for this B-ZIP member in Il12b-negative regulation (supplemental Fig. S2A). We also tested whether C/EBPδ was required for regulating p40 expression in response to IL-10. Similar to our findings with Cebpb−/− macrophages, C/EBPδ did not have an essential role in IL-10-mediated inhibition of IL-12p40 transcription (supplemental Fig. S2B). At this stage it remains to be established whether C/EBPβ and C/EBPδ could have a redundant inhibitory role in the effects of IL-10 on Il12b expression, as it is plausible that multiple B-ZIP proteins could compete for the same sites in the enhancer.
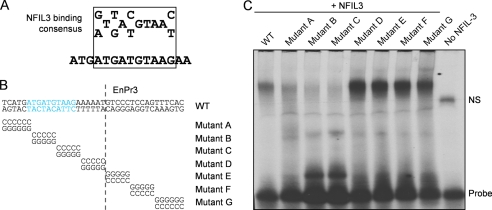
We next examined the sequences around mutations h and i in the Il12b enhancer more closely and found a putative NFIL3 consensus binding site (45) within the IL-10 target sequence. Using IVTT-generated NFIL3 coupled with mutagenesis of the IL-10 target sequence, we found that the 10-bp sequence ATGATGTAAG was specifically bound by NFIL3 but not by C/EBPβ (Fig. 3, A–C, and supplemental Fig. S3A). Furthermore, mutants in either the DNA binding or leucine zipper dimerization domains reduced binding (supplemental Fig. S3B). Collectively, these bioinformatic, expression profiling, and biochemical data suggested that NFIL3 was involved in the signaling network activated by IL-10 to block IL-12p40 production.
FIGURE 3.
A NFIL3 binding site is identified in the enhancer region of Il12b. A, sequence of the consensus NFIL3 binding site. B and C, binding of NFIL3 to the enhancer. B, the sequence of gel-shift oligonucleotides used is shown, with each sequential 5- or 6-bp mutation (Mutants A–G). C, IVTT NFIL3 was generated and bound to 32P-labeled oligonucleotide probes and then complexes were resolved by electrophoresis. Binding data are representative of three experiments.
NFIL3 Is Sufficient to Block Il12b Transcription
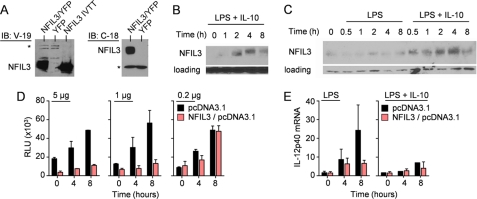
We next attempted to overexpress NFIL3 in primary macrophages by transducing bone marrow stem cells with a retrovirus encoding NFIL3. Although we were able to establish retroviral producer lines that expressed NFIL3 (Fig. 4A), we observed that macrophages grown in (colony-stimulating factor-1) following stem cell transduction lost YFP expression and did not express NFIL3 mRNA or protein. These results were consistent with immunoblotting experiments in primary macrophages, which showed that NFIL3 is a relatively low abundance protein whose induced expression declines after 8 h of LPS plus IL-10 stimulation (Fig. 4, B and C). Collectively, we concluded that NFIL3 amounts in macrophages are tightly regulated and that constitutive overexpression may be toxic. We therefore turned to the RAW264.7 macrophage cell line, reasoning that this immortalized line may be permissive to enforced NFIL3 expression. Using transient overexpression, we determined that NFIL3 suppressed the activity of the EnPr2 reporter and suppressed the LPS-induced IL-12p40 mRNA production in a dose-dependent way (Fig. 4, D and E). NFIL3 is therefore capable of blocking Il12b transcription.
FIGURE 4.
Overexpression of NFIL3 inhibits Il12b transcription. A, specificity of anti-NFIL3 antibodies. NFIL3 retroviral producer cell lines were analyzed by immunoblot with two anti-NFIL3 antibodies, V-19 and C-18. Empty vector YFP+ lines were used as a negative control, and NFIL3 IVTT was used as a positive control. The asterisk denotes a nonspecific band used as a loading control. B and C, BMDMs from Il10−/− (B) and C57BL/6 (C) mice were stimulated with LPS with or without IL-10. Protein lysates were immunoblotted for NFIL3 or Grb2 as a loading control. D, RAW264.7 macrophages were transiently cotransfected with 0.2 μg, 1 μg, or 5 μg of NFIL3/pcDNA3.1 and 5 μg of the EnPr2 luciferase reporter. Cells were stimulated in duplicate with LPS, and luciferase activity was measured at the times indicated. Data are presented as the mean relative luciferase units (RLU) plus the S.D. and are representative of two experiments. E, RAW264.7 macrophages were transiently transfected with 2 μg of NFIL3/pcDNA3.1, stimulated with LPS or LPS plus IL-10 in triplicate for the indicated times, and then IL-12p40 mRNA was detected by qRT-PCR. Data are presented as the mean IL-12p40 expression relative to GAPDH expression plus the S.D. Data are representative of two experiments.
Macrophages Lacking NFIL3 Overproduce IL-12p40
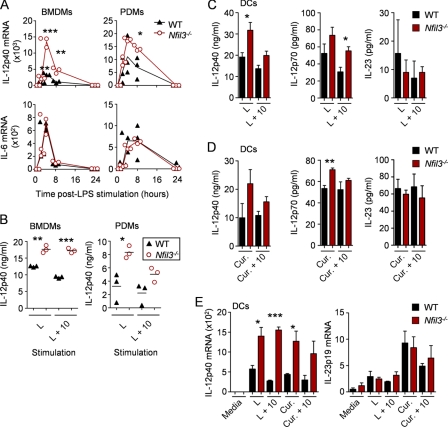
NFIL3-deficient mice have recently been independently generated by four groups and were shown to have deficiencies in NK (natural killer) cells and IgE production (30, 46–48). We isolated BMDMs and PDMs from Nfil3−/− mice and stimulated these cells with LPS. We found that Nfil3−/− macrophages overproduced IL-12p40 mRNA and secreted excess IL-12p40 protein relative to control macrophages (Fig. 5A and B). Furthermore, the ability of IL-10 to suppress IL-12p40 in the absence of NFIL3 was reduced but not completely ablated, suggesting that NFIL3 is a component of the IL-10 anti-inflammatory mechanism. It is important to mention that in this experimental scenario the effects of IL-10 are never complete because the added IL-10 is consumed from the media over the assay time. In our experiments on primary macrophages, we did not observe substantial differences in IL-6 (Fig. 5, A and B), IL-10, or IL-1α (data not shown) amounts in Nfil3−/− macrophages relative to controls, indicating that the inhibitory effect of NFIL3 was specific to IL-12p40. We also performed experiments to test if IL-12 and IL-23 were also increased in the absence of NFIL3. We reasoned that the observed increases in p40 could translate to elevated IL-12 and/or IL-23. We used CD11c+ in vitro-generated DCs stimulated with LPS or curdlan, a potent inducer of IL-23p19, to test this hypothesis. We found that in NFIL3-deficient CD11c+ DCs, LPS and curdlan stimulation caused an increase in IL-12p40 and IL-12p70 secretion relative to controls (Fig. 5, C and D). Under these conditions, however, increased IL-23 was not observed in the NFIL3-deficient DCs, even though Il23a transcription was induced by curdlan (Fig. 5E), suggesting the possibility that NFIL3 has a more complex role in regulating the IL-12 and IL-23 balance than stochastic inhibition of the quantity of p40 available to bind p35 and p19.
FIGURE 5.
NFIL3 is required for inhibition of IL-12p40. A, BMDMs and PDMs from Nfil3−/− mice or littermate WT controls (n = 3) were stimulated with LPS, and RNA was analyzed by qRT-PCR for expression of IL-12p40 and IL-6. Data are presented as the individual mRNA levels, including the means, normalized to GAPDH. B, supernatants from cells stimulated for 8 h with LPS (L) and with LPS plus IL-10 (L + 10) were analyzed for secreted IL-12p40 by ELISA. Data are presented as the individual protein levels (solid lines depict the respective means). C and D) CD11c+ DCs from Nfil3−/− and littermate WT controls (n = 3) were stimulated with LPS (C) or curdlan (Cur.) (D) with and without IL-10 for 24 h. Secreted IL-12p40, IL-12p70, and IL-23 amounts were determined by ELISA. E, DCs from above were stimulated for 4 h with LPS or curdlan alone or plus IL-10, and qRT-PCR was used to measure IL-12p40 and IL-23p19 transcripts. Data from C–E are presented as the mean protein levels or mRNA levels normalized to GAPDH plus the S.E. All data are representative of at least two experiments unless otherwise noted. *, **, and *** represent significance, where p < 0.05, < 0.01, and < 0.001, respectively, by Student's t test.
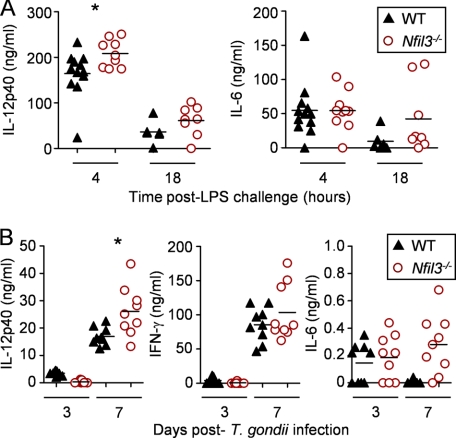
Because Nfil3−/− macrophages produced increased IL-12p40, we next extended our investigation to in vivo models of systemic inflammation linked to IL-12 production. First, we challenged Nfil3−/− mice or controls with a sublethal dose of E. coli LPS (20 mg/kg). In independent experiments, we observed that Nfil3−/− mice were sensitive to LPS, and most mice died within 18 h, whereas the majority of control animals recovered from the transient systemic shock. Furthermore, the absence of NFIL3 caused a significant increase in circulating IL-12p40, without increased IL-6 (Fig. 6A). We also used T. gondii infection as a probe for in vivo IL-12 production because this protozoan parasite is one of the strongest known inducers of IL-12 (49), and the source of IL-12 in this infection is exclusively from myeloid lineages (50). We infected Nfil3−/− or matched littermate control mice with 1000 cysts of the attenuated type II T. gondii strain, N28E2 (41), by the intraperitoneal route. We found that infected Nfil3−/− mice became lethargic and lost weight, and they were sacrificed after 7 days. We tested systemic cytokine production in the infected animals and found a significant increase in IL-12p40 amounts and a trend to increased IFN-γ in the blood of the Nfil3−/− mice (Fig. 5B). Because NK cells are absent in the NFIL3-deficient mice (30, 46), it is important to note that these experiments do not necessarily indicate that the increase in IL-12p40 alone is the cause of disease in these mice. Further experiments with conditional Nfil3 knockout mice will be necessary to establish the cell-specific contributions of NFIL3 to the overall immune response to T. gondii. Collectively, however, our in vivo data, when combined with the in vitro stimulation experiments, argue that NFIL3 is one essential control point in IL-12 production.
FIGURE 6.
NFIL3 regulates systemic IL-12p40 levels in vivo. A, Nfil3−/− and littermate WT controls were administered a sublethal LPS challenge (20 mg/kg), and serum IL-12p40 and IL-6 protein levels were detected by ELISA at 4 and 18 h post-challenge. Data are presented as the individual serum protein levels, including the means. B, Nfil3−/− and littermate WT controls were infected with T. gondii. Serum IL-12p40, IFN-γ, and IL-6 protein levels were detected by ELISA 3 and 7 days post-infection. Data are presented as the individual serum protein levels, including the means. Data are representative of three independent experiments. *, p < 0.05 by Student's t test.
DISCUSSION
The underlying mechanisms involved in the IL-10-mediated anti-inflammatory effect have been sought since the discovery of IL-10 in 1991. Herein we established that the IL-10 pathway activates the expression of the B-ZIP protein NFIL3, which is required to inhibit transcription of the gene encoding IL-12p40. We found that NFIL3 targets a distal enhancer in Il12b previously shown to be essential for the TLR-mediated expression of IL-12p40. Our data suggest that NFIL3 is one component of a complex mechanism elicited by IL-10 to suppress cytokine production. Although we found that NFIL3 is necessary for the full suppressive effects of IL-10 on IL-12p40, IL-10 can partially inhibit IL-12p40 production in Nfil3−/− macrophages, suggesting that additional mechanisms intersect with NFIL3 for the full inhibitory effect.
Anti-inflammatory signal transduction from the IL-10R requires the STAT3-mediated expression of downstream target genes and their products (27, 39, 51). Abundant evidence from both murine and human systems has shown that STAT3 is necessary and sufficient for all the known anti-inflammatory effects of IL-10 on myeloid lineage cells (35, 39, 52). Furthermore, the anti-inflammatory signaling system operates independent of direct inhibitory effects on the NF-κB or the MAP kinase pathways (27, 42). Instead, STAT3 induces the expression of a specific group of genes, including Nfil3, whose protein products interfere with the transcriptional programs of key proinflammatory cytokines (39). These data raise a quandary because other cytokines activate STAT3 in macrophages (e.g. IL-6) but cannot activate the anti-inflammatory effect. Inhibition of anti-inflammatory signaling from the IL-6R is relieved when SOCS3 is absent, suggesting that mechanisms exist to actively suppress the IL-10-like STAT3 signal from non-IL-10 STAT3-activating receptors (22, 53). Indeed, we have shown that it is possible to convert any cytokine receptor into an IL-10-like receptor by ensuring STAT3 activation but in the absence of SOCS3-mediated inhibition (35). We interpret these data to suggest that the signal from the IL-10R needs to be activated by this receptor alone, whereas activation of the same pathway by other receptors is blocked. How does the IL-10R control inflammatory gene expression? It is important to mention that the anti-inflammatory effects of IL-10 are gene-specific and not directed at all primary and secondary genes induced by TLR signaling (39). Instead, IL-10 targets specific genes, including primary genes such as Tnf and Cxcl1, and numerous secondary genes, including Il6 and Il12b. Other canonical TLR-induced genes such as Ikba or Tnfaip3 are unaffected by IL-10 in both mouse and human primary macrophages (39, 42). The net result of this inhibitory pathway is immune homeostasis at the level of myeloid cells, as loss of normal IL-10 function in both mice and humans leads to excessive inflammation.
How the downstream target genes of STAT3 mediate gene-specific effects on inflammatory genes remain largely unknown. Using microarray approaches that involved restimulation of IL-10-deficient macrophages in the presence of LPS, we have determined the identity of many of the IL-10-induced genes (35, 39, 42, 44). Because the primary target of the effects of IL-10 is gene transcription (27), we have focused on IL-10-induced genes implicated in transcriptional regulation (Fig. 2). Four of the IL-10-induced transcription factors are B-ZIP transcription factors that can homo- and heterodimerize, depending on the specific factor. In the present study, C/EBPβ and C/EBPδ were candidate factors previously established in the TLR pathway and more specifically involved in Il12b transcription. However, macrophages lacking either factor show no differences in IL-10-mediated inhibition of Il12b (supplemental Fig. S2). Instead, we found that NFIL3, was up-regulated by IL-10, bound to the Il12b enhancer, and repressed Il12b transcription when overexpressed (Figs. 2–4).
Macrophages from NFIL3-deficient mice overproduced IL-12p40 and, most importantly, had defects in the IL-10-mediated repression of IL-12p40 production (Fig. 5). These data suggest that NFIL3 is a component of the IL-10 anti-inflammatory signaling pathway. We found that LPS stimulation alone induced NFIL3 transcription (Fig. 2, A and C). We attribute this effect to the production and autocrine-paracrine signaling of endogenous IL-10 from LPS-stimulated bone marrow-derived macrophages. Indeed, NFIL3 expression was ablated when we stimulated STAT3-deficient BMDMs with LPS (Fig. 2C). By contrast, thioglycolate-elicited peritoneal-derived macrophages produce lower amounts of IL-10 and hence exhibit little or no induction of NFIL3 transcript following LPS stimulation (Fig. 2B). Only when exogenous IL-10 is added to LPS-stimulated PDMs is there an induction of NFIL3 (Fig. 2B). For this reason, macrophages from Nfil3−/− mice have increased production of IL-12p40 following LPS stimulation with or without the addition of exogenous IL-10.
The effects of NFIL3 appear to be specific for Il12b because we have not observed any defects in IL-6, TNF-α, or IL-1α expression in TLR-stimulated Nfil3−/− macrophages. However, a recent publication has shown that IL-6, in addition to IL-12p40, is disregulated in the absence of NFIL3 (54). Further studies will be necessary to establish the full transcriptional program regulated by NFIL3 in macrophages and the extent to which IL-10 regulated NFIL3 diverges relative to other pathways. In this regard it is worth noting that the NFIL3-deficient mice have recently been reported to multiple phenotypes, including an absence of CD8+ DCs, and defects in T cell cytokine production in addition to the initially reported defects in NK development (30, 46–48, 55, 56). Therefore, considerable caution is warranted in the interpretation of whole animal experiments meant to perturb immune responses.
The inhibitory effect of IL-10 on IL-12p40 expression forms a key pathway of immune homeostasis, especially in the intestine. Mice lacking IL-10 suffer from a profound breakdown of intestinal homeostasis driven by gut flora (57), and humans lacking components of the IL-10R have an extreme form of intestinal inflammation that can be relieved by bone marrow transplantation (23). The source of IL-10 required to maintain normal intestinal homeostasis is an active area of research and involves multiple T cell subsets, including Tregs, and also myeloid cells within the intestine (57). Because STAT3 is necessary and sufficient for IL-10 signaling in macrophages, the phenotypic similarity of intestinal disease in myeloid lineage-specific STAT3 conditional knockout mice was anticipated (51). However, a detailed genetic investigation of the underlying driver cytokines that promulgate intestinal disease in the myeloid-specific STAT3 knockouts showed that loss of TLR-driven IL-12p40 but not TNF-α or IFN signaling could rescue the disease (25). These insights, coupled with extensive investigation into the effects of IL-10 on suppressing IL-12p40 argue that this regulatory pathway is central to restraining IL-12p40 and thereby IL-12 and IL-23 production by activated myeloid cells.
It is worth noting that the production of IL-12 and IL-23 by activated myeloid cells is regulated at different levels. First, Il12b, Il23a, and Il12a are all controlled at the transcriptional level by combinations of pathways activated by the TLR and IFN signaling pathways (2). For example, Il12b expression after TLR activation can increase hundreds-fold and requires different transcription factors to assemble at the promoter and enhancer (2, 33). Microbial compounds differentially induce Il23a, including preferential activation by fungal cell walls, accounting for the differential IL-23:IL-12 ratio observed in response to fungal infection (38). A second major regulatory layer controls the assembly of the heterodimers in the endoplasmic reticulum. Because IL-12p40 is made in excess to the p19 and p35 subunits, the final production of mature IL-12 and IL-23 is limited by the amount of partner p19 and p35. Such a scenario is potentially another way to ensure that the correct amounts of IL-12 and IL-23 are made. A variant in the coding region of Il12b in SJL mice that enhances heterodimer formation in the endoplasmic reticulum has been discovered recently (58). It is possible that the increased amounts of IL-12 and IL-23 made in SJL mice accounts for the susceptibility of this strain to the trinitrobenzene sulfonic acid colitis model. In the case of our in vitro experiments, we did not observe substantial effects of the loss of NFIL3 on IL-23 amounts in in vitro experiments. Further work on in vivo T cell polarization will be necessary to establish whether NFIL3 has more complex effects on IL-12 and IL-23 beyond regulation of IL-12p40 amounts.
In summary, we have established that IL-10 regulates Il12b transcription through inhibitory effects on a distal enhancer, independent of the Il12b promoter. The underlying biochemical mechanism of the inhibitory effect involved IL-10-mediated expression of NFIL3, which targets the enhancer. Thus, NFIL3 is a key component of the STAT3-mediated anti-inflammatory mechanism.
Supplementary Material
Acknowledgments
We thank Tak Mak, Jillian Haight, and Thomas Look for the gift of NFIL3-deficient mice; Andy High for mass spectrometry; and Hugh Brady and Scott Plevy for discussions.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program, National Cancer Institute, Center for Cancer Research (to P. F. J.). This work was also supported by National Institutes of Health Grant F32 CA138064 (to J. E. Q.), The Hartwell Foundation (to P. J. M.), CORE grant P30 CA21765, and the American Lebanese Syrian Associated Charities.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- TLR
- Toll-like receptor
- C/EBP
- CCAAT/enhancer binding protein
- DC
- dendritic cell
- qRT-PCR
- quantitative RT-PCR
- IVTT
- in vitro transcription-translation
- BMDM
- bone marrow-derived macrophage
- PDM
- peritoneal-derived macrophage
- IL-10R
- IL-10 receptor.
REFERENCES
- 1. Lyakh L., Trinchieri G., Provezza L., Carra G., Gerosa F. (2008) Immunol. Rev. 226, 112–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Trinchieri G. (2003) Nat. Rev. Immunol. 3, 133–146 [DOI] [PubMed] [Google Scholar]
- 3. McGeachy M. J., Chen Y., Tato C. M., Laurence A., Joyce-Shaikh B., Blumenschein W. M., McClanahan T. K., O'Shea J. J., Cua D. J. (2009) Nat. Immunol. 10, 314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahern P. P., Schiering C., Buonocore S., McGeachy M. J., Cua D. J., Maloy K. J., Powrie F. (2010) Immunity 33, 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muñoz M., Heimesaat M. M., Danker K., Struck D., Lohmann U., Plickert R., Bereswill S., Fischer A., Dunay I. R., Wolk K., Loddenkemper C., Krell H. W., Libert C., Lund L. R., Frey O., Hölscher C., Iwakura Y., Ghilardi N., Ouyang W., Kamradt T., Sabat R., Liesenfeld O. (2009) J. Exp. Med. 206, 3047–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Teng M. W., Andrews D. M., McLaughlin N., von Scheidt B., Ngiow S. F., Möller A., Hill G. R., Iwakura Y., Oft M., Smyth M. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8328–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Teng M. W., von Scheidt B., Duret H., Towne J. E., Smyth M. J. (2011) Cancer Res. 71, 2077–2086 [DOI] [PubMed] [Google Scholar]
- 8. Franke A., McGovern D. P., Barrett J. C., Wang K., Radford-Smith G. L., Ahmad T., Lees C. W., Balschun T., Lee J., Roberts R., Anderson C. A., Bis J. C., Bumpstead S., Ellinghaus D., Festen E. M., Georges M., Green T., Haritunians T., Jostins L., Latiano A., Mathew C. G., Montgomery G. W., Prescott N. J., Raychaudhuri S., Rotter J. I., Schumm P., Sharma Y., Simms L. A., Taylor K. D., Whiteman D., Wijmenga C., Baldassano R. N., Barclay M., Bayless T. M., Brand S., Büning C., Cohen A., Colombel J. F., Cottone M., Stronati L., Denson T., de Vos M., D'Inca R., Dubinsky M., Edwards C., Florin T., Franchimont D., Gearry R., Glas J., Van Gossum A., Guthery S. L., Halfvarson J., Verspaget H. W., Hugot J. P., Karban A., Laukens D., Lawrance I., Lemann M., Levine A., Libioulle C., Louis E., Mowat C., Newman W., Panés J., Phillips A., Proctor D. D., Regueiro M., Russell R., Rutgeerts P., Sanderson J., Sans M., Seibold F., Steinhart A. H., Stokkers P. C., Torkvist L., Kullak-Ublick G., Wilson D., Walters T., Targan S. R., Brant S. R., Rioux J. D., D'Amato M., Weersma R. K., Kugathasan S., Griffiths A. M., Mansfield J. C., Vermeire S., Duerr R. H., Silverberg M. S., Satsangi J., Schreiber S., Cho J. H., Annese V., Hakonarson H., Daly M. J., Parkes M. (2010) Nat. Genet. 42, 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGovern D. P., Gardet A., Törkvist L., Goyette P., Essers J., Taylor K. D., Neale B. M., Ong R. T., Lagacé C., Li C., Green T., Stevens C. R., Beauchamp C., Fleshner P. R., Carlson M., D'Amato M., Halfvarson J., Hibberd M. L., Lördal M., Padyukov L., Andriulli A., Colombo E., Latiano A., Palmieri O., Bernard E. J., Deslandres C., Hommes D. W., de Jong D. J., Stokkers P. C., Weersma R. K., Sharma Y., Silverberg M. S., Cho J. H., Wu J., Roeder K., Brant S. R., Schumm L. P., Duerr R. H., Dubinsky M. C., Glazer N. L., Haritunians T., Ippoliti A., Melmed G. Y., Siscovick D. S., Vasiliauskas E. A., Targan S. R., Annese V., Wijmenga C., Pettersson S., Rotter J. I., Xavier R. J., Daly M. J., Rioux J. D., Seielstad M. (2010) Nat. Genet. 42, 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mizuki N., Meguro A., Ota M., Ohno S., Shiota T., Kawagoe T., Ito N., Kera J., Okada E., Yatsu K., Song Y. W., Lee E. B., Kitaichi N., Namba K., Horie Y., Takeno M., Sugita S., Mochizuki M., Bahram S., Ishigatsubo Y., Inoko H. (2010) Nat. Genet. 42, 703–706 [DOI] [PubMed] [Google Scholar]
- 11. Zhernakova A., van Diemen C. C., Wijmenga C. (2009) Nat. Rev. Genet. 10, 43–55 [DOI] [PubMed] [Google Scholar]
- 12. Engel D. R., Koscielny A., Wehner S., Maurer J., Schiwon M., Franken L., Schumak B., Limmer A., Sparwasser T., Hirner A., Knolle P. A., Kalff J. C., Kurts C. (2010) Nat. Med. 16, 1407–1413 [DOI] [PubMed] [Google Scholar]
- 13. Strober W., Fuss I., Mannon P. (2007) J. Clin. Invest. 117, 514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buonocore S., Ahern P. P., Uhlig H. H., Ivanov II, Littman D. R., Maloy K. J., Powrie F. (2010) Nature 464, 1371–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hue S., Ahern P., Buonocore S., Kullberg M. C., Cua D. J., McKenzie B. S., Powrie F., Maloy K. J. (2006) J. Exp. Med. 203, 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Izcue A., Hue S., Buonocore S., Arancibia-Cárcamo C. V., Ahern P. P., Iwakura Y., Maloy K. J., Powrie F. (2008) Immunity 28, 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamada N., Hisamatsu T., Okamoto S., Chinen H., Kobayashi T., Sato T., Sakuraba A., Kitazume M. T., Sugita A., Koganei K., Akagawa K. S., Hibi T. (2008) J. Clin. Invest. 118, 2269–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mizoguchi A., Ogawa A., Takedatsu H., Sugimoto K., Shimomura Y., Shirane K., Nagahama K., Nagaishi T., Mizoguchi E., Blumberg R. S., Bhan A. K. (2007) J. Clin. Invest. 117, 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elliott M., Benson J., Blank M., Brodmerkel C., Baker D., Sharples K. R., Szapary P. (2009) Ann. N.Y. Acad. Sci. 1182, 97–110 [DOI] [PubMed] [Google Scholar]
- 20. Mannon P. J., Fuss I. J., Mayer L., Elson C. O., Sandborn W. J., Present D., Dolin B., Goodman N., Groden C., Hornung R. L., Quezado M., Yang Z., Neurath M. F., Salfeld J., Veldman G. M., Schwertschlag U., Strober W. (2004) N. Engl. J. Med. 351, 2069–2079 [DOI] [PubMed] [Google Scholar]
- 21. Olleros M. L., Vesin D., Martinez-Soria E., Allenbach C., Tacchini-Cottier F., Pache J. C., Marchal G., Rahman J., Fernández C., Izui S., Garcia I. (2007) Immunology 122, 350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Shea J. J., Murray P. J. (2008) Immunity 28, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Glocker E. O., Kotlarz D., Boztug K., Gertz E. M., Schäffer A. A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D., Hätscher N., Pfeifer D., Sykora K. W., Sauer M., Kreipe H., Lacher M., Nustede R., Woellner C., Baumann U., Salzer U., Koletzko S., Shah N., Segal A. W., Sauerbrey A., Buderus S., Snapper S. B., Grimbacher B., Klein C. (2009) N. Engl. J. Med. 361, 2033–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamada N., Hisamatsu T., Okamoto S., Sato T., Matsuoka K., Arai K., Nakai T., Hasegawa A., Inoue N., Watanabe N., Akagawa K. S., Hibi T. (2005) J. Immunol. 175, 6900–6908 [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi M., Kweon M. N., Kuwata H., Schreiber R. D., Kiyono H., Takeda K., Akira S. (2003) J. Clin. Invest. 111, 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murray P. J. (2006) Curr. Opin. Pharmacol. 6, 379–386 [DOI] [PubMed] [Google Scholar]
- 27. Murray P. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8686–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou L., Nazarian A. A., Smale S. T. (2004) Mol. Cell. Biol. 24, 2385–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aste-Amezaga M., Ma X., Sartori A., Trinchieri G. (1998) J. Immunol. 160, 5936–5944 [PubMed] [Google Scholar]
- 30. Kamizono S., Duncan G. S., Seidel M. G., Morimoto A., Hamada K., Grosveld G., Akashi K., Lind E. F., Haight J. P., Ohashi P. S., Look A. T., Mak T. W. (2009) J. Exp. Med. 206, 2977–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sterneck E., Tessarollo L., Johnson P. F. (1997) Genes Dev. 11, 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sterneck E., Paylor R., Jackson-Lewis V., Libbey M., Przedborski S., Tessarollo L., Crawley J. N., Johnson P. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10908–10913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou L., Nazarian A. A., Xu J., Tantin D., Corcoran L. M., Smale S. T. (2007) Mol. Cell. Biol. 27, 2698–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pauleau A. L., Rutschman R., Lang R., Pernis A., Watowich S. S., Murray P. J. (2004) J. Immunol. 172, 7565–7573 [DOI] [PubMed] [Google Scholar]
- 35. El Kasmi K. C., Holst J., Coffre M., Mielke L., de Pauw A., Lhocine N., Smith A. M., Rutschman R., Kaushal D., Shen Y., Suda T., Donnelly R. P., Myers M. G., Jr., Alexander W., Vignali D. A., Watowich S. S., Ernst M., Hilton D. J., Murray P. J. (2006) J. Immunol. 177, 7880–7888 [DOI] [PubMed] [Google Scholar]
- 36. El Kasmi K. C., Qualls J. E., Pesce J. T., Smith A. M., Thompson R. W., Henao-Tamayo M., Basaraba R. J., König T., Schleicher U., Koo M. S., Kaplan G., Fitzgerald K. A., Tuomanen E. I., Orme I. M., Kanneganti T. D., Bogdan C., Wynn T. A., Murray P. J. (2008) Nat. Immunol. 9, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qualls J. E., Neale G., Smith A. M., Koo M. S., DeFreitas A. A., Zhang H., Kaplan G., Watowich S. S., Murray P. J. (2010) Sci. Signal. 3, ra62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. LeibundGut-Landmann S., Gross O., Robinson M. J., Osorio F., Slack E. C., Tsoni S. V., Schweighoffer E., Tybulewicz V., Brown G. D., Ruland J., Reis e Sousa C. (2007) Nat. Immunol. 8, 630–638 [DOI] [PubMed] [Google Scholar]
- 39. Lang R., Patel D., Morris J. J., Rutschman R. L., Murray P. J. (2002) J. Immunol. 169, 2253–2263 [DOI] [PubMed] [Google Scholar]
- 40. Gingras S., Parganas E., de Pauw A., Ihle J. N., Murray P. J. (2004) J. Biol. Chem. 279, 54702–54707 [DOI] [PubMed] [Google Scholar]
- 41. Craver M. P., Rooney P. J., Knoll L. J. (2010) Mol. Biochem. Parasitol 169, 120–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smallie T., Ricchetti G., Horwood N. J., Feldmann M., Clark A. R., Williams L. M. (2010) J. Exp. Med. 207, 2081–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams L. M., Ricchetti G., Sarma U., Smallie T., Foxwell B. M. (2004) Immunology 113, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. El Kasmi K. C., Smith A. M., Williams L., Neale G., Panopoulos A. D., Panapoulos A., Watowich S. S., Häcker H., Foxwell B. M., Murray P. J. (2007) J. Immunol. 179, 7215–7219 [DOI] [PubMed] [Google Scholar]
- 45. Zhang W., Zhang J., Kornuc M., Kwan K., Frank R., Nimer S. D. (1995) Mol. Cell. Biol. 15, 6055–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gascoyne D. M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., Brady H. J. (2009) Nat. Immunol. 10, 1118–1124 [DOI] [PubMed] [Google Scholar]
- 47. Kashiwada M., Levy D. M., McKeag L., Murray K., Schröder A. J., Canfield S. M., Traver G., Rothman P. B. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Motomura Y., Kitamura H., Hijikata A., Matsunaga Y., Matsumoto K., Inoue H., Atarashi K., Hori S., Watarai H., Zhu J., Taniguchi M., Kubo M. (2011) Nat Immunol 12, 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gazzinelli R. T., Wysocka M., Hayashi S., Denkers E. Y., Hieny S., Caspar P., Trinchieri G., Sher A. (1994) J. Immunol. 153, 2533–2543 [PubMed] [Google Scholar]
- 50. Hou B., Benson A., Kuzmich L., A L. D., Yarovinsky F. (2010) Proc. Natl. Acad. Sci. U.S.A. 108, 278–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takeda K., Clausen B. E., Kaisho T., Tsujimura T., Terada N., Förster I., Akira S. (1999) Immunity 10, 39–49 [DOI] [PubMed] [Google Scholar]
- 52. Williams L., Bradley L., Smith A., Foxwell B. (2004) J. Immunol. 172, 567–576 [DOI] [PubMed] [Google Scholar]
- 53. Murray P. J. (2007) J. Immunol. 178, 2623–2629 [DOI] [PubMed] [Google Scholar]
- 54. Kobayashi T., Matsuoka K., Sheikh S. Z., Elloumi H. Z., Kamada N., Hisamatsu T., Hansen J. J., Doty K. R., Pope S. D., Smale S. T., Hibi T., Rothman P. B., Kashiwada M., Plevy S. E. (2011) J. Immunol. 186, 4649–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kashiwada M., Cassel S. L., Colgan J. D., Rothman P. B. (2011) EMBO J. 30, 2071–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kashiwada M., Pham N. L., Pewe L. L., Harty J. T., Rothman P. B. (2011) Blood 10.1182/blood-2010-07-295873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barnes M. J., Powrie F. (2009) Immunity 31, 401–411 [DOI] [PubMed] [Google Scholar]
- 58. Zwiers A., Fuss I. J., Seegers D., Konijn T., Garcia-Vallejo J. J., Samsom J. N., Strober W., Kraal G., Bouma G. (2011) J. Immunol. 186, 3572–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.