SUMMARY
Nonsense-mediated decay (NMD) degrades both normal and aberrant transcripts harboring stop codons in particular contexts. Mutations that perturb NMD cause neurological disorders in humans, suggesting that NMD has roles in the brain. Here, we identify a brain-specific microRNA—miR-128—that represses NMD and thereby controls batteries of transcripts in neural cells. miR-128 represses NMD by targeting the RNA helicase UPF1 and the exon-junction complex core component MLN51. The ability of miR-128 to regulate NMD is a conserved response occuring in frogs, chickens, and mammals. miR-128 levels are dramatically increased in differentiating neuronal cells and during brain development, leading to repressed NMD and upregulation of mRNAs normally targeted for decay by NMD; overrepresented are those encoding proteins controlling neuron development and function. Together, these results suggest the existence of a conserved RNA circuit linking the microRNA and NMD pathways that induces cell type-specific transcripts during development.
INTRODUCTION
Nonsense-mediated decay (NMD) was originally identified as a RNA surveillance mechanism that rapidly degrades aberrant mRNAs harboring premature termination codons (PTCs) (Bhuvanagiri et al., 2010; Chang et al., 2007). Subsequently, it was discovered that NMD also regulates many normal transcripts. Microarray analysis revealed that ~3 to 10% of mRNAs are misregulated when NMD is perturbed in eukaryotes ranging from yeast to humans (Chan et al., 2007; Mendell et al., 2004; Rehwinkel et al., 2006). While the “NMD-inducing features” responsible for the regulation of normal transcripts have not all been elucidated, in general, they place a stop codon in a context that is recognized as premature (Rebbapragada and Lykke-Andersen, 2009). For example, one NMD-inducing feature is an exon-exon junction downstream of the stop codon. This triggers NMD because RNA splicing recruits a set of proteins called the exon-junction complex (EJC) that interacts with proteins recruited during translation termination, including the RNA helicase UPF1, a key factor essential for NMD (Bhuvanagiri et al., 2010; Chang et al., 2007). mRNAs that harbor a stop codon in the final exon avoid NMD because the EJCs recruited upstream of the stop codon are probably displaced by the translocating ribosome during the first round of translation (Bhuvanagiri et al., 2010; Chang et al., 2007). Many NMD-targeted mRNAs are normally spliced, thereby generating a normal protein, but NMD substrates are also commonly generated by alternative splicing events that place a stop codon upstream of an exon-exon junction. Alternative polyadenylation can also elicit NMD by generating a long 3′ UTR, which triggers NMD by a UPF1-dependent mechanism (Hogg and Goff, 2010).
The discovery that NMD regulates large batteries of normal transcripts raises the question of its physiological relevance. This has proven difficult to address given that NMD is a dual mechanism that not only regulates normal gene expression but also functions as a quality control mechanism. For example, while knockout mice studies have revealed that NMD factors are required for early embryonic development (McIlwain et al., 2010; Medghalchi et al., 2001; Weischenfeldt et al., 2008), it remains to be determined whether this is the result of a defect in NMD’s gene regulatory role, its RNA surveillance role, or both. We elected to investigate the functional relevance of NMD in controlling normal gene expression by first addressing whether NMD itself is regulated. In particular, we examined the possibility that NMD is regulated by microRNAs (miRNAs). These are small (~22-nt) non-coding RNAs that bind and repress the expression of specific target mRNAs through translational repression and/or rapid mRNA decay (Bartel, 2009). We focused on miRNAs because many are expressed in a cell type-specific and developmentally regulated manner. We postulated that if we identified a regulated miRNA that controlled NMD, that it would drive a highly directed NMD response that could potentially regulate specific biological events. Here, we provide evidence that we have indeed identified such a miRNA/NMD regulatory circuit.
RESULTS
miR-128 Regulates NMD
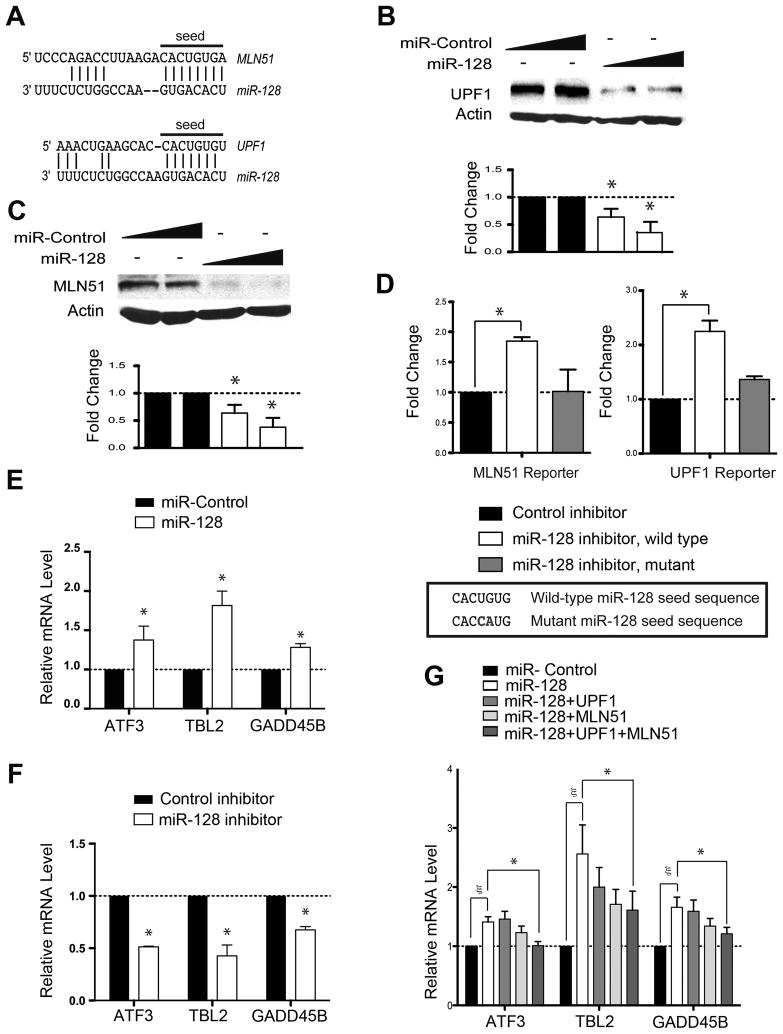
To identify miRNAs that have the potential to regulate NMD, we performed an in silico screen for conserved miRNAs targeting NMD factors using the miRNA algorithm TargetScan (http://www.targetscan.org/). Through this analysis, we identified miR-128-1 and miR-128-2 as potential regulators of two NMD factors: UPF1 and MLN51 (also known as CASC3 or BTZ) (Figure 1A). UPF1 is a highly conserved RNA helicase essential for NMD (Chang et al., 2007), while MLN51 is a core component of the EJC that strongly promotes NMD in mammals (Chang et al., 2007). miR-128-1 and -2 are in introns in the R3HDM1 and RCS (ARPP-21) genes, respectively. Because both miR-128-1 and miR-128-2 are processed to generate mature miRNAs with identical sequences (Hsu et al., 2006), we will refer to them both as miR-128.
Figure 1. miR-128 Regulates NMD.
(A) Predicted base pairing of mature miR-128 seed sequences in the 3′ UTRs of MLN51 and UPF1 (http://www.targetscan.org).
(B, C) Western blot analysis of endogenous UPF1 and MLN51 protein levels in HEK293 cells 24 h after transfection with a mature miR-128 mimic or a random negative-control miRNA mimic (Ambion/ABI, Inc.). β-actin is the internal control. Bottom panels show the mean of three experiments; error bars represent standard deviation (SD).
(D) Luciferase expression in HeLa cells transfected for 24 h with firefly luciferase pmiR (Ambion/ABI, Inc.) reporters containing MLN51 or UPF1 3′ UTR sequences, including the putative miR-128 target sites shown in panel A. The mutants have the indicated 2-nt mutation in the target sequence. The cells were cotransfected with a miR-128-specific inhibitor or a negative-control inhibitor (Ambion/ABI, Inc.). A Renilla luciferase construct was co-transfected to control for transfection efficiency.
(E, F) Quantitative polymerase chain reaction (qPCR) analysis of HeLa cells transfected for 24 h with the indicated miRNA mimics (panel E) or LNA inhibitor (Exiqon, Inc; panel F). Shown are the results from three independent experiments, normalized to GAPDH mRNA levels.
(G) qPCR analysis of HEK293 cells transfected with the miR-128 or control mimic in combination with UPF1, MLN51, and/or empty expression plasmids. Quantification was conducted as in panel E.
For all panels, error bars indicate standard deviation. Asterisks and symbol (ζ) denote statistically significant differences; P < 0.05 (paired Student’s t-test).
To determine whether miR-128 represses the expression of UPF1 and MLN51, we transfected a “miR-128 mimic” (a double-stranded 21-nt form of miR-128 that is processed into a functional single-stranded miRNA in cells), into HEK293 cells, which normally express relatively low levels of endogenous mature miR-128 (Figure S1A). The miR-128 mimic decreased UPF1 and MLN51 protein levels by ~70% (Figures 1B and 1C). In a complementary experiment, we transfected a sequence-specific miR-128 inhibitor in HeLa cells, which expresses high levels of miR-128 (Figure S1A). The miR-128 inhibitor decreased endogenous mature miR-128 level and increased UPF1 and MLN51 protein levels (Figures S1B-D). As evidence for specificity, sequence-specific inhibitors of other miRNAs did not have a significant effect on UPF1 levels (Figure S1E). To determine whether miR-128 directly targets UPF1 and MLN51, we inserted their putative target sequence (Figure 1A), along with surrounding 3′ UTR sequences, into a reporter vector. Co-transfection of the sequence-specific miR-128 inhibitor increased luciferase activity from both the UPF1- and the MLN51–3′ UTR reporter vectors (Figures 1D and S1F). Introduction of a two-nucleotide mutation in the miR-128 seed sequence in either the UPF1 3′ UTR or the MLN51 3′ UTR abrogated this regulation (Figure 1D).
The discovery that miR-128 regulates UPF1 and MLN51 suggested the possibility that NMD itself is regulated by miR-128. If true, this would suggest the existence of a gene control mechanism bestowing upon a miRNA the ability to upregulate—rather than downregulate—physiologically important transcripts. The latter follows from the recent discovery that NMD triggers the rapid decay of mRNAs encoding functional proteins, as described in the Introduction. By inhibiting NMD, miR-128 would upregulate the transcripts that would normally be rapidly degraded. To investigate this possibility, we first examined whether miR-128 upregulates ATF3, TBL2, and GADD45B transcripts, all of which have been shown to be NMD substrates (Mendell et al., 2004). GADD45B mRNA has an intron in its 3′ UTR, which recruits an EJC downstream of its stop codon and thereby triggers NMD; ATF3 mRNA is targeted for decay by NMD because it harbors a premature termination codon introduced by alternative splicing; and TBL2 mRNA has a long 3′ UTR (~2.0 kb), a feature recently shown to trigger mammalian NMD (Rebbapragada and Lykke-Andersen, 2009). We found that all three of these known NMD substrates were upregulated by the miR-128-mimic (Figure 1E), while the miR-128 inhibitor decreased their level (Figure 1F), indicating that miR-128 regulates NMD. To determine whether miR-128 regulates NMD through its ability to repress UPF1 and MLN51 expression, we assessed whether forced expression of UPF1 and MLN51 from constructs lacking their native 3′ UTRs reversed miR-128’s inhibitory effect on NMD. Indeed, we found that UPF1 and/or MLN51 expression partially or fully reversed the upregulation of NMD substrates by the miR-128 mimic (Figures 1G and S1G), indicating that miR-128 acts through MLN51 and UPF1 to regulate NMD in mammalian cells.
The miR-128-NMD Circuit is Conserved
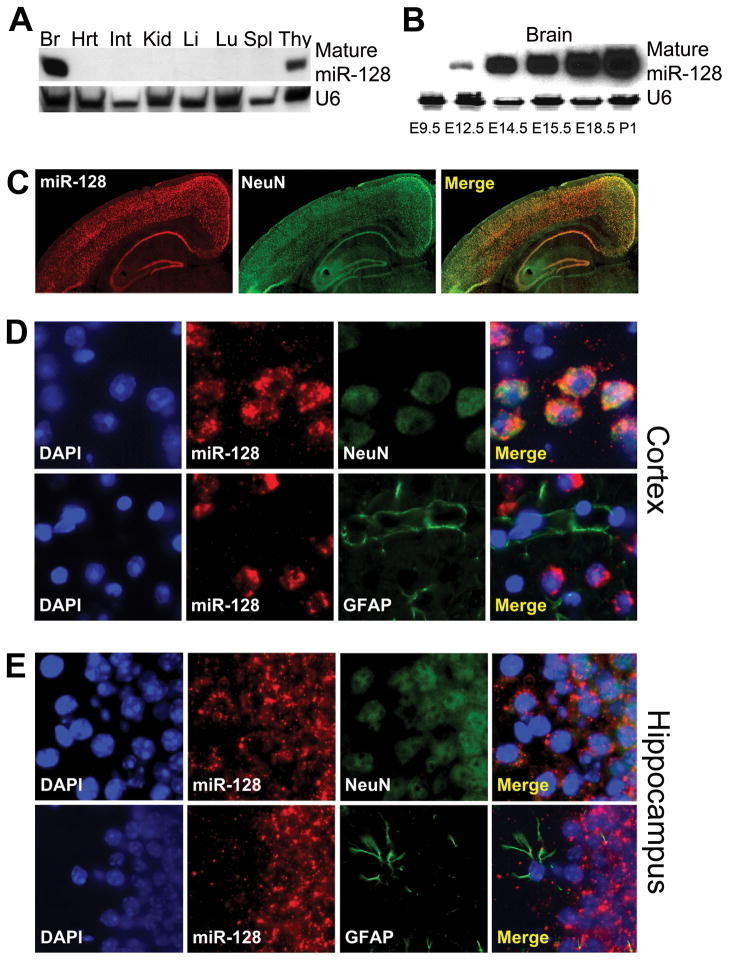
miR-128 and its NMD factor targets are remarkably conserved. Two loci encoding identical copies of mature miR-128 (miR-128-1 and miR-128-2) are present in a wide variety of vertebrates, including frogs, chickens, mice, and humans (Figures S2A and S2B). In these same species, the miR-128 target, UPF1, has a 7-nt region in the 3′ UTR that is 100% complementary to the miR-128 seed sequence (Figures 2A and S2C). Consistent with the EJC having a role in NMD in mammals, but not many lower species (Rehwinkel et al., 2006), mammalian MLN51, but not chicken or frog MLN51, have 3′ UTR target sites exactly matching the miR-128 seed (Figures 2A and S2D).
Figure 2. The miR-128/NMD Regulatory Circuit Is Conserved.
(A) miR-128-1/-2 and its conserved targets in the UPF1 and MLN51 3′ UTRs; human (Hsa), mouse (Mmu), chicken (Gga), and frog (Xla).
(B) miR-128 levels in chick neural tubes assessed by Taqman-qPCR analysis. Exogenous miR-128 (40 μM miR-128 mimic, Ambion Inc.) was electroporated 60 hours after fertilization and RNA was harvested 84 h after fertilization (n=3). miR-128 levels were normalized to U6 snRNA.
(C) qPCR analysis of the indicated transcripts in chick neural tube electroporated with the miR-128 or control miRNA mimic and harvested as described in panel B.
(D) miR-128 levels in X. laevis embryos assessed by Taqman-qPCR analysis. Exogenous miR-128 (5 pmoles of miR-128 mimic, Ambion Inc.) was injected into two-cell embryos and RNA was prepared at the time points indicated (n=5). miR-128 levels were normalized to L19 RNA.
(E) qPCR analysis of the indicated transcripts from stage (st) 10.5 embryos injected as described in panel D. TIAR and DKK1 transcripts are putative NMD substrates, based on their having NMD-inducing features (http://www.ncbi.nlm.nih.gov).
For all panels, experiments were performed in triplicate, error bars indicate standard deviation, and asterisks indicate statistically significant differences (P<0.05; paired Student’s t-test).
The conservation of miR-128 and its target, UPF1, in birds and amphibians raised the possibility that miR-128 might also repress NMD in these organisms. To directly test this hypothesis in birds, we used chick neural tubes, which we found exhibited a ~10-fold induction of miR-128 between 72 and 120 h post-fertilization (Figure 2B). In ovo electroporation of miR-128 mimic 60 h post-fertilization (when endogenous miR-128 levels are low) repressed chick UPF1 mRNA expression and upregulated three NMD substrates (Humphrey et al., 2008; Le Guiner et al., 2003; Pacheco et al., 2004) 24 h later (Figure 2C). Of note, repression of chick NMD was achieved even with only a modest increase miR-128 level (~3-fold; Figure 2B). Consistent with the fact that the chick MLN51 3′ UTR does not exhibit complementarity with the miR-128 seed sequence, chick MLN51 mRNA levels were not significantly reduced in response to forced miR-128 expression (Figure 2C).
To test whether miR-128 represses NMD in amphibians, we assayed the effect of the miR-128 mimic on NMD in Xenopus laevis embryos, which initially lack detectable miR-128 transcripts and then dramatically induce them after stage 17 (the late neuronal fold stage; Figure 2D). Injection of modest amounts of the miR-128 mimic in two-cell embryos (Figure 2D) repressed X. laevis UPF1 mRNA expression and upregulated X. laevis transcripts harboring NMD-inducing features (Figure 2E). As with chick embryos, X. laevis embryos responded to miR-128 by lowering their level of UPF1 mRNA, not MLN51 mRNA. We conclude that the ability of miR-128 to suppress NMD is a conserved response that extends to at least mammals, birds, and amphibians.
miR-128 is a Brain-Enriched miRNA that Promotes Neural Differentiation
To define the biological context under which miR-128 might repress NMD and thereby upregulate NMD target transcripts, we examined the expression pattern of miR-128 in mice in vivo. Northern blot analysis revealed that mature miR-128 is most highly expressed in the adult brain, at lower levels in the adult thymus, and undetectable in the other adult tissues we tested (Figure 3A). To assess the developmental window when miR-128 might function, we assayed miR-128 levels during mouse embryonic and postnatal brain development. Mature miR-128 level in the mouse brain dramatically increases between embryonic day 9.5 (E9.5) and E14.5, continues to increase during postnatal development, and persists at high levels in adulthood (Figures 3B and S3A). Locked-nucleic acid (LNA) in situ hybridization of adult mouse brain sections revealed that miR-128 is highly expressed in specific regions, including the cerebral cortex, hippocampus, and the olfactory bulb (Figures 3C and S3B). To determine which cell types express miR-128 in these brain regions, we used a method that permits analysis of single tissue sections by LNA in situ hybridization followed by immunofluorescence analysis. This approach revealed that miR-128 is specifically expressed in cells positive for the neuron-specific marker NeuN, but not in NeuN-negative cells, including GFAP-positive glial cells (Figures 3D, 3E, and S3C). The expression of miR-128 in neurons, not glial cells, is consistent with previous results obtained from primary mouse neuronal and glial cell cultures (Smirnova et al., 2005). As a negative control, we used a probe against the testis-specific microRNA miR-883 and observed no detectable hybridization (Figure S3D). We conclude that miR-128 is selectively expressed in neurons within the cerebral cortex, hippocampus, and the olfactory bulb.
Figure 3. miR-128 is a Brain-Enriched, Developmentally Regulated, Neuron-Specific miRNA.
(A) Northern blot analysis of adult mouse tissues. U6 small nuclear RNA (snRNA) is the loading control. Brain (Br), heart (Ht), Intestine (Int), liver (Li), lung (Lu), spleen (Spl), and thymus (Thy).
(B) Northern blot analysis of mouse brains from the indicated embryonic (E) and postnatal (P) time points. U6 snRNA is the loading control.
(C) miR-128 LNA in situ hybridization followed by immunofluorescence analysis of the neuronal marker NeuN in a section of adult mouse brain containing the cerebral cortex and hippocampus (low magnification view).
(D, E) miR-128 LNA in situ hybridization followed by immunofluorescence analysis of adult cerebral cortex (D) and hippocampus (E) (both high magnification views). DAPI staining indicates location of cell nuclei (note the distinct glial cell morphologies revealed by GFAP staining).
We examined miR-128 expression in the rat and found that, like in mice, miR-128 is expressed in the adult cerebral cortex and hippocampus (Figure S4A). In primary rat hippocampal neuronal cells undergoing differentiation in vitro we observed an upregulation of mature miR-128 transcripts in the absence of a change in level of primary (pri) miR-128 transcript (Figure S4B), suggesting that miR-128 expression is upregulated by a negative posttranscriptional mechanism that is relieved during neuronal maturation.
The discovery that mature miR-128 transcripts are dramatically induced during brain development in vivo (Figures 3B and S3A) and during neuron maturation in vitro (Figure S4B) raised the possibility that miR-128 promotes neuron differentiation. To address this, we used the P19 stem cell line, which differentiates into neuron-like cells in response to retinoic acid (RA). We found that undifferentiated P19 cells lack detectable mature miR-128, but when induced to differentiate in response to RA, they dramatically upregulate mature miR-128 transcripts (Figure S4C and S4D). To determine whether induced miR-128 has a functional role in P19 cells, we transfected them with the sequence-specific miR-128 inhibitor described above. This inhibitor largely prevented the upregulation of Tuj-1 (Tubb3) and Map2 mRNA (neural markers) and the downregulation of Oct4 mRNA (stem cell marker) in response to RA (Figure S4E). In contrast, the miR-128 inhibitor did not significantly affect the expression of these three genes in undifferentiated P19 cells (Figure S4E), consistent with the fact that undifferentiated P19 cells do not detectably express mature miR-128 transcripts (Figures S1A, S4C, and S4D). Together, these data indicated that miR-128 has a role in promoting gene expression events closely associated with neural differentiation. To assess whether miR-128 is sufficient to induce neural differentiation, we transfected P19 cells with the miR-128 mimic. This triggered the same response as that elicited by RA: upregulation of Tuj-1 and Map2 mRNA levels and downregulation of Oct4 mRNA level (Figure S4F). Of note, this response was elicited with a concentration of miR-128 mimic that approximated a physiological level of mature miR-128 (8.1-fold less than in RA-differentiated P19 cells [SD ±2.3, n=3]). The downregulation of Oct4 is unlikely to result from the direct action of miR-128, as miRNA-target prediction algorithms (including targetscan.org and microrna.org) did not identify Oct4 as a direct miR-128 target. Instead, we interpret the downregulation of Oct4 as meaning that miR-128 triggers the differentiation of neural stem cells. The following lines of evidence suggested that miR-128 elicits this differentiation response in P19 cells through its ability to repress NMD. First, the same response was elicited by knockdown of the miR-128 target UPF1 (Figure S4F). Second, the miR-128 mimic downregulated the levels of Upf1- and Mln51–3′ UTR reporters (Figure S4G). Third, the miR-128 mimic upregulated the NMD substrate Arc mRNA (Giorgi et al., 2007), directly demonstrating that miR-128 represses NMD in P19 cells (Figure S4F). Fourth, RA downregulated Upf1 mRNA (Figure S4F), a response largely prevented by the miR-128 inhibitor (Figure S4E). Finally, the miR-128 inhibitor blunted the ability of RA to suppress NMD, as judged by the NMD substrate Arc (Figure S4E).
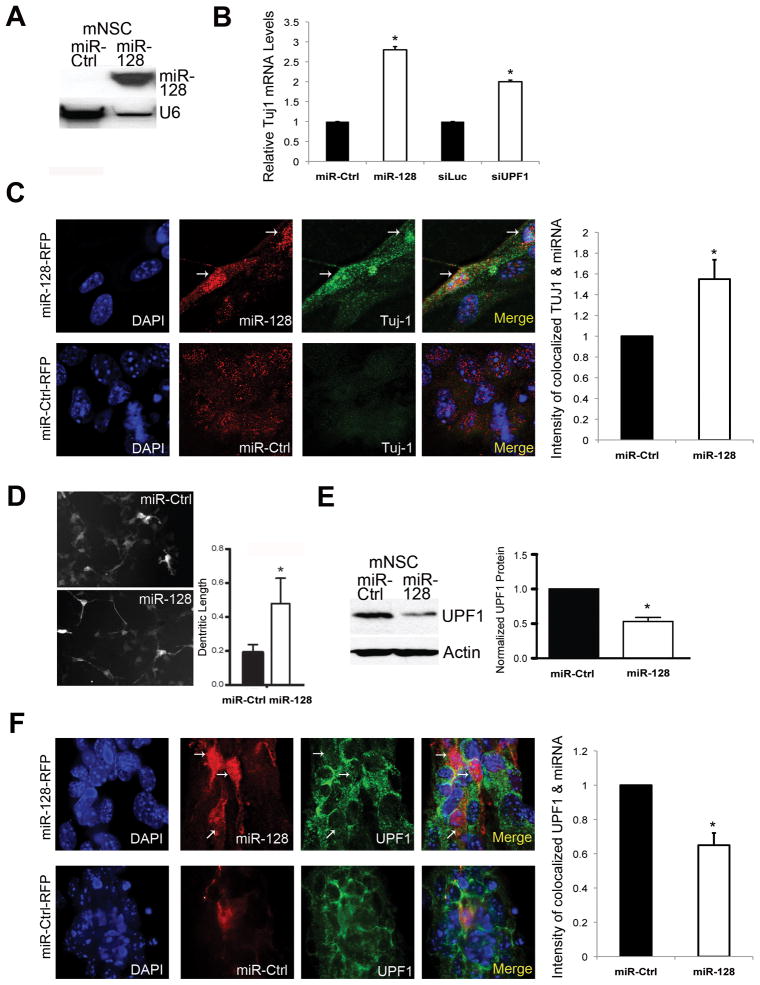
As an independent cell system to examine the ability of miR-128 to promote neural differentiation, we used mouse primary embryonic neural stem cells (NSCs), which, like P19 cells, lack detectable mature miR-128 transcripts (Figure 4A). We asked whether miR-128 is sufficient to promote neural differentiation-associated events in NSCs when they are grown under conditions that do not normally promote their differentiation (i.e., as neurospheres). We found that infection of NSCs with a lentivirus vector expressing physiological levels of miR-128 (6.3-fold less than postnatal day-1 brain [SD +0.07, n=3]) was sufficient to increase the expression of Tuj-1 mRNA (Figure 4B) and TUJ-1 protein expression (Figure 4C). Forced miR-128 expression in NSCs also increased the average dendritic spine length of NSCs (Figure 4D). Two lines of evidence suggested that miR-128 promotes TUJ-1 expression in NSCs through its ability to downregulate the NMD factor UPF1. First, forced expression of physiological levels of miR-128 reduced the expression of UPF1 in NSCs, as shown by both Western blot analysis (Figure 4E) and co-immunofluorescence quantitative analyses (Figure 4F). Second, depletion of UPF1 by RNAi was sufficient to upregulate Tuj-1 expression in NSCs (Figure 4B).
Figure 4. miR-128 Promotes a Neural Differentiation Phenotype and Inhibits UPF1 Expression in Neural Stem Cells.
(A) Northern blot analysis of mouse NSCs grown as neurospheres for 3 days under proliferating conditions and transfected with either the miR-128 mimic (miR-128) or control miRNA mimic (miR-Ctrl) (Ambion/ABI, Inc.). Shown is mature miR-128; U6 snRNA is the loading control.
(B) qPCR analysis of NSCs grown as described in panel A and transfected with miR-128 mimic, miR-Control, a siRNA specific for UPF1 (siUPF1), or luciferase siRNA (siLUC) as a negative control. Tuj-1 mRNA levels were normalized against L19 mRNA.
(C) Deconvolution microscopic analyses of TUJ-1 protein level in NSCs grown as described in panel A, infected with either miR-128-RFP- or miR-Ctrl-RFP-lentivirus, and cultured for 3 days. The images on the top left are of miR-128-RFP-lentivirus-infected cells; arrows mark the two cells expressing miR-128; the images on the bottom left are of miR-Ctrl-RFP-lentivirus-infected cells. DAPI staining shows location of all cell nuclei. The relative values shown on the right were determined using colocalization software and are the mean of three independent experiments. The intensity of colocalization of TUJ-1 and the miR-Ctrl is arbitrarily set to a value of 1.
(D) Mouse NSCs grown under adherent conditions were cotransfected with a GFP expression plasmid and either the miR-128 mimic or miR-Ctrl, followed by culture for 24 h. The graph shows the quantification of dendritic length in GFP-positive cells determined using ImageJ software.
(E) Left: Western blot analysis of endogenous UPF1 protein levels in mouse NSCs cultured and transfected as described in panel A. Right: quantification of 3 independent experiments using β-actin as a normalization control.
(F) Deconvolution microscopic analyses of UPF1 protein level in NSCs cultured, infected, and analyzed as described in panel C.
For all panels, error bars represent standard deviation and asterisks indicate statistically significant differences (P<0.05; paired Student’s t-test).
miR-128 Elicits Widespread Upregulation of Neural-Related Transcripts in Neural Stem Cells
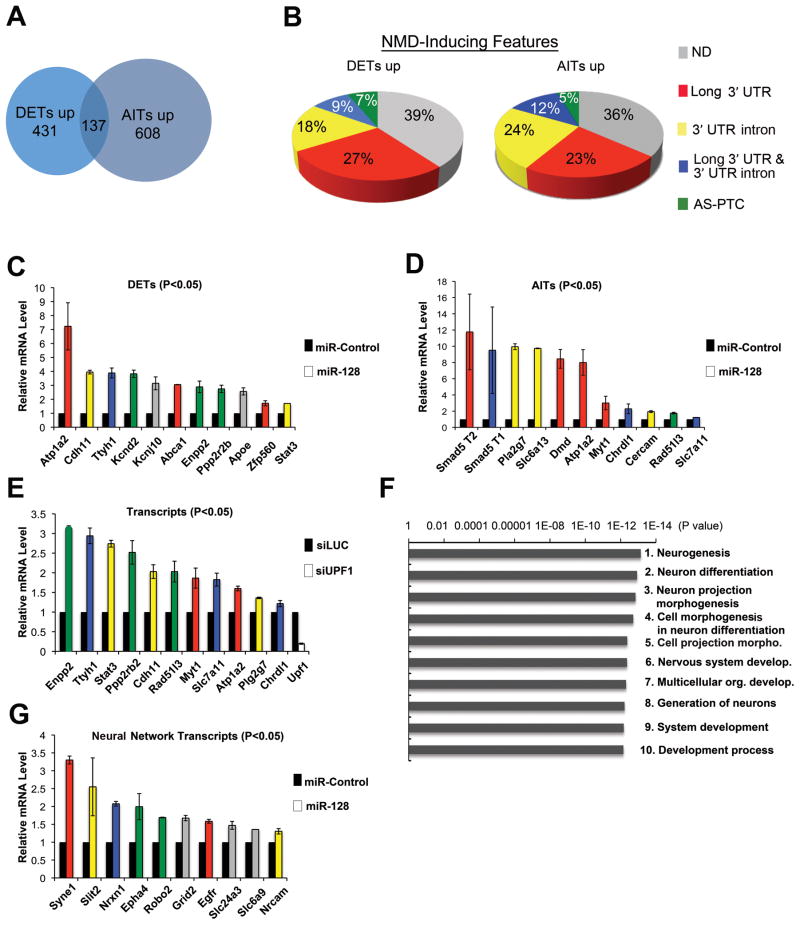
Together, the results described above suggested that miR-128 induces a gene expression program that promotes neural differentiation. To further assess this possibility, we used microarray analysis. We employed exon arrays for this analysis, as this platform detects expression levels of individual exons and thus allows detection of not only differentially expressed transcripts (DETs), but also alternative isoform transcripts (AITs). The latter is particularly relevant to our study because alternative RNA processing events (e.g., RNA splicing, alternative promoter usage, and alternative polyadenylation-site usage) often place a translation termination codon in a premature context and thereby trigger NMD (Rebbapragada and Lykke-Andersen, 2009). Exon array analysis identified 568 DETs and 745 AITs significantly upregulated by miR-128 in NSCs; 137 transcripts were in both the DET and AIT categories (Figure 5A). Tables S1 and S2 list the upregulated DETs and AITs, respectively. Downregulated DETs and AITs are also listed in these tables.
Figure 5. miR-128 Elicits Widespread Upregulation of Neural-Related Transcripts in Neural Stem Cells.
(A) VENN diagram of transcripts significantly upregulated (up) (p<0.05) in response to miR-128 in NSCs cultured and transfected as described in Figure 4A. The transcripts were identified using the Affymetrix mouse 1.0 ST exon array, which covers most of the known exons in 17,000 mouse protein-coding genes. Differentially expressed transcripts (DETs) and alternative isoform transcripts (AITs) are defined in the text.
(B) NMD-inducing features in the 100 most upregulated DETs and AITs, as determined using the SpliceMiner program: http://www.tigerteamconsulting.com/SpliceMiner/.
(C, D) Verification of DETs and AITs by qPCR analysis, performed on NSCs cultured and transfected as in panel A. mRNA levels were normalized to L19 RNA levels. The colors of the bars represent the known NMD-inducing feature of each transcript (see panel B).
(E) qPCR analysis of NSCs cultured as described in panel C and transfected with a siRNA specific for UPF1 (siUPF1) to inhibit NMD or a Luciferase siRNA (siLUC) as a negative control. Quantification and bar colors are as in panel C. We tested all transcripts in panels C and D for responsiveness to UPF1 knockdown; all those not shown in panel E were not statistically upregulated (p<0.05).
(F) Gene ontology (GO) processes most statistically overrepresented in DETs upregulated in response to forced miR-128 expression in NSCs, as identified using MetaCore™ software (GeneGo, San Diego, CA).
(G) Validation of selected transcripts in the neuronal GO categories “neurogenesis,” “neuron differentiation,” and “neuron projection morphogenesis.” qPCR analysis was performed on total cellular RNA from NSCs treated as described in panel C. Quantification and bar colors are as in panel C.
For all panels, experiments were performed in triplicate and error bars represent standard deviation. All transcripts shown in panels C, D, E, and G were significantly upregulated compared to controls (P<0.05; paired Student’s t-test).
If miR-128 represses NMD, this predicts that a significant proportion of the transcripts upregulated by miR-128 will have NMD-inducing features. Thus, we evaluated the upregulated DETs and AITs for three NMD-inducing features: (i) long 3′ UTR (>2 kb), (ii) 3′ UTR intron, and (iii) open reading frame (ORF) interrupted by a premature termination codon (PTC) because of alternative splicing (AS-PTC). We found that 61% and 64% of the 100 most strongly upregulated DETs and AITs, respectively, had NMD-inducing features (Figure 5B; listed in Tables S3 and S4). This is a conservative estimate given that databases are incomplete and it is likely that NMD can be induced by features besides the three we examined. As a control, we examined downregulated DETs and AITs and found that they had significantly (p<0.05) less NMD-inducing features than the upregulated DETs and AITs (Figure S5A). A long 3′ UTR was the most enriched NMD-inducing feature in miR-128-upregulated transcripts (>3-fold more frequent in upregulated [Figure 5B] than in downregulated [Figure S5A] DETs and AITs).
To validate the microarray results, we used qPCR analysis. Figure 5C shows that all of the most upregulated DETs identified by microarray analysis in response to miR-128 were verified by qPCR. Nine of these 11 validated upregulated DETs had known NMD-inducing features (indicated by the red, yellow, blue, and green colors, whose meanings are defined in Figure 5B). Remarkably, most of these DETs encode proteins with known functions in the nervous system: brain lipid metabolism (Enpp2, Apoe, and Abca1), synapse formation and function (Cdh11), ion channel function (Atp1a2, Kcnj10, Kcnd2 and Ttyh1), and cell signaling (Ppp2rb2 and Stat3). The upregulated AITs with highest statistical significance (p<0.00005) were confirmed by qPCR analysis with primers specific for the particular alternative transcript upregulated by miR-128 (Figure 5D). All 11 of the AITs we tested had NMD-inducing features (Figure 5D) and, like the verified DETs, most encode proteins with neural functions, including regulation of neural cell proliferation and differentiation (Smad5, Chrdl1, and Myt1), brain development (Pla2g7 and Cercam), and synapse functions (Dmd, Slc6a13, and Atp1a2). Of note, all of these “neural AITs” are predicted to encode the same protein as the major non-alternative isoform because the NMD-inducing feature is in the 3′ UTR, not the coding region. The most upregulated AIT, Smad5 mRNA, has two upregulated isoforms, T1 and T2, which have a 3′ UTR intron and a long 3′ UTR, respectively (Figure 5D).
The finding that many of the AITs and DETs have NMD-inducing features suggests that many are direct NMD targets. To directly assess this possibility, we determined whether they were upregulated in response to depletion of the NMD factor UPF1. Figure 5E shows AITs and DETs significantly upregulated in response to UPF1 depletion, providing strong evidence that these particular transcripts are direct NMD targets. However, some of the AITs and DETs with NMD-inducing features were not significantly upregulated after UPF1 depletion (see Figure 5E legend). While some of these may not be direct NMD targets, it is also possible that they are regulated by a branch of the NMD pathway that only requires trace amounts of UPF1 or is independent of UPF1. In this regard, we previously identified NMD mRNA substrates that are not upregulated when UPF1 is depleted by ~90% using RNAi (Chan et al., 2007).
To statistically categorize the miR-128-upregulated transcripts, we performed Gene Ontology (GO) process analysis on all 568 DETs and 745 AITs significantly upregulated (p<0.05) by miR-128. This GO analysis revealed that neural processes were overrepresented in both upregulated DET (Figure 5F) and AITs (Figure S5B). Remarkably, for the upregulated DETs, six of the ten most significantly overrepresented GO processes were neural categories; the four other categories also involve neural processes (Figure 5F). To verify regulation of the transcripts with “neural activities,” we performed qPCR analysis on DETs in 3 neural GO categories: “neurogenesis,” “neuron differentiation,” and “neuron projection morphogenesis.” As shown in Figure 5G, these “neural network” DETs were indeed upregulated by miR-128. These transcripts encode proteins involved in axon guidance (Slit2, Robo2, and Epha4), synapse function (Syne1, Nrxn1, Slc6a9, and Grid2), and neural differentiation and survival (Egfr, Nrcam, and Slc24a3). In striking contrast to the upregulated DETs and AITs, downregulated DETs and AITs were not overrepresented in any GO category specifically involving the nervous system (Figures S5C and S5D). Among the GO process categories overrepresented for downregulated DETs and AITs were several involved in cell proliferation (e.g., “cell cycle”) and RNA metabolism (e.g., “mRNA splice site selection”, “ribonucleoprotein complex biogenesis” and “RNA processing”).
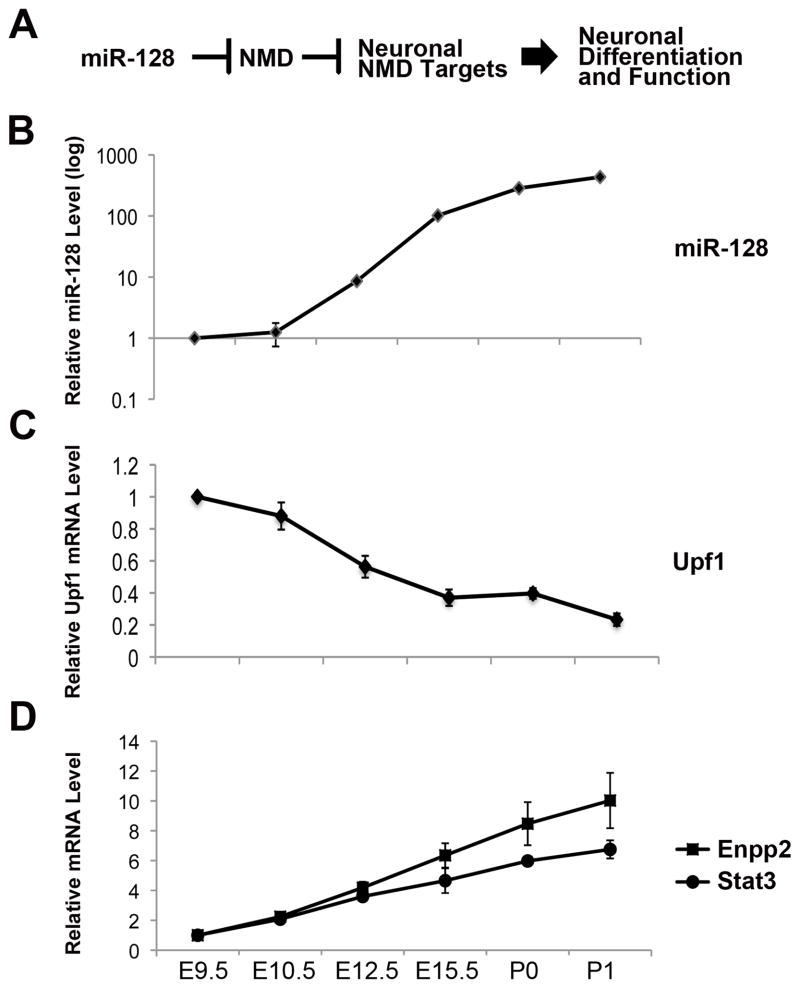
The data in our study led us to posit the model pictured in Figure 6A. Central to this model is the induction of miR-128 during brain development, which we showed to be the case by qPCR analysis (~430-fold increase in mature miR-128 levels between E9.5 and P1; Figure 6B), consistent with what we observed by Northern analysis (Figure 3B). Our model predicts that this induction of miR-128 will depress the level of one or both of the NMD factors it targets. Consistent with this prediction, we found that Upf1 mRNA levels decreased by ~5-fold during brain development (Figure 6C). In contrast, Mln51 mRNA level did not significantly decrease (Figure S6). Mln51 may not be downregulated for several reasons, including regulation by inputs in addition to NMD and/or region-specific NMD regulation that is masked by whole-brain analysis. A final prediction of our model is that the reduction in Upf1 level will lead to an upregulation of NMD substrates. Indeed, we observed that 4 of the 5 NMD/miR-128-regulated transcripts that we tested were upregulated during brain development (Figures 6D and S6). Among these upregulated transcripts are Enpp2, which encodes a protein controlling brain lipid metabolism (Nakanaga et al., 2010), and Stat3, which encodes a transcription factor that controls NSC differentiation (Cheng et al., 2011).
Figure 6. The Dramatic Upregulation of miR-128 During Brain Development is Accompanied by Upregulation of NMD Substrates.
(A) Model.
(B–D) qPCR analysis of pooled mouse brains from the indicated embryonic (E) and postnatal (P) time points. Mature miR-128 levels (B) were determined by Taqman-qPCR analysis and normalized against U6 snRNA levels. Upf1 mRNA (C) and NMD substrate (D) mRNA levels were determined using qPCR analysis and normalized against L19 mRNA levels. Error bars represent standard deviation.
DISCUSSION
We report the identification of a regulatory circuit that upregulates gene expression in a tissue-specific and developmentally regulated manner. This microRNA/NMD circuit acts post-transcriptionally and appears to be relatively ancient, as its components are highly conserved. In mice, this circuit acts in the nervous system, leading to the upregulation of transcripts encoding proteins involved in neural differentiation, maturation, and function. We suspect that this circuit regulates the nervous system in many other species, as the microRNA involved, miR-128, exhibits a brain-selective pattern of expression in several vertebrate species (Bak et al., 2008; Kapsimali et al., 2007; Sempere et al., 2004; Smirnova et al., 2005). In addition, several recent studies have indicated that NMD functions in brain development and/or neurological function. For example, knockdown of NMD factors in zebrafish causes disrupted brain patterning during embryonic development (Anastasaki et al., 2011; Wittkopp et al., 2009) and knockout of the NMD gene, Smg1, in mice prevents formation of the three primary embryonic brain vesicles (McIlwain et al., 2010). Adult mice heterozygous for a null mutation in the EJC factor gene, Magoh, have reduced brain size and defects in the cortex, including disordered cortical layering and few neurons (Silver et al., 2010). Likewise, knockdown of Magoh and other EJC factors in Xenopus laevis embryos elicits defects in sensory neurons and causes full-body paralysis (Haremaki et al., 2010).
NMD also appears to be important for modulating mature neuron functions. For example, knockdown of the EJC factor eIF4AIII in rat neurons activates their excitatory synaptic strength (Giorgi et al., 2007). A recent genetic screen in D. melanogaster revealed that mutations in several NMD factor genes disrupt neuromuscular junction synapses, neurotransmission responses, and synaptic vesicle cycling (Long et al., 2010). In humans, germline mutations in the NMD gene, UPF3B, cause intellectual disability (Tarpey et al., 2007) and recent studies have shown that mutations in UPF3B are also associated with autism, attention deficit hyperactivity disorder, and schizophrenia (Addington et al., 2010; Laumonnier et al., 2010). An interesting future question is why UPF3B is unique among NMD genes in not eliciting embryonic lethality when mutated (McIlwain et al., 2010; Medghalchi et al., 2001; Silver et al., 2010; Weischenfeldt et al., 2008). Two possible explanations are that UPF3B is selectively required for only a branch of the NMD pathway (Chan et al., 2007) and its loss is buffered by a compensatory response involving its paralog UPF3A (Chan et al., 2009)
We obtained several lines of evidence that suggest that NMD is progressively inhibited as neural development proceeds. While this serves the purpose of upregulating transcripts important for neural development and function, it brings up the possibility that NMD’s quality control role is sub-optimal in mature neurons. If so, perhaps mature neurons have less of a need for RNA surveillance than other cell types. Another possibility is that mature neurons actually have a magnitude of NMD similar to most other cell types, but that the immature neural cells that they come from have high-efficiency NMD.
Like most miRNAs, miR-128 has relatively modest effects on its targets, which we suggest is optimal for its role in negatively regulating an essential mechanism. Thus, by repressing, not eliminating NMD, miR-128 modulates the levels of a broad array of transcripts without unduly compromising the quality control function of NMD. Furthermore, because miR-128 is developmentally regulated and expressed in a cell type- and tissue-specific manner (Figures 3 and S3), its negative effects on NMD are confined to particular contexts. We suggest that the expression of miR-128 in the brain and its ability to fine-tune (not turn on-and-off) NMD makes it an optimal mechanism to serve as a homeostatic mechanism to maintain normal brain function. Indeed, there is growing evidence that the brain has several homeostatic mechanisms in place that, if perturbed, lead to specific brain aberrations, including autism, schizophrenia, attention deficit disorders, and intellectual disability (Ramocki and Zoghbi, 2008). Given that loss of the NMD factor UPF3B appears to cause these same conditions (Addington et al., 2010; Tarpey et al., 2007), it is tempting to speculate that NMD has important roles in brain homeostatic mechanisms. In support of this, the NMD target transcript, Arc, has recently been shown to participate in a homeostatic mechanism that controls neural activity (Beique et al., 2011).
We posit that miR-128 upregulates waves of transcripts with different properties and functions. We suggest that the first wave is dominated by mRNAs directly regulated by NMD. Some of these first-wave transcripts will encode proteins that induce secondary waves of mRNAs, most of which will not be NMD substrates but will have important roles in amplifying and refining neural responses. Another category of transcripts that might contribute to miR-128’s effects in the nervous system is direct miR-128 targets other than UPF1 and MLN51. Little is known about direct miR-128 targets (Godlewski et al., 2008; Zhang et al., 2009), but it is possible that a subset of them will directly impinge on neural development and/or function.
In summary, our findings demonstrate that the convergence of two post-transcriptional pathways—NMD and microRNAs—can have a profound impact on gene expression in the nervous system (Figures 6A). This may have clinical relevance, as there is increasing evidence linking both NMD and miR-128 to human neurological diseases. For example, as described above, humans with mutations that cripple NMD have intellectual disability and psychological disorders (Addington et al., 2010; Tarpey et al., 2007). In the case of miR-128, its levels have been reported to be deregulated in autism (Abu-Elneel et al., 2008), prion-induced neurodegeneration (Saba et al., 2008), Huntington’s disease (Lee et al., 2011), Parkinson’s disease (Kim et al., 2007), and Alzhemier’s disease (Lukiw, 2007). Together, these clinical findings suggest the possibility that the defects in the miR-128/NMD circuit result in changes in gene expression that contribute to a subset of human psychiatric and neurodegenerative disorders.
EXPERIMENTAL PROCEDURES
Mammalian cell culture and transfections
HEK293, HeLa, P19, primary rat neuronal cells, and mNSCs were cultured as described in the Supplement. Methods for their transient transfection and infection with lentiviruses are also described in the Supplement.
Chick neural tube electroporations and X. laevis embryo microinjections
X. laevis embryos were cultured and microinjected, as described (Patil et al., 2006). Chick stage-13 neural tubes were electroporated in ovo, as described (Cao et al., 2007).
Assays
The Supplement explains the assays and reagents used for RNA (qPCR, TaqMan-qPCR, Northern blotting, in situ hybridization, and microarray) and protein (Western blotting and immunofluorescence) analyses.
Supplementary Material
Acknowledgments
We are grateful to Jens Lykke-Anderson (UCSD) and Catherine Tomasseto (Institut de Genetique, Strasbourg, France) for expression vectors and antiserum. We thank Kim Tolias (Baylor College of Medicine) and Sadhan Majumder (M. D. Anderson Cancer Center) for providing primary cells, Howard Gutstein (M. D. Anderson Cancer Center) for providing rat brain tissue sections, Christopher Kinter (Salk Institute) for providing X. laevis eggs, Benjamin Yu and Christopher Cowing-Zitron (UCSD) for bioinformatic support, and Gilbert Cote, Angela Bhalla, and Miriam Buttigieg (M. D. Anderson Cancer Center) for their experimental assistance and/or intellectual contributions to various portions of this manuscript. Finally, we thank Fred “Rusty” Gage, Ahmed Denli, and Leah Boyer (Salk Institute, La Jolla) for antibodies and helpful advice. This work was supported by National Institutes of Health grant GM058595.
Footnotes
Supplemental data include 6 figures, 5 supplemental tables, experimental procedures and supplementary references.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, Lao K, Kosik KS. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, Miller R, Tossell J, Bakalar J, Germain G, et al. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasaki C, Longman D, Capper A, Patton EE, Caceres JF. Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beique JC, Na Y, Kuhl D, Worley PF, Huganir RL. Arc-dependent synapse-specific homeostatic plasticity. Proc Natl Acad Sci U S A. 2011;108:816–821. doi: 10.1073/pnas.1017914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem J. 2010;430:365–377. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Cheng X, Jin G, Zhang X, Tian M, Zou L. Stage-dependent STAT3 activation is involved in the differentiation of rat hippocampus neural stem cells. Neurosci Lett. 2011 doi: 10.1016/j.neulet.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Haremaki T, Sridharan J, Dvora S, Weinstein DC. Regulation of vertebrate embryogenesis by the exon junction complex core component Eif4a3. Dev Dyn. 2010;239:1977–1987. doi: 10.1002/dvdy.22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Goff SP. Upf1 senses 3′UTR length to potentiate mRNA decay. Cell. 2010;143:379–389. doi: 10.1016/j.cell.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PW, Huang HD, Hsu SD, Lin LZ, Tsou AP, Tseng CP, Stadler PF, Washietl S, Hofacker IL. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey BD, Kirsch S, Morris D. Molecular cloning and characterization of the chicken cationic amino acid transporter-2 gene. Comp Biochem Physiol B Biochem Mol Biol. 2008;150:301–311. doi: 10.1016/j.cbpb.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T, Briault S, Fryns JP, Hamel B, Chelly J, et al. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol Psychiatry. 2010;15:767–776. doi: 10.1038/mp.2009.14. [DOI] [PubMed] [Google Scholar]
- Le Guiner C, Gesnel MC, Breathnach R. TIA-1 or TIAR is required for DT40 cell viability. J Biol Chem. 2003;278:10465–10476. doi: 10.1074/jbc.M212378200. [DOI] [PubMed] [Google Scholar]
- Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, et al. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Long AA, Mahapatra CT, Woodruff EA, 3rd, Rohrbough J, Leung HT, Shino S, An L, Doerge RW, Metzstein MM, Pak WL, et al. The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J Cell Sci. 2010;123:3303–3315. doi: 10.1242/jcs.069468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Nakanaga K, Hama K, Aoki J. Autotaxin--an LPA producing enzyme with diverse functions. J Biochem. 2010;148:13–24. doi: 10.1093/jb/mvq052. [DOI] [PubMed] [Google Scholar]
- Pacheco TR, Gomes AQ, Barbosa-Morais NL, Benes V, Ansorge W, Wollerton M, Smith CW, Valcarcel J, Carmo-Fonseca M. Diversity of vertebrate splicing factor U2AF35: identification of alternatively spliced U2AF1 mRNAS. J Biol Chem. 2004;279:27039–27049. doi: 10.1074/jbc.M402136200. [DOI] [PubMed] [Google Scholar]
- Patil SS, Alexander TB, Uzman JA, Lou CH, Gohil H, Sater AK. Novel gene ashwin functions in Xenopus cell survival and anteroposterior patterning. Dev Dyn. 2006;235:1895–1907. doi: 10.1002/dvdy.20834. [DOI] [PubMed] [Google Scholar]
- Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Raes J, Izaurralde E. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw HJ, Myung K, Walsh CA, Gaiano N, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, Gong Y, Yin B, Qiang B, Zhao J, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med. 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.