Abstract
Background
Infections with the opisthorchid liver flukes Clonorchis sinensis, Opisthorchis viverrini, and O. felineus cause severe health problems globally, particularly in Southeast Asia. Early identification of the infection is essential to provide timely and appropriate chemotherapy to patients.
Results
In this study we evaluate a PCR-based molecular identification method, Multiplex Ligation-dependent Probe Amplification (MLPA), which allows rapid and specific detection of single nucleotide acid differences between Clonorchis sinensis, Opisthorchis viverrini and O. felineus. Three probe pairs were derived from the Internally Transcribed Spacer 1 (ITS1) of three opisthorchid liver flukes using a systematic phylogenetic analysis. Specific loci were detected in all three species, yielding three amplicons with 198,172 and 152 bp, respectively, while no cross reactions were observed. A panel of 66 C. sinensis isolates was screened using MLPA. All species were positively identified, and no inhibition was observed. The detection limit was 103 copies of the ITS gene for the three liver flukes, or about 60 pg genomic DNA for Clonorchis sinensis. Amplification products can be detected by electrophoresis on agarose gel or in a capillary sequencer. In addition, genomic DNA of Clonorchis sinensis in fecal samples of infected rats was positively amplified by MLPA.
Conclusion
The flexibility and specificity make MLPA a potential tool for specific identification of infections by opisthorchid liver flukes in endemic areas.
Background
Clonorchis sinensis (C. sinensis), Opisthorchis viverrini (O. viverrini) and Opisthorchis felineus (O. felineus) (Opisthorchiidae) are among the most frequent endemic food-borne liver flukes, causing severe clonorchiasis and opisthorchiasis. Humans contract the disease through consumption of raw or inadequately cooked freshwater fish containing the infective metacercariae. About 35 million people are infected with C. sinensis globally. Main endemic areas are located in southern Asia including China, Korea, Japan, Taiwan and Vietnam [1]. In China the estimated infection by C. sinensis is 15 million [2,3]. Approximately 9 million people are infected with O. viverrini in Thailand, Cambodia, and Laos [4-6]. In eastern Europe 1.2 to 1.5 million patients are infected with O. felineus [7]. In recent years, endemic areas of liver flukes are expanding to North America and Europe due to fish import and immigration [7-9].
Current clinical diagnosis of liver fluke infection is by direct microscopy of eggs in feces. However, this procedure is time-consuming and inaccurate, resulting in false-negatives due to the difficulty to distinguish eggs from each other or from those of closely related heterophyides [10-12]. As a result, appropriate chemotherapy may be delayed. Hence there is an urgent need for a novel tool to diagnose the infection.
A number of methods have been developed to identify or detect liver flukes using DNA, mRNA or protein. Among PCR-based molecular methods, nested-PCR [13] and loop-mediated isothermal amplification (LAMP) [14] are particularly promising [15-19]. However, simple PCR amplification carries the risk of false-negative data, due to PCR inhibitors involved in complex samples [20,21]. Moreover, mixed liver fluke infection may also hamper the application of simple PCR [22]. As an alternative, multiplex PCR may achieve high efficiency with simultaneous amplification of two or more genetic loci in one reaction, while this also may reduce the number of false-negative or false-positive results [23]. Although multiplex PCR has been reported to discriminate between C. sinensis and O. viverrini [24], it is technically difficult to optimize PCRs for amplification of multiple genes or loci, because each primer pair requires a different optimal combination of reagents and annealing temperatures. To overcome this problem we developed a multiplex PCR amplification technique using multiplex ligation-dependent probe amplification (MLPA).
MLPA is a simple, robust and fast method designed for simultaneous detection of specific genomic sequences targeting multiple mutations to amplify specific MLPA probes rather than target DNA [25]. In MLPA (Figure 1), two oligonucleotides (up to 45 pairs in one reaction) hybridize immediately on the target DNA. In addition to a target-specific sequence, each of the oligonucleotides contains one of two sequences recognized by a universal PCR primer pair. After denaturing and hybridization, two parts of each MLPA probe are ligated by a specific ligase enzyme, followed by PCR amplification using a universal primer pair. Non-hybridized probes are not removed, enabling a high throughput 'one-tube' method. MLPA probes are designed in such a way that each amplification product is identified by size using separation by capillary electrophoresis. Differences in relative probe signals between samples reflect differences in the probe target sequences. MLPA is widely used to identify point mutations [26], insertions [27], deletions [28], duplications [29], and recombination events [30] and is also applied for quantification of mRNAs [31] and determination of methylation status of CpG islands [32].
Figure 1.
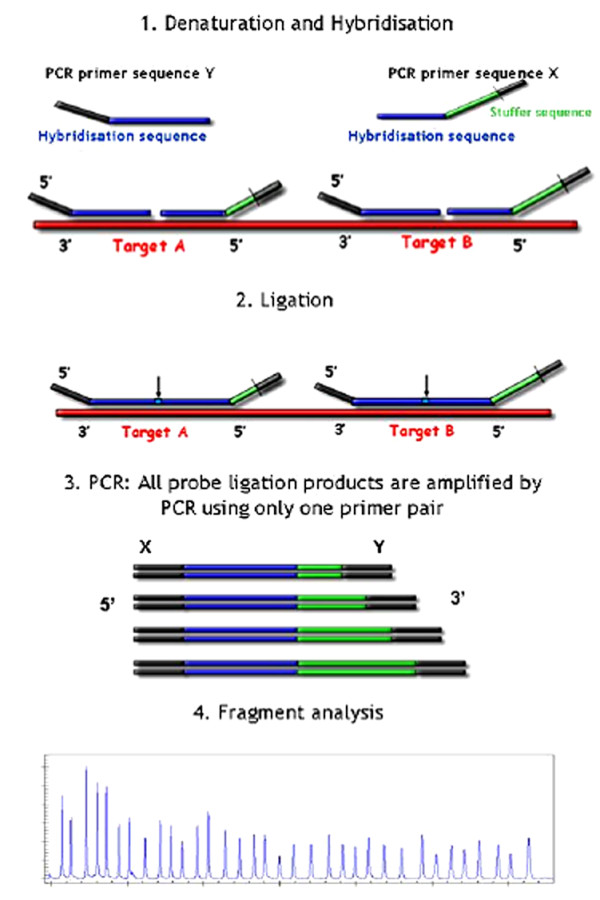
Outline of the MLPA technique (http://www.mlpa.com). After hybridisation to their target sequence in the sample DNA, the probe oligonucleotides are enzymatically ligated. One probe oligonucleotide contains a non-hybridising stuffer sequence of variable length. Ligation products can be amplified using PCR primer sequences X and Y amplification product of each probe has a unique length (130-480 nt). Amplification products are separated by electrophoresis.
The elegance and simplicity of MLPA makes it applicable to various types of clinical samples, such as blood [33], amniotic fluid [34] or tumor tissue [35]. In pathogen detection, MLPA has only been applied to Mycobacterium tuberculosis [36], bacterial species in oral biofilms [37], respiratory viruses [38] and Penicillium marneffei [39], but not to parasites. In this study, we evaluate MLPA for the rapid identification of the opisthorchid liver flukes C. sinensis, O. viverrini and O. felineus, and establish specificity of the method to discriminate these three liver flukes in a single-tube reaction.
Results
In this study, the MLPA assay was adapted to identify C. sinensis, and discriminate with other opisthorchid liver flukes, O. viverrini, and O. felineus. We first performed a haplotype analysis of the three liver flukes and of phylogenetically related species to search for unique loci for MLPA probe design (Figure 2). Three specific loci were selected for designing species-specific pairs of oligonucleotide probes for MLPA (Table 1). Web-based BLAST analysis showed a low degree of similarity of the specific probes compared with other parasites. Three artificial templates of C. sinensis, O. viverrini and O. felineus were used to evaluate the specificity and sensitivity of the MLPA assay. MLPA reactions without artificial templates were used as negative controls. Artificial templates of padlock probes were used to evaluate the detection limits of the MLPA assay. These pairs of probes allowed specific amplification of the ITS1 gene of C. sinensis, O. felineus and O. viverrini, and yielded three amplicon of sizes 198, 170, and 152 bp, respectively (Figure 3). The amplicons were 100% consistent with the initial DNA used in each MLPA reaction. The detection limit of the MLPA assay for artificial DNA was found to be approximately 103 copies of the ITS gene for the three liver flukes analyzed (Figure 4) or 60 picogram genomic DNA for C. sinensis (data not shown) after the amplification products were visualized by electrophoresis on a 5% agarose gel. A total of 66 different DNA samples of adult liver flukes were tested and the data showed that the pairs of MLPA probes were able to amplify all loci presented in the samples (Table 2). No inhibition was observed after the addition of the same concentration of Opisthorchis viverrini or Opisthorchis felineus artificial template DNA (Table 2). The MLPA products could be successfully detected using a capillary sequencer (Figure 5). Moreover, specific amplification was also achieved by the use of fecal samples from infected rats (Table 3).
Figure 2.
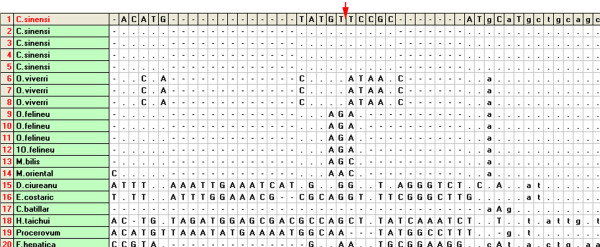
The multiple alignment analysis of ITS1 gene of C.sinensis, O.viverrini, O.felineus and closely species in taxonomy using DNAsp software (partially). The arrow show the positions within nucleotides of three padlock probes designed in this study. The dots represent identical nucleotides as in the C.sinensis sequence given at the top, the dashes indicate deletions.
Table 1.
Probes and primers used in this study
| Primer | Oligonucleotides Sequences(5`-3`) |
|---|---|
| CsPLaw | PaACATACATGTAGAACATGACGGGTTCGGGCAATTCGTTATTGGCCCTATAGTGAGGTCTTCTCTATTGTCACCGTATGCACATCTCGGAATCAAGCTGg |
| CsPLfw | hACTGGATTCAGGTTCACGAAGCTGCATTATCGATCAGTACCAGTGTAGTACAGCGCCGGTGAAATTATCGCCACAGGCCTTTCTGCAGCATGCATGCGGAb |
| OfPLaw | PcCTATACATGTAGAATATGACGGAGTTCGGGCAATTCGTTATTGGCCCTATAGTGAGGTCTTCTCTATTGTCACCGTATGCACATCTCGGAATCAAGCTGg |
| OfPLfw | hACTGGATTCAGGTTCACGAAGCTGCATTATCGATCAGTTATCGCCACAGGCCTTTAACGCTGCAGCATGTATGTd |
| OvPLaw | PeTTATACATGTAGGTTATACATGACGTTCGGGCAATTCGTTATTGGCCCTATAGTGAGGTCTTCTCTATTGTCACCGTATGCACATCTCGGAATCAAGCTGg |
| OvPLfw | hACTGGATTCAGGTTCACGAAGCTCAGGCCTTTAACGCTGCAGCATGTATGGf |
| pF3 | Fam-CAGCTTAGTTCCGAGATGT |
| pB3 | ACTGGATTCAGGTTCACGA |
| Cs-T | CCGTCATGTTCTACATGTATGTTCCGCATGCATGCTGCAG |
| Of-T | TCCGTCATATTCTACATGTATAGACATACATGCTGCAGCGTT |
| Ov-T | GTCATGTATAACCTACATGTATAACCATACATGCTGCAGCGTT |
| ITS1f | CGATTCTAGTTCCGTCATCT |
| ITS1r | CCGCTCAGAGTTGTACTCAT |
a-f: complementary sequence of ITS loci in padlock probes, denoted with italics and bold.
g: pF3 binding sequence in underline and bold.
h: pB3 binding sequence in underline.
P: phosphorylated
Fam: Fam fluorescent labled at the 5' end.
Figure 3.
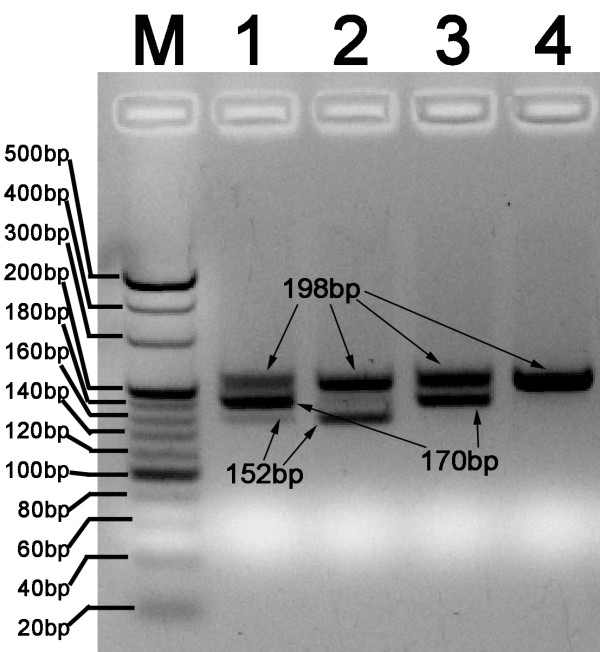
Specificity analysis of three MLPA probes on agarose electrophoresis. All three probe pairs (CsPL for C.sinensis, OfPL for O.felineus, OvPL for O.viverrini) were added, different combined artificial DNA (Cs-T for C.sinensis, Ov-T for O.viverrini, Of-T for O.felineus) used as templates. The MLPA amplicons were separated in 5% agarose gel by electrophoresis and the image was taken under UV light. Fragment sizes: C.sinensis(198bp), O.felineus (170bp), O.viverrini(152bp). Lane M, 20 bp DNA ladder; Lane1, Cs-T+Ov-T+Of-T; Lane 2, Cs-T+Ov-T; lane 3, Cs-T+ Of-T; lane 4, Cs-T.
Figure 4.
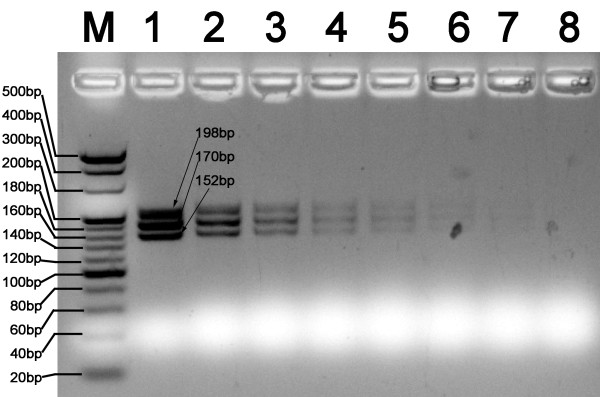
Analytical sensitivity of MLPA assay in detection of the artificial template of the ITS1 gene. DNA samples from these artificial templates were subjected to MLPA analysis and MLPA products were separated in 5% agarose gel by electrophoresis. M, 20 bp DNA ladder; Lanes 1 to 8, 2 × 109, 2 × 108, 2 × 107, 2 × 106, 25 × 105, 2 × 104, 2 × 103 and 2 × 102 copies/reaction, respectively.
Table 2.
Evaluation of the three probe pairs in 66 different strains of C.sinensis isolates
| No. | GenBank | Source | CsPL+OfPL+OvPL+Ov-T+Of-T | CsPL+OfPL+OvPL+Of-T | CsPL+OfPL+OvPL | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 198bp | 170bp | 152bp | 198bp | 170bp | 152bp | 198bp | 170bp | 152bp | |||
| 1 | HQ874538 | Cat, Anhui, China | + | + | + | + | + | - | + | - | - |
| 2 | HQ874523 | Cat, Anhui, China | + | + | + | + | + | - | + | - | - |
| 3 | HQ874584 | Cat, Anhui, China | + | + | + | + | + | - | + | - | - |
| 4 | HQ874537 | Cat, Anhui, China | + | + | + | + | + | - | + | - | - |
| 5 | HQ874599 | Cat, Anhui, China | + | + | + | + | + | - | + | - | - |
| 6 | HQ874585 | Cat, Anhui, China | + | + | + | + | + | - | + | - | - |
| 7 | HQ874586 | Cat, Anhui, China | + | + | + | + | + | - | + | - | - |
| 8 | HQ874588 | Cat, Anhui, China | + | + | + | + | + | - | + | - | - |
| 9 | HQ874540 | Cat, Guangdong, China | + | + | + | + | + | - | + | - | - |
| 10 | HQ874535 | Cat, Guangdong, China | + | + | + | + | + | - | + | - | - |
| 11 | HQ874541 | Cat, Guangdong, China | + | + | + | + | + | - | + | - | - |
| 12 | HQ874602 | Cat, Guangdong, China | + | + | + | + | + | - | + | - | - |
| 13 | HQ874587 | Cat, Guangdong, China | + | + | + | + | + | - | + | - | - |
| 14 | HQ874532 | Cat, Guangdong, China | + | + | + | + | + | - | + | - | - |
| 15 | HQ874581 | Cat, Guangdong, China | + | + | + | + | + | - | + | - | - |
| 16 | HQ874582 | Cat, Guangdong, China | + | + | + | + | + | - | + | - | - |
| 17 | HQ874542 | Cat, Guangxi, China | + | + | + | + | + | - | + | - | - |
| 18 | HQ874536 | Cat, Guangxi, China | + | + | + | + | + | - | + | - | - |
| 19 | HQ874543 | Cat, Guangxi, China | + | + | + | + | + | - | + | - | - |
| 20 | HQ874529 | Cat, Guangxi, China | + | + | + | + | + | - | + | - | - |
| 21 | HQ874580 | Cat, Guangxi, China | + | + | + | + | + | - | + | - | - |
| 22 | HQ874533 | Cat, Guangxi, China | + | + | + | + | + | - | + | - | - |
| 23 | HQ874525 | Cat, Guangxi, China | + | + | + | + | + | - | + | - | - |
| 24 | HQ874579 | Cat, Guangxi, China | + | + | + | + | + | - | + | - | - |
| 25 | HQ874544 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 26 | HQ874545 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 27 | HQ874593 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 28 | HQ874578 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 29 | HQ874539 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 30 | HQ874592 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 31 | HQ874546 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 32 | HQ874547 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 33 | HQ874524 | Cat, Hubei, China | + | + | + | + | + | - | + | - | - |
| 34 | HQ874601 | Cat, Henan, China | + | + | + | + | + | - | + | - | - |
| 35 | HQ874550 | Cat, Henan, China | + | + | + | + | + | - | + | - | - |
| 36 | HQ874597 | Cat, Henan, China | + | + | + | + | + | - | + | - | - |
| 37 | HQ874595 | Cat, Henan, China | + | + | + | + | + | - | + | - | - |
| 38 | HQ874573 | Cat, Henan, China | + | + | + | + | + | - | + | - | - |
| 39 | HQ874572 | Cat, Henan, China | + | + | + | + | + | - | + | - | - |
| 40 | HQ874571 | Cat, Henan, China | + | + | + | + | + | - | + | - | - |
| 41 | HQ874589 | Cat, Henan, China | + | + | + | + | + | - | + | - | - |
| 42 | HQ874598 | Cat, Hunan, China | + | + | + | + | + | - | + | - | - |
| 43 | HQ874590 | Cat, Hunan, China | + | + | + | + | + | - | + | - | - |
| 44 | HQ874591 | Cat, Hunan, China | + | + | + | + | + | - | + | - | - |
| 45 | HQ874551 | Cat, Hunan, China | + | + | + | + | + | - | + | - | - |
| 46 | HQ874534 | Cat, Hunan, China | + | + | + | + | + | - | + | - | - |
| 47 | HQ874552 | Cat, Hunan, China | + | + | + | + | + | - | + | - | - |
| 48 | HQ874553 | Cat, Hunan, China | + | + | + | + | + | - | + | - | - |
| 49 | HQ874554 | Cat, Hunan, China | + | + | + | + | + | - | + | - | - |
| 50 | HQ874555 | Dog, Jilin, China | + | + | + | + | + | - | + | - | - |
| 51 | HQ874556 | Dog, Jilin, China | + | + | + | + | + | - | + | - | - |
| 52 | HQ874557 | Dog, Jilin, China | + | + | + | + | + | - | + | - | - |
| 53 | HQ874570 | Dog, Jilin, China | + | + | + | + | + | - | + | - | - |
| 54 | HQ874528 | Dog, Jilin, China | + | + | + | + | + | - | + | - | - |
| 55 | HQ874527 | Dog, Jilin, China | + | + | + | + | + | - | + | - | - |
| 56 | HQ874558 | Cat, Jiangsu, China | + | + | + | + | + | - | + | - | - |
| 57 | HQ874566 | Cat, Jiangsu, China | + | + | + | + | + | - | + | - | - |
| 58 | HQ874559 | Cat, Jiangsu, China | + | + | + | + | + | - | + | - | - |
| 59 | HQ874530 | Cat, Jiangsu, China | + | + | + | + | + | - | + | - | - |
| 60 | HQ874583 | Cat, Jiangsu, China | + | + | + | + | + | - | + | - | - |
| 61 | HQ874569 | Cat, Jiangsu, China | + | + | + | + | + | - | + | - | - |
| 62 | HQ874604 | Cat, Jiangsu, China | + | + | + | + | + | - | + | - | - |
| 63 | HQ874565 | Cat, Jiangxi, China | + | + | + | + | + | - | + | - | - |
| 64 | HQ874560 | Cat, Jiangxi, China | + | + | + | + | + | - | + | - | - |
| 65 | HQ874561 | Cat, Jiangxi, China | + | + | + | + | + | - | + | - | - |
| 66 | HQ874564 | Cat, Jiangxi, China | + | + | + | + | + | - | + | - | - |
| 67 | Negative control | - | - | - | - | - | - | - | - | - | |
CsPL: Clonorchis sinensis Padlock probe pairs
OfPL: Opisthorchis felineus Padlock probe pairs
OvPL: Opisthorchis viverrini Padlock probe pairs
Cs-T: Artificial template of Clonorchis sinensis Padlock probe pairs
Ov-T: Artificial template of Opisthorchis viverrini Padlock probe pairs
Of-T: Artificial template of Opisthorchis felineus Padlock probe pairs
Figure 5.
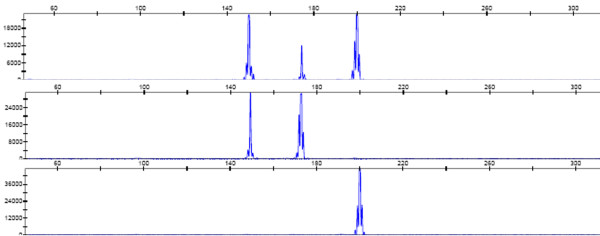
Electropherogram showing peaks generated by MLPA. Template DNA from individual species is used in the reaction. The MLPA reaction included all the 3 probes designed, Rox 500 as internal molecular standerd. Fragment sizes (bp) correspond to: 198 = C.sinensis, 170 = O.felineus, 152 = O.viverrini
Table 3.
Fecal samples of infected rats and data from the three different detection methods
| Detection method | ||||||
|---|---|---|---|---|---|---|
| Fecal Samples no. | Microscopy detection | ITS PCR | MLPA | |||
| First Run | Second Run | CsPL | OvPL | OfPL | ||
| 1 | + | - | + | + | - | - |
| 2 | + | - | + | + | - | - |
| 3 | + | - | + | + | - | - |
| 4 | + | - | + | + | - | - |
| 5 | + | - | + | + | - | - |
| 6 | + | - | + | + | - | - |
| 7 | + | - | + | + | - | - |
| 8 | + | - | + | + | - | - |
| 9 | + | - | + | + | - | - |
| 10 | + | - | + | + | - | - |
| 11 | + | - | + | + | - | - |
| 12 | + | - | + | + | - | - |
| 13 | + | - | + | + | - | - |
| 14 | + | - | + | + | - | - |
| 15 | + | - | + | + | - | - |
| 16 | + | - | + | + | - | - |
| 17 | + | - | + | + | - | - |
| 18 | + | - | + | + | - | - |
| 19 | + | - | + | + | - | - |
| 20 | + | - | + | + | - | - |
| 21 | + | - | + | + | - | - |
| 22 | + | - | + | + | - | - |
| 23 | + | - | + | + | - | - |
| 24 | + | - | + | + | - | - |
| 25 | + | - | + | + | - | - |
| 26 | + | - | + | + | - | - |
| 27 | + | - | + | + | - | - |
| 28 | + | - | + | + | - | - |
| 29 | + | - | + | + | - | - |
| 30 | + | - | + | + | - | - |
| 31 | + | - | + | + | - | - |
| 32 | + | - | + | + | - | - |
| 33 | + | - | + | + | - | - |
| 34 | + | - | + | + | - | - |
| 35 | + | - | + | + | - | - |
| 36 | + | - | + | + | - | - |
| 37 | - | - | - | - | - | - |
| 38 | - | - | - | - | - | - |
| 39 | - | - | - | - | - | - |
| 40 | - | - | - | - | - | - |
| 41 | - | - | - | - | - | - |
| 42 | - | - | - | - | - | - |
| 43 | - | - | - | - | - | - |
| 44 | - | - | - | - | - | - |
| 45 | - | - | - | - | - | - |
| 46 | - | - | - | - | - | - |
| 47 | - | - | - | - | - | - |
| 48 | - | - | - | - | - | - |
CsPL: Clonorchis sinensis Padlock probe pairs
OfPL: Opisthorchis felineus Padlock probe pairs
OvPL: Opisthorchis viverrini Padlock probe pairs
Discussion
The use of ligated oligonucleotide probes in MLPA, and a number of other methods, allows specific detection of changes in single nucleic acids of targeted genes [40-42]. These probes hybridize to and capture these areas, which are then enriched through rolling circle amplification or by PCR. Using these probes we were able to simultaneously detect gene loci and characterize different strains in a single reaction. In the present study we evaluate the MLPA assay to identify and discriminate three opisthorchid liver flukes. The sensitivity and specificity of MLPA highlights that the method is a useful tool for prompt and accurate diagnosis, pathogen characterization, and epidemiological studies of parasite infections.
In our initial test, we evaluated the MLPA assay to identify and discriminate three liver flukes using artificial templates. All of the three probe pairs used allow specific amplification of the ITS1 locus of each respective species. The MLPA reaction was sensitive enough to detect 103 copies of artificial template DNA. This is consistent with previous studies on MLPA in oral biofilm, where DNA was detected at picogram levels [37]. The size of the C. sinensis genome varies from 500 to 700 Mbp (Wang et al., unpublished data), and 1 pg of DNA is equal to 978 Mbp of genomic DNA [43]. The weight of C. sinensis DNA is approximately 0.511-0.716 pg and the 103 copies of C. sinensis DNA detectable by MLPA is then roughly equivalent to 0.5-70 pg of genomic DNA. These results were consistent with the 60 pg genomic DNA of C. sinensis mentioned above. Results were comparable to a previous study indicating that the sensitivity of MLPA is equivalent to real-time PCR [38], while similar results were obtained in some studies focusing on O. viverrini [24] and C. sinensis [18]. However, our results deviate from a previous report where with simple PCR a detection limit of 10-12 ng was obtained [17]. This might be explained by the use of different target genes or by different copy numbers of target genes in the genome.
Furthermore, the results of the inhibition test indicated that the MLPA assay was not inhibited by the presence of non-target DNA. These data demonstrate a considerable potential for MLPA in future clinical applications, which normally involve complex DNA mixtures. Although enhancement of sensitivity requires further optimization to capture low copy numbers of template DNA, the alternative strategy would be to increase the efficiency of the MLPA reaction, or the employment of more sensitive detection equipment. The first option might be achieved by the addition of more efficient amplification facilitators such as dimethyl sulfoxide [44], dithiothreitol [45], betaine [46], bovine serum albumin and single-stranded DNA binding T4 gene 32 protein (gp32) [47]. For the later option, a real time detector could be used to track the limited fluorescent-labeled amplicons [38]. The results would be comparable with those of capillary electrophoresis or of fragment analysis of fluorescent-labeled amplicons. However, the electrophoresis maybe the optimized method to detect MLPA products for unequipped laboratory or lab of local hospital [25,48].
Conclusion
In the current study the MLPA assay was adapted to identify and discriminate three liver flukes in a 'one-tube' reaction, which was proven to be a sensitive and specific tool with high efficiency. Multiplex amplification makes this assay useful for high through-put analysis of pathogens in large clinical or ecological samples [48]. The flexible arms of the probes allow for minimal inflorescent labeling. The advantages of this method have a potential for wider application, e.g. to the detection of other parasites or to diagnostics of mixed infections in severely ill patients.
Material and Methods
Ethical Standards
All animals were handled in strict accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies. Procedures involving vertebrate animals were reviewed and approved by Sun Yat-Sen University's Animal Care and Use Committee.
Parasite sampling and genomic DNA extraction
Sixty-six C. sinensis individuals were collected from infected cats or dogs, the most common reservoir hosts, in 9 provinces in China mainland (Table 4). Genomic DNA from adult worms was extracted using a commercial DNA extraction kit (Dong sheng Biocompany, Guangdong, China) according to manual instruction. Briefly as: single adult was ground in a 1.5 ml microcentrifuge tube containing 200 μl of extraction buffer I, after shortly homogenizing, proteinase K(New England Biolabs, U.K.) and RNase A(New England Biolabs, U.K.) were added to final concentrations of 100 μg/ml and 20 μg/ml, respectively, and incubated for 3 h at 37°C. Following this, 200 μl Buffer II was added to the mixture, and incubated for 10 min at 65°C. Then, 200 μl ethanol was added to the mixture. Totally mixture was moved into the spin column after tightly vortex, after spin for 1 min at 8000 rpm, extra protein was removed using Buffer III, and then the column was washed twice with 70% ethanol, followed by centrifuge at 12000 rpm for 2 min to remove extra ethanol, DNA was recovery using 50 μl buffer EB. RNAse (5 μl each, 10 mg/ml in pH7.4 NaAC) treatment was performed at 37°C for 30 min. The DNA quantification was done at 260 nm in a UV spectrophotometer (Shimadzu, Japan).
Table 4.
C.sinensis isolates and strain information of reference ITS gene sequences in this study
| Species | GenBank | Source |
|---|---|---|
| Clonorchis sinensis | EU038112 | Shenyang, China |
| EU038113 | Shenyang, China | |
| EU038114 | Shenyang, China | |
| EU038115 | Shenyang, China | |
| EU038116 | Shenyang, China | |
| EU038117 | Shenyang, China | |
| EU038118 | Shenyang, China | |
| EU038119 | Shenyang, China | |
| EU038120 | Gimhae-si, Gyeongsangnam-do, South Korea | |
| EU038121 | Gimhae-si, Gyeongsangnam-do, South Korea | |
| EU038122 | Gimhae-si, Gyeongsangnam-do, South Korea | |
| EU038123 | Gurye-gun, Jeollanam-do, SouthKorea | |
| EU038124 | Gurye-gun, Jeollanam-do, SouthKorea | |
| EU038125 | Gurye-gun, Jeollanam-do, SouthKorea | |
| EU038126 | Jinju-si, Gyeongsangnam-do, South Korea | |
| EU038127 | Jinju-si, Gyeongsangnam-do, South Korea | |
| EU038128 | Jinju-si, Gyeongsangnam-do, South Korea | |
| EU038129 | Jinju-si, Gyeongsangnam-do, South Korea | |
| EU038130 | Jinju-si, Gyeongsangnam-do, South Korea | |
| EU038131 | Jinju-si, Gyeongsangnam-do, South Korea | |
| HQ874538 | Cat, Anhui, China | |
| HQ874523 | Cat, Anhui, China | |
| HQ874584 | Cat, Anhui, China | |
| HQ874537 | Cat, Anhui, China | |
| HQ874599 | Cat, Anhui, China | |
| HQ874585 | Cat, Anhui, China | |
| HQ874586 | Cat, Anhui, China | |
| HQ874588 | Cat, Anhui, China | |
| HQ874540 | Cat, Guangdong, China | |
| HQ874535 | Cat, Guangdong, China | |
| HQ874541 | Cat, Guangdong, China | |
| HQ874602 | Cat, Guangdong, China | |
| HQ874587 | Cat, Guangdong, China | |
| HQ874532 | Cat, Guangdong, China | |
| HQ874581 | Cat, Guangdong, China | |
| HQ874582 | Cat, Guangdong, China | |
| HQ874542 | Cat, Guangxi, China | |
| HQ874536 | Cat, Guangxi, China | |
| HQ874543 | Cat, Guangxi, China | |
| HQ874529 | Cat, Guangxi, China | |
| HQ874580 | Cat, Guangxi, China | |
| HQ874533 | Cat, Guangxi, China | |
| HQ874525 | Cat, Guangxi, China | |
| HQ874579 | Cat, Guangxi, China | |
| HQ874544 | Cat, Hubei, China | |
| HQ874545 | Cat, Hubei, China | |
| HQ874593 | Cat, Hubei, China | |
| HQ874578 | Cat, Hubei, China | |
| HQ874539 | Cat, Hubei, China | |
| HQ874592 | Cat, Hubei, China | |
| HQ874546 | Cat, Hubei, China | |
| HQ874547 | Cat, Hubei, China | |
| HQ874524 | Cat, Hubei, China | |
| HQ874601 | Cat, Henan, China | |
| HQ874550 | Cat, Henan, China | |
| HQ874597 | Cat, Henan, China | |
| HQ874595 | Cat, Henan, China | |
| HQ874573 | Cat, Henan, China | |
| HQ874572 | Cat, Henan, China | |
| HQ874571 | Cat, Henan, China | |
| HQ874589 | Cat, Henan, China | |
| HQ874598 | Cat, Hunan, China | |
| HQ874590 | Cat, Hunan, China | |
| HQ874591 | Cat, Hunan, China | |
| HQ874551 | Cat, Hunan, China | |
| HQ874534 | Cat, Hunan, China | |
| HQ874552 | Cat, Hunan, China | |
| HQ874553 | Cat, Hunan, China | |
| HQ874554 | Cat, Hunan, China | |
| HQ874555 | Dog, Jilin, China | |
| HQ874556 | Dog, Jilin, China | |
| HQ874557 | Dog, Jilin, China | |
| HQ874570 | Dog, Jilin, China | |
| HQ874528 | Dog, Jilin, China | |
| HQ874527 | Dog, Jilin, China | |
| HQ874558 | Cat, Jiangsu, China | |
| HQ874566 | Cat, Jiangsu, China | |
| HQ874559 | Cat, Jiangsu, China | |
| HQ874530 | Cat, Jiangsu, China | |
| HQ874583 | Cat, Jiangsu, China | |
| HQ874569 | Cat, Jiangsu, China | |
| HQ874604 | Cat, Jiangsu, China | |
| HQ874565 | Cat, Jiangxi, China | |
| HQ874560 | Cat, Jiangxi, China | |
| HQ874561 | Cat, Jiangxi, China | |
| HQ874564 | Cat, Jiangxi, China | |
| Opisthorchis felineus | EU038134 | Novosibirsk, Russia |
| EU038135 | Novosibirsk, Russia | |
| EU038136 | Novosibirsk, Russia | |
| EU038137 | Novosibirsk, Russia | |
| EU038138 | Novosibirsk, Russia | |
| EU038139 | Novosibirsk, Russia | |
| EU038140 | Novosibirsk, Russia | |
| Opisthorchis viverrini | EU038150 | Vientiane, Laos |
| EU038151 | Vientiane, Laos | |
| EU038152 | Vientiane, Laos | |
| EU038153 | Vientiane, Laos | |
| EU038141 | Khammouan, Laos | |
| EU038142 | Khammouan, Laos | |
| EU038143 | Khammouan, Laos | |
| EU038144 | Savannakhet, Laos | |
| EU038145 | Savannakhet, Laos | |
| EU038146 | Savannakhet, Laos | |
| EU038147 | Savannakhet, Laos | |
| EU038148 | Savannakhet, Laos | |
| Metorchis bilis | EU038154 | Spain |
| Metorchis orientalis | HM347228 | Pseudorasbora parva, China |
| Dexiogonimus ciureanus | AY245702 | Israel |
| Euryhelmis costaricensis | AB521800 | Aomori, Nishimeya Village, Japan |
| Procerovum sp. | AB536892 | fish-metacercaria, Nakorn Pathom, Thailand |
| Haplorchis taichui | AB536889 | fecal sample, Savannakhet, Laos |
| Cercaria batillariae | AY626543 | Miyagi, Japan |
| Fasciola hepatica | FJ756394 | Iran |
Fecal sampling, DNA extraction and qualification
Thirty-six fecal samples were collected from 36 infected rats at 8 weeks after infection with metacercariae. Twelve fecal samples from 12 uninfected rats were used as control. The feces of rats were firstly examined by FECT methods[49]. One gram of feces was taken for FECT. The pellet after centrifugation was resuspended with 1 ml of 10% formalin and 20 μl of suspension was used for microscopy detection.
DNA was extracted from fecal samples as described previously[50]. Briefly, 800 mg feces were washed twice with 1 ml PBS. After centrifugation, the pellet was resuspended into 200 μl of 2% polyvinylpolypyrolidone (PVPP, Sigma, St. Louis, MO) and heated for 10 min at 100°C. After sodium dodecyl sulfate-proteinase K treatment (2 h at 55°C), DNA was isolated using QIAamp Tissue Kit spin columns (QIAgen, Hilden, Germany), and eluted using 100 μl of elution buffer.
The quality of the DNA of C.sinensis was confirmed by successful PCR amplification with universal fungal primers ITS1F and ITS1R [51]. The first run of PCR to detect fungal DNA was performed as follows: an initial 95°C for 5 min and then 25 cycles of 95°C for 30 s, 62°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 7 min. Then one microliter amplicons of first run was used as the PCR template for the second run under the same reaction program. Amplicons were analyzed by electrophoresis (Bio-Rad, Hercules, CA) on 2% agarose gels (NuSieve, Rockland, ME).
Nucleotide polymorphism analysis of ITS gene of C.sinensis isolates
ITS1 rDNA regions of C.sinensis were amplified using primers ITS1F and ITS1R [51]. PCR was performed as follows: 95°C for 5 min; 25 cycles of 95°C for 30 s, 62°C for 30 s, 72°C for 1 min, with final extension at 72°C for 7 min. Amplicons were detected by electrophoresis (Bio-Rad, California, U.S.A.) on 2% agarose gel (NuSieve, Rockland, ME U.S.A.), then amplicons were sequenced with primer ITS1Fand ITS1R by Invitrogen company (Invitrogen, Shanghai, China). Sequences were edited using SEQMAN in the Lasergene software (DNASTAR, Wisconsin, U.S.A.) and submitted to GenBank. Sequences of ITS1 of O. viverrini, O.felineus and other references strains closely to liver flukes in phylogenetic were down load from GenBank. All sequences aligned with Bionumerics version 4.61 (Applied Maths, Kortrijk, Belgium). Single-nucleotide polymorphism of ITS1 gene were analyzed by DNaSP4 software (Universitat de Barcelona; Software for Population Genetics and Molecular Evolution analyse; 4.20.0002).
Probe design
DNA sequences of C.sinensis, O. viverrini, O.felineus and reference strains were aligned automatically and adjusted manually in BioNumerics v. 4.61 (Applied Maths, Kortrijk, Belgium) to identify informative nucleotide polymorphisms. The design of the MLPA probes was performed as described [25]. Three pairs of completely synthetic MLPA probes targeting the ITS region were designed with an annealing temperature >65°C according to the RAW program (http://www.mlpa.com/WebForms/WebFormMain.aspx ) and no secondary structures according to the mFOLD server (http://www.bioinfo.rpi.edu/applications/mfold). For probe sequences were listed in Table 2. Specificity of the probes was confirmed by BLAST analysis in GenBank.
MLPA analysis
MLPA reactions were performed referring to the standard protocol on http://www.mlpa.com/WebForms/WebFormMain.aspx?Tag=wl2zCji\rCGANQgZPuTixtCplCA1mmwJoFo/xHPnTgc| with some modifications. Briefly as: Routinely 500 pg of DNA from pure culture was used. All the MLPA reagents come from MRC-Holland (Amsterdam the Netherlands). The hybridization and ligation of probes were performed in Biosystems 2720 thermal cycler according to the standard MLPA protocol. pF3 (Fam fluorescent labeled at 5' end) and pB3 were used as universal PCR primers in the ligated probes amplification. PCR amplification was performed for 25 cycles (30 s 95°C, 30 s 55°C and 1 min 72°C) with a denature at 95°C for 5 min and a extension step at 72°C for 7 min.
Specificity and validation of signal quantification
Genome DNA of 66 C.sinensis and 3 artificial template of C.sinensis, O. viverrini and O.felineus were used as templates to evaluate the specificity and sensitivity of the MLPA assay. MLPA reaction without C.sinensis DNA or artificial templates were used as negative controls. Artificial template of padlock probe was used to evaluate the detection limit of the MLPA assay. Two microlitres of each 10-fold serial diluted artificial template mixture and genome DNA of C.sinensis was used as templates for MLPA reaction. Amplified products were analyzed by electrophoresis on 5% agarose gels, stained with ethidium bromide and photographed. 20bp DNA ladder was used as molecular weight standard.
Results detection using capillary sequencer
One microliter of the products was dissolved in 9 μl of deionized formamide, 0.2 nM Gene-Scan®-ROX 500 size standards, and 0.5 μl loading dye (all from Applied Biosystems, Foster City, CA, USA) and denatured for five minutes at 95°C. The products were electrophoresed on an ABI Prism® 3730XL Genetic Analyzer model capillary sequencer (Applied Biosystems) in the GeneScan mode. Analysis of the products was performed using Gene-Scan 3.7 and Genotyper® 3.7 software (Applied Biosystems) consecutively.
Evaluation of MLPA in fecal samples of infected rats
Crude-extracted DNA of 2 μl each from 48 fecal samples was used as a template for MLPA assays. The amplified products were analyzed by electrophoresis.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
XY, JX and JS designed the present experiments. JS carried out these experiments and drafted the manuscript. PL, QM and CL collected the isolates using in this study. YH, XL and CD give crucial reviews of this manuscript, GSdH give crucial English revision to this manuscript. All authors read and approved the final version of the manuscript.
Contributor Information
Jiufeng Sun, Email: sunjiuf@mail.sysu.edu.cn.
Jin Xu, Email: xujinteam@163.com.
Pei Liang, Email: 365129706@qq.com.
Qiang Mao, Email: maoqiang0903@126.com.
Yan Huang, Email: biohy@yahoo.com.cn.
Xiaoli Lv, Email: unkindlxl@163.com.
Chuanhuan Deng, Email: dengchuanhuan@163.com.
Chi Liang, Email: liangchi@mail.sysu.edu.cn.
G S de Hoog, Email: s.hoog@cbs.knaw.nl.
Xinbing Yu, Email: yuhxteam@163.com.
Acknowledgements and funding
This work is supported by the National S & T Major Program (Grant No. 2008ZX10004-011), and National Basic Research and Development Key Project of China (973 project, No.2010CB530000).
References
- FAO. Assessment and management of seafood quality and safety. Fisheries technical paper. 2004. p. 444.
- Lun ZR, Gasser RB, Lai DH, Li AX, Zhu XQ, Yu XB, Fang YY. Clonorchiasis: a key foodborne zoonosis in China. Lancet Infect Dis. 2005;5:31–41. doi: 10.1016/S1473-3099(04)01252-6. [DOI] [PubMed] [Google Scholar]
- WHO. Control of food borne trematode infections: report of a WHO study group. WHO, Geneva, Switzerland. 1995. WHO Technical Report Series No.849.
- Sayasone S, Odermatt P, Phoumindr N, Vongsaravane X, Sensombath V, Phetsouvanh R, Choulamany X, Strobel M. Epidemiology of Opisthorchis viverrini in a rural district of southern Lao PDR. Trans R Soc Trop Med Hyg. 2007;101:40–47. doi: 10.1016/j.trstmh.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Sriamporn S, Pisani P, Pipitgool V, Suwanrungruang K, Kamsa-ard S, Parkin DM. Prevalence of Opisthorchis viverrini infection and incidence of cholangiocarcinoma in Khon Kaen, Northeast Thailand. Trop Med Int Health. 2004;9:588–594. doi: 10.1111/j.1365-3156.2004.01234.x. [DOI] [PubMed] [Google Scholar]
- Sripa B, Sithithaworn P, Sirisinha S. Opisthorchis viverrini and opisthorchiasis: the 21st century review. Acta Trop. 2003;88:169–170. doi: 10.1016/j.actatropica.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Sithithaworn P, Haswell-Elkins M. Epidemiology of Opisthorchis viverrini. Acta Trop. 2003;88:187–194. doi: 10.1016/j.actatropica.2003.02.001. [DOI] [PubMed] [Google Scholar]
- Yossepowitch O, Gotesman T, Assous M, Marva E, Zimlichman R, Dan M. Opisthorchiasis from imported raw fish. Emerg Infect Dis. 2004;10:2122–2126. doi: 10.3201/eid1012.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer WM, Sellman JS, Walker PF. Biliary liver flukes (Opisthorchiasis and Clonorchiasis) in immigrants in the United States: often subtle and diagnosed years after arrival. J Travel Med. 2004;11:157–159. [PubMed] [Google Scholar]
- Tesana S, Srisawangwonk T, Kaewkes S, Sithithaworn P, Kanla P, Arunyanart C. Eggshell morphology of the small eggs of human trematodes in Thailand. Southeast Asian J Trop Med Public Health. 1991;22:631–636. [PubMed] [Google Scholar]
- Lee SH, Hwang SW, Chai JY, Seo BS. Comparative Morphology Of Eggs Of Heterophyids And Clonorchis Sinensis Causing Human Infections In Korea. Kisaengchunghak Chapchi. 1984;22:171–180. doi: 10.3347/kjp.1984.22.2.171. [DOI] [PubMed] [Google Scholar]
- Zhou XN, Lv S, Yang GJ, Kristensen TK, Bergquist NR, Utzinger J, Malone JB. Spatial epidemiology in zoonotic parasitic diseases: insights gained at the 1st International Symposium on Geospatial Health in Lijiang, China, 2007. Parasites & Vectors. 2009;2:10. doi: 10.1186/1756-3305-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi A, Sanath Kumar H, Kenchanna Prakasha B, Lu J, Xu X, Hu W, Feng Z, Karunasagar I. Clonorchis sinensis: development and evaluation of a nested polymerase chain reaction (PCR) assay. Exp Parasitol. 2007;115:291–295. doi: 10.1016/j.exppara.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Cai XQ, Xu MJ, Wang YH, Qiu DY, Liu GX, Lin A, Tang JD, Zhang RL, Zhu XQ. Sensitive and rapid detection of Clonorchis sinensis infection in fish by loop-mediated isothermal amplification (LAMP) Parasitol Res. pp. 1379–1383. [DOI] [PubMed]
- Kang S, Sultana T, Loktev VB, Wongratanacheewin S, Sohn WM, Eom KS, Park JK. Molecular identification and phylogenetic analysis of nuclear rDNA sequences among three opisthorchid liver fluke species (Opisthorchiidae: Trematoda) Parasitol Int. 2008;57:191–197. doi: 10.1016/j.parint.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Sato M, Thaenkham U, Dekumyoy P, Waikagul J. Discrimination of O. viverrini, C. sinensis, H. pumilio and H. taichui using nuclear DNA-based PCR targeting ribosomal DNA ITS regions. Acta Trop. 2009;109:81–83. doi: 10.1016/j.actatropica.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Parvathi A, Umesha KR, Kumar S, Sithithaworn P, Karunasagar I. Development and evaluation of a polymerase chain reaction (PCR) assay for the detection of Opisthorchis viverrini in fish. Acta Trop. 2008;107:13–16. doi: 10.1016/j.actatropica.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Traub RJ, Macaranas J, Mungthin M, Leelayoova S, Cribb T, Murrell KD, Thompson RC. A new PCR-based approach indicates the range of Clonorchis sinensis now extends to Central Thailand. PLoS Negl Trop Dis. 2009;3:e367. doi: 10.1371/journal.pntd.0000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B, Schmidt J, Mehlhorn H. Sensitive and species-specific detection of Clonorchis sinensis by PCR in infected snails and fishes. Parasitol Res. 2007;100:911–914. doi: 10.1007/s00436-006-0322-9. [DOI] [PubMed] [Google Scholar]
- Meine MH, Zanotelli ML, Neumann J, Kiss G, de Jesus Grezzana T, Leipnitz I, Schlindwein ES, Fleck A Jr, Gleisner AL, de Mello Brandao A. et al. Randomized clinical assay for hepatic grafts preservation with University of Wisconsin or histidine-tryptophan-ketoglutarate solutions in liver transplantation. Transplant Proc. 2006;38:1872–1875. doi: 10.1016/j.transproceed.2006.06.071. [DOI] [PubMed] [Google Scholar]
- Abu Al-Soud W, Radstrom P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J Clin Microbiol. 2000;38:4463–4470. doi: 10.1128/jcm.38.12.4463-4470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai JY, Park JH, Han ET, Guk SM, Shin EH, Lin A, Kim JL, Sohn WM, Yong TS, Eom KS. et al. Mixed infections with Opisthorchis viverrini and intestinal flukes in residents of Vientiane Municipality and Saravane Province in Laos. J Helminthol. 2005;79:283–289. doi: 10.1079/JOH2005302. [DOI] [PubMed] [Google Scholar]
- Harper GL, King RA, Dodd CS, Harwood JD, Glen DM, Bruford MW, Symondson WO. Rapid screening of invertebrate predators for multiple prey DNA targets. Mol Ecol. 2005;14:819–827. doi: 10.1111/j.1365-294X.2005.02442.x. [DOI] [PubMed] [Google Scholar]
- Le TH, Van De N, Blair D, Sithithaworn P, McManus DP. Clonorchis sinensis and Opisthorchis viverrini: development of a mitochondrial-based multiplex PCR for their identification and discrimination. Exp Parasitol. 2006;112:109–114. doi: 10.1016/j.exppara.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogervorst FB, Nederlof PM, Gille JJ, McElgunn CJ, Grippeling M, Pruntel R, Regnerus R, van Welsem T, van Spaendonk R, Menko FH. et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- Stern RF, Roberts RG, Mann K, Yau SC, Berg J, Ogilvie CM. Multiplex ligation-dependent probe amplification using a completely synthetic probe set. Biotechniques. 2004;37:399–405. doi: 10.2144/04373ST04. [DOI] [PubMed] [Google Scholar]
- Volikos E, Robinson J, Aittomaki K, Mecklin JP, Jarvinen H, Westerman AM, de Rooji FW, Vogel T, Moeslein G, Launonen V. et al. LKB1 exonic and whole gene deletions are a common cause of Peutz-Jeghers syndrome. J Med Genet. 2006;43:e18. doi: 10.1136/jmg.2005.039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstman JA, Jalali GR, Rappaport EF, Hacker AM, Scott C, Emanuel BS. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum Mutat. 2006;27:814–821. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting SM, Snijders PJ, Meijer GA, Ylstra B, van den Ijssel PR, Snijders AM, Albertson DG, Coffa J, Schouten JP, van de Wiel MA. et al. Increased gene copy numbers at chromosome 20q are frequent in both squamous cell carcinomas and adenocarcinomas of the cervix. J Pathol. 2006;209:220–230. doi: 10.1002/path.1966. [DOI] [PubMed] [Google Scholar]
- Eldering E, Spek CA, Aberson HL, Grummels A, Derks IA, de Vos AF, McElgunn CJ, Schouten JP. Expression profiling via novel multiplex assay allows rapid assessment of gene regulation in defined signalling pathways. Nucleic Acids Res. 2003;31:e153. doi: 10.1093/nar/gng153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluwe L, Nygren AO, Errami A, Heinrich B, Matthies C, Tatagiba M, Mautner V. Screening for large mutations of the NF2 gene. Genes Chromosomes Cancer. 2005;42:384–391. doi: 10.1002/gcc.20138. [DOI] [PubMed] [Google Scholar]
- Slater HR, Bruno DL, Ren H, Pertile M, Schouten JP, Choo KH. Rapid, high throughput prenatal detection of aneuploidy using a novel quantitative method (MLPA) J Med Genet. 2003;40:907–912. doi: 10.1136/jmg.40.12.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsham MJ, Pals G, Schouten JP, Van Spaendonk RM, Concus A, Carey TE, Benninger MS. Delineating genetic pathways of disease progression in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2003;129:702–708. doi: 10.1001/archotol.129.7.702. [DOI] [PubMed] [Google Scholar]
- Bergval IL, Vijzelaar RN, Dalla Costa ER, Schuitema AR, Oskam L, Kritski AL, Klatser PR, Anthony RM. Development of multiplex assay for rapid characterization of Mycobacterium tuberculosis. J Clin Microbiol. 2008;46:689–699. doi: 10.1128/JCM.01821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terefework Z, Pham CL, Prosperi AC, Entius MM, Errami A, van Spanning RJ, Zaura E, Ten Cate JM, Crielaard W. MLPA diagnostics of complex microbial communities: relative quantification of bacterial species in oral biofilms. J Microbiol Methods. 2008;75:558–565. doi: 10.1016/j.mimet.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Reijans M, Dingemans G, Klaassen CH, Meis JF, Keijdener J, Mulders B, Eadie K, van Leeuwen W, van Belkum A, Horrevorts AM, Simons G. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol. 2008;46:1232–1240. doi: 10.1128/JCM.02294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JM, Sun JF, Feng PY, Li XQ, Lu CM, Lu S, Cai WY, Xi LY, de Hoog GS. Rapid identification and characterization of Penicillium marneffei using multiplex ligation-dependent probe amplification (MLPA) in paraffin-embedded tissue samples. J Micro Method. 2011;85:33–39. doi: 10.1016/j.mimet.2011.01.022. [DOI] [PubMed] [Google Scholar]
- Faruqi AF, Hosono S, Driscoll MD, Dean FB, Alsmadi O, Bandaru R, Kumar G, Grimwade B, Zong Q, Sun Z. et al. High-throughput genotyping of single nucleotide polymorphisms with rolling circle amplification. BMC Genomics. 2001;2:4. doi: 10.1186/1471-2164-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M. Lock and roll: single-molecule genotyping in situ using padlock probes and rolling-circle amplification. Histochem Cell Biol. 2006;126:159–164. doi: 10.1007/s00418-006-0213-2. [DOI] [PubMed] [Google Scholar]
- Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Bartos J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry A. 2003;51:127–128. doi: 10.1002/cyto.a.10013. author reply 129. [DOI] [PubMed] [Google Scholar]
- Pomp D, Medrano JF. Organic solvents as facilitators of polymerase chain reaction. Biotechniques. 1991;10:58–59. [PubMed] [Google Scholar]
- Nagai M, Yoshida A, Sato N. Additive effects of bovine serum albumin, dithiothreitol, and glycerol on PCR. Biochem Mol Biol Int. 1998;44:157–163. doi: 10.1080/15216549800201172. [DOI] [PubMed] [Google Scholar]
- Henke W, Herdel K, Jung K, Schnorr D, Loening SA. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P, Nygren AO, Schouten JP, Jacobs H. Rapid and quantitative detection of homologous and non-homologous recombination events using three oligonucleotide MLPA. Nucleic Acids Res. 2005;33:e188. doi: 10.1093/nar/gni187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ST H. In: International handbook of foodborne pathogens. Miliotis MD, Bier JW, editor. Marcel Dekker, New York; 2003. Clonorchis sinensis; pp. 581–592. [Google Scholar]
- Verweij JJ, Pit DS, van Lieshout L, Baeta SM, Dery GD, Gasser RB, Polderman AM. Determining the prevalence of Oesophagostomum bifurcum and Necator americanus infections using specific PCR amplification of DNA from faecal samples. Trop Med Int Health. 2001;6:726–731. doi: 10.1046/j.1365-3156.2001.00770.x. [DOI] [PubMed] [Google Scholar]
- Liu WQ, Liu J, Zhang JH, Long XC, Lei JH, Li YL. Comparison of ancient and modern Clonorchis sinensis based on ITS1 and ITS2 sequences. Acta Trop. 2007;101:91–94. doi: 10.1016/j.actatropica.2006.08.010. [DOI] [PubMed] [Google Scholar]