Abstract
Spermatogonial differentiation is orchestrated by the precise control of gene expression involving retinoic acid signaling. MicroRNAs have emerged as important regulators of spermatogenesis, and here we show that the Mirlet7 family miRNAs are expressed in mouse spermatogonia and spermatocytes. Retinoic acid significantly leads to the induction of Mirlet7 miRNAs through suppression of Lin28. We further confirmed both in vitro and in vivo that expressions of Mycn, Ccnd1, and Col1a2, which are targets of Mirlet7, were downregulated during spermatogonial differentiation. These results suggest that Mirlet7 family miRNAs play a role in retinoic acid-induced spermatogonial differentiation.
Keywords: Lin28, microRNAs, Mirlet7 family, retinoic acid, spermatogonial differentiation
Upregulation of Mirlet7 family miRNAs by retinoic acid potentially contributes to retinoic acid-induced spermatogonial differentiation.
INTRODUCTION
Spermatogenesis is a highly coordinated and complex process during which mature sperm originate from a common spermatogonial stem cell (SSC) [1]. In the adult mammals, SSCs either enter a self-renewal pathway or undergo repeated mitotic divisions to sequentially produce A paired (Apr) and A aligned (Aal) spermatogonia, which are committed to further development [1]. The Aal spermatogonia or undifferentiated spermatogonia subsequently differentiate into A1 spermatogonia in a process that does not include a proliferative division. The A1 spermatogonia, or differentiating spermatogonia, further form, successively, A2, A3, A4, In (intermediate), and type B spermatogonia through a series of mitotic divisions. The type B spermatogonia divide into preleptotene spermatocytes that subsequently enter into meiosis to yield haploid step 1 spermatids, which undergo spermiogenesis, resulting in the production of spermatozoa [1].
The differentiation of Aal into A1 spermatogonia is a key step in spermatogenesis [2]. Retinoic acid (RA), an active metabolite of vitamin A, is critical in this process [3, 4]. For example, spermatogonial differentiation is blocked at the Aal to A1 transition in vitamin A-deficient (VAD) rats and mice. Administration of retinol or RA to the VAD rat or mouse reinitiates spermatogenesis in a synchronous fashion by releasing the block on spermatogonial differentiation [4]. Several lines of evidence suggest that RA directly induces spermatogonial differentiation via the expression of numerous RA-targeted genes encoding proteins such as Stra8, Kit, Ccnd2, etc. [5–7]. However, the mechanisms by which RA affects spermatogonial differentiation remain largely unclear, and it is likely that the expression of some genes during RA-induced spermatogonial differentiation undergo posttranscriptional regulation.
MicroRNAs (miRNAs) are small noncoding single-stranded conserved regulatory RNA molecules approximately 22 nucleotides long. They are initially generated from long primary transcripts (pri-miRNA) that have an imperfectly matched stem-loop structure [8]. These pri-miRNAs are first processed by the nuclear RNase III DROSHA and its partner, DGCR8, to produce precursor miRNAs (pre-miRNAs), which are then transported into the cytoplasm and further processed by another cytoplasmic RNase III DICER to yield short double-stranded miR-miR* duplexes. One strand of miR is subsequently incorporated into a miRNA-induced silencing complex (miRISC) [8]. The mature miRNAs direct the miRISC to interact with the seed sequence (nucleotides 2–8 at its 5′ end of miRNA) matched recognition sites, generally in the 3′ untranslated region (UTR) of target mRNA. These interactions inhibit the expression of the target genes at the posttranscriptional level through mRNA decay and translational inhibition [8]. A recent study indicated that more than 60% of human protein-coding genes carry 3′ UTR miRNA target sites, suggesting a potential global role for miRNAs in the regulation of gene expression [9]. Indeed, miRNAs have been shown to play critical roles in a wide spectrum of biological processes, including cell proliferation, differentiation, and apoptosis [8]. Emerging evidence has revealed that miRNAs are present in abundance in male germ cells [10–16] and that miRNAs could play an important role during spermatogenesis [17–23].
The lethal-7 (let-7) gene is one of the first two miRNAs identified in Caenorhabditis elegans [24]. Mature Mirlet7 miRNAs play an important role in mammals in the differentiation of stem cells and tumor cells [25]. In this current study, we used both an in vitro and an in vivo system to test the hypothesis that the level of Mirlet7 family miRNAs is dramatically altered in RA-induced spermatogonial differentiation and likely plays a key role in this process.
MATERIALS AND METHODS
Animals and Treatments
Animal experiments were conducted in accordance with the “Guidelines for the Care and Use of Research Animals of the National Institutes of Health” and were approved by the Institutional Animal Care and Use Committee of Washington State University. The C57BL/6 (B6) mice were obtained from the Jackson Laboratory and maintained in a standard animal facility with free access to food and water. To generate VAD mice, B6 female mice were fed a VAD diet (Teklad Trucking) for at least 4 wk and were bred with B6 males. The males born to these dams received this diet until they became VAD. At 14 wk of age, when body weight was slightly decreased, animals were injected with all-trans-retinoic acid (ATRA; 1 mg of ATRA per animal in 100 μl of 90% sesame oil and 10% ethanol, i.p., twice for 24 h; Sigma) or injected with vehicle as controls twice for 24 h before being euthanized. Immediately after the mice were euthanized, testes were removed and were either used for total RNA extraction using Trizol (Invitrogen) or processed for histological studies.
Cell Cultures
Isolation of THY+ spermatogonia from 5- to 7-day-old mice on a B6 background was accomplished using magnetic-activated cell sorting as described previously [6]. The purity of isolated spermatogonia was identified by POU5F1 staining, and about 85% of cells were POU5F1 positive (data not shown). Cells were cultured under feeder cell-free and serum-free conditions for 24 h with 0.7 μM ATRA or vehicle at 37°C in an atmosphere of 5% CO2 in air.
Monolayer cultures of P19 cells (mouse embryonal carcinoma cells; ATCC) were grown in α-minimal essential medium (α-MEM; Invitrogen) containing 10% fetal bovine serum. Cells were treated with ethanol or ATRA for the indicated periods of time.
In Situ miRNA Hybridization with Locked Nucleic Acid Probes
In situ miRNA hybridization (ISH) procedure was performed to examine the spatial expression of Mirlet7a, Mirlet7c, and Mirlet7e in 20-day-old and adult mouse testis based on a published protocol with modifications [26]. Briefly, the testes from adult and 20-day-old B6 mice were fixed for 24 h in 10% neutral-buffered formalin or 4% paraformaldehyde and then dehydrated through a graded ethanol series before paraffin embedding and sectioning at 5 μm. Sections were deparaffinized in two consecutive xylene baths for 5 min each, then 5 min each in serial dilutions of ethanol (100%, 100%, 95%, and 75%), followed by an incubation for 20 min in 0.2 N HCl and two washes with diethyl pyrocarbonate (DEPC)-treated water. Slides were digested with 1 μg/ml proteinase K (Roche) at 37°C for 30 min. The reaction was stopped in 0.2% glycine for 10 min, and the slides were washed twice in DEPC-treated water, immersed in 100% ethanol for 30 sec, and air dried. Slides were then hybridized in an incubation chamber overnight at 37°C using 200 nM digoxigenin (DIG)-labeled locked nucleic acid (LNA) probes (Exiqon) diluted with Enzo ISH buffer (Enzo Diagnostic). After hybridization, slides were washed in 0.2× SSCs with 2% bovine serum albumin at 4°C for 5 min. An anti-DIG/alkaline phosphatase (Roche) antibody at a 1:100 dilution in PBS was applied to the slides for 30 min at 37°C. Slides were washed with detection buffer for 5 min at room temperature and incubated with nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt solution (Roche) at 37°C under monitoring. Sections were then counterstained with nuclear fast red (Vector Laboratories) for 2 min, washed in water, and rinsed in 100% ethanol for 2 min and xylene for 5 min. Sections were digitally photographed on a Nikon Microphoto-FX microscope (Meridian Instrument Company Inc.) with an Olympus OLY-200 digital camera (Olympus America Inc.). Sections from at least two B6 mice were analyzed for miRNA localization.
Quantitative RT-PCR Assays
Total RNA was isolated using Trizol reagent per the manufacturer's instructions, treated with DNaseI (Ambion) to remove possible contaminating genomic DNA, and quantified in an ND-1000 Spectrometer (Thermo Scientific). Only RNA samples with a value of ≥1.8 on 260:280 ratios were used for subsequent quantitative RT-PCR (qRT-PCR) analyses. For miRNA qRT-PCR, miRNA expression was determined using Mir-X miRNA First-Strand Synthesis Kit and SYBR Advantage qPCR Premix (Clontech) on the Applied Biosystems 7500 Fast system according to the supplier's protocol. Relative miRNA expression was normalized to the U6 small nuclear RNA (snRNA) according to the ΔCT model [27]. For mRNA and pri-miRNA qRT-PCR, 200 ng of total RNA from each sample was reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad). Real-time PCR was performed with Fast SYBR Green PCR Mastermix (Applied Biosystems) on the Applied Biosystems 7500 Fast system. Relative expression of Lin28, pri-Mirlet7g, pri-Mirlet7d/a/f, Col1a2, Ccnd1, and Mycn was normalized to the ribosomal protein S2 (Rps2) in each sample. Quantitative comparison of a given RNA between RA treatment and vehicle was determined using the comparative CT method (2−ΔΔCT) [27]. Primer pairs used are listed in Supplemental Table S1 (all Supplemental Data are available online at www.biolreprod.org).
Western Blot Analysis
Tissues or cells were lysed and homogenized at 4°C in radioimmunoprecipitation assay buffer (50 mM Tris buffer containing 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 0.5% deoxycholic acid, and 1 mM ethylenediaminetetraacetic acid [EDTA; pH 8.0]) in the presence of a protease inhibitor cocktail (Roche). The homogenate was centrifuged at 12 000 × g for 2 min, and the resulting supernatant was used for Western blot analysis. Concentrations of the protein samples were determined by the DC protein assay kit (Bio-Rad). Proteins were separated on 10% SDS polyacrylamide gels and transferred onto nitrocellulose membranes (Bio-Rad). The membranes were blocked in 5% nonfat milk (Bio-Rad) and then probed with antibodies against LIN28 (1:400; R&D Systems Inc.), N-MYC (1:200; Santa Cruz Biotechnology), and CCND1 (1:200; Santa Cruz Biotechnology). In most cases, the membranes were stripped after use and reprobed with a β-ACTIN antibody (1:5000; Sigma) to confirm equal protein loading. Immunoreactive proteins were visualized using the Western Lighting ECL detection system (PerkinElmer Inc.).
Chromatin Immunoprecipitation
After treatment with ATRA or EtOH for 24 h, P19 cells were cross-linked with 1% formaldehyde/α-MEM for 10 min at room temperature and quenched by adding glycine to a final concentration of 0.25 M for 5 min. Cells were collected and washed twice with cold PBS containing 1× protease inhibitor cocktail (Roche). Cell pellets were lysed in 1 ml of lysis buffer (1% SDS, 50 mM Tris-HCl [pH 8.0], 10 mM EDTA, and 1× protease inhibitor cocktail) for 10 min on ice and sonicated on ice to obtain a chromatin size of 200 to 1000 bp. After preclearing with protein A/G agarose beads (Upstate), an equivalent amount of sheared chromatin was immunoprecipitated with antibody overnight at 4°C, followed by incubation with protein A/G for 1 h. Antibodies used in chromatin immunoprecipitation (ChIP) assays were anti-RARA (kindly provided by Dr. Rochette-Egly, Institut de Génétique et de Biologie Moléculaire et Cellulaire CNRS/INSERM/ULP), anti-RARG (Santa Cruz Biotechnology), and immunoglobulin G (IgG) control (Santa Cruz Biotechnology). Protein-DNA complexes were eluted in fresh 1% SDS/0.1 M NaHCO3. Cross-linking was reversed, and protein was removed. DNA was recovered and purified for PCR using specific primers. The specific primer sequences were the following: Lin28, forward 5′-tggagattgaggcatccagt-3′ and reverse 5′-cgctgtccaatcagaaacac-3′; and Pou5f1, forward 5′-cctggggtcccgtcctaagg-3′ and reverse 5′-cctggtggaaagacggctca-3′.
Statistical Analyses
For all analyses, data were statistically processed using a Student t-test for all pairs computed by SigmaStat 3.0 (SPSS, Chicago, IL). A P value of ≤0.05 was considered significant. Data represent mean ± SD.
RESULTS
Expression of Mirlet7 Family miRNAs in Spermatogonia
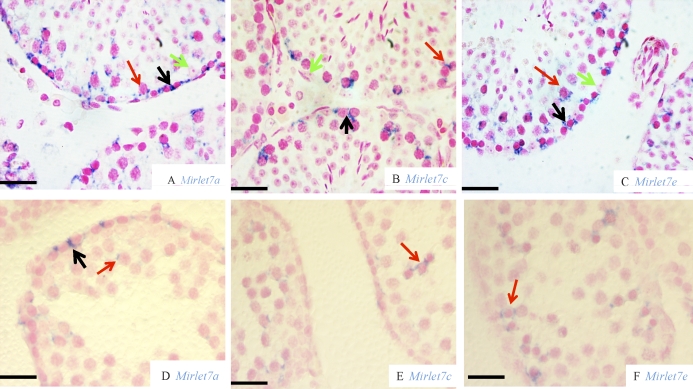
We performed miRNA ISH with LNA probes to localize the different Mirlet7 family miRNAs in mouse testis. We chose Mirlet7a, Mirlet7c, and Mirlet7e as representative members of the family. In the adult testis, Mirlet7a staining was mainly observed in the cytoplasm of type A spermatogonia, whereas no Mirlet7a staining was seen in Sertoli cells (Fig. 1A). Furthermore, Mirlet7a was also present in some spermatocytes (Fig. 1A). A few interstitial cells showed a weak Mirlet7a staining (Fig. 1A). Similarly to Mirlet7a, ISH staining for both Mirlet7c (Fig. 1B) and Mirlet7e (Fig. 1C) was seen in spermatogonia and spermatocytes. In a testis from a 20-day-old mouse, Mirlet7a staining was present in some spermatogonia, and a weaker staining was observed in a few spermatocytes (Fig. 1D). Both Mirlet7c (Fig. 1E) and Mirlet7e (Fig. 1F) staining was seen in spermatocytes; however, very weak, if any, Mirlet7c and Mirlet7e staining was seen in spermatogonia. No staining was detected in Sertoli cells for any Mirlet7a, Mirlet7c, and Mirlet7e probes. No staining for the scramble miRNA was seen in either adult or 20-wk-old testis (data not shown). Together, these results indicate that Mirlet7 family miRNAs are mainly expressed in premeiotic and meiotic germ cells, suggesting that they may play critical roles in spermatogonial differentiation.
FIG. 1.
In situ hybridization of Mirlet7 family miRNAs in mouse testis. Analysis of Mirlet7a (adult [A] and 20-day-old [D]), Mirlet7c (adult [B] and 20-day-old [E]), and Mirlet7e (adult [C] and 20-day-old [F]) is shown. Cells with positive signal are colored blue. Nucleus was counterstained with fast red. Black arrow, spermatogonia; red arrow, spermatocytes; green arrow, Sertoli cells. Bars = 20 μm.
Expression of Mirlet7 Family Members Induced by RA Signaling
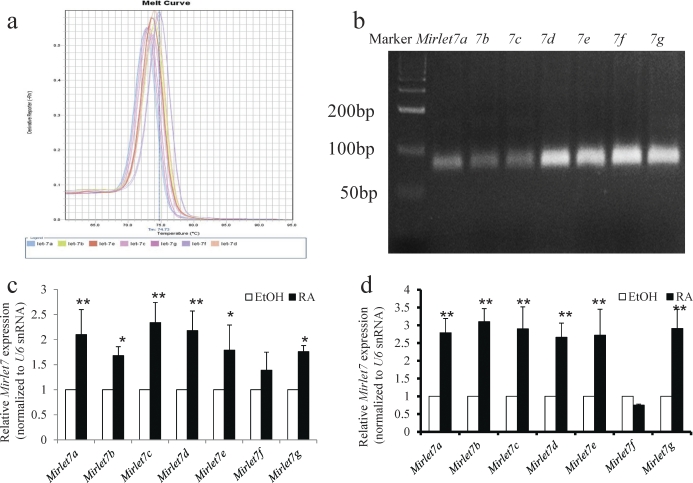
Because RA is known to induce spermatogonial differentiation, we next examined whether RA signaling regulates the expression of Mirlet7 family members. We used both an in vitro model of mouse spermatogonial differentiation and an in vivo model composed of VAD mice. The levels of mature Mirlet7 family members were determined by miRNA qRT-PCR. Following RA treatment and before miRNA qRT-PCR, we verified that the tissue responded to RA by checking the levels of the RA-responsive gene Stra8 (also a marker for spermatogonial differentiation). The expected RA induction of Stra8 was confirmed in all RA-treated samples (data not shown). The melting curve showed a unique peak from the qPCR amplification of each Mirlet7 family miRNA (Fig. 2a). This demonstrated the specificity of the amplification, whereas the correct product sizes of ∼70 bp were verified by gel electrophoresis (Fig. 2b). We found that RA treatment significantly increased the expression of six members of the Mirlet7 family miRNAs, including Mirlet7a, Mirlet7b, Mirlet7c, Mirlet7d, Mirlet7e, and Mirlet7g, in both in vitro (Fig. 2c) and in vivo (Fig. 2d) experiments. The RA similarly increased the level of Mirlet7 family miRNAs in P19 cells (Supplemental Fig. S1a). These data strongly indicate that the Mirlet7 family miRNAs are regulated by RA signaling during RA-induced spermatogonial differentiation.
FIG. 2.
Retinoic acid signaling induces expression of the Mirlet7 family miRNAs. a) Melt curves of qRT-PCR amplification of Mirlet7a, Mirlet7b, Mirlet7c, Mirlet7d, Mirlet7e, Mirlet7f, and Mirlet7g. b) The qRT-PCR products for Mirlet7a, Mirlet7b, Mirlet7c, Mirlet7d, Mirlet7e, Mirlet7f, and Mirlet7g resolved on 3.5% agarose gel. c) Retinoic acid-inducible expression of the Mirlet7 family miRNAs in isolated THY+ spermatogonia. THY+ spermatogonia from 8 to 10 mice in each experiment were pooled and were treated with ethanol or RA (0.7 μM) for 24 h in vitro, and miRNAs were subjected to qRT-PCR. U6 snRNA was used for normalization between samples (mean ± SD, *P < 0.05, **P < 0.001, ethanol control versus RA treatment for 24 h, n = 3; Student t-test). d) Treatment of VAD male mice with RA for 24 h significantly increased the expression of the Mirlet7a, Mirlet7b, Mirlet7c, Mirlet7d, Mirlet7e, and Mirlet7g in testes (mean ± SD, **P < 0.001, sesame oil control versus RA treatment for 24 h, n = 6; Student t-test).
Posttranscriptional Regulation of RA-Induced Mirlet7 Family Members by LIN28
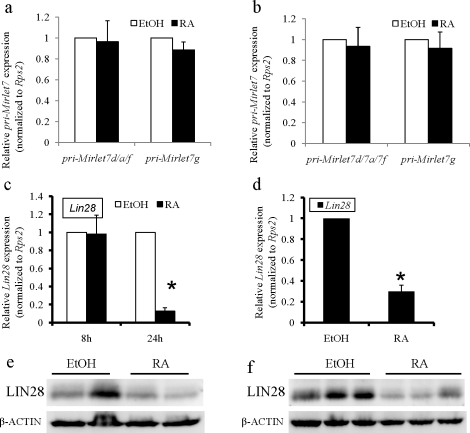
The six members of the mature Mirlet7 family miRNAs upregulated by RA signaling were produced from six different transcription units (Mirlet7a-1/7f-1/7d, Mirlet7g, Mir100/Mirlet7a-2/MiR125b-1, Mirlet7a-3/Mirlet7b, Mir99a/Mirlet7c/Mir-125b-2, and Mir99b/Mirlet7e/Mir125a). To investigate the mechanisms by which RA signaling regulates these miRNAs, we examined the levels of their primary transcripts in both in vitro and in vivo experimental systems. We chose pri-Mirlet7a-1/7f-1/7d and pri-Mirlet7g as representative members and found that the abundance of both the pri-Mirlet7a-1/7f-1/7d and pri-Mirlet7g were not induced by RA (Fig. 3, a and b), suggesting that RA signaling upregulated multiple Mirlet7 family miRNAs through a posttranscriptional pathway. Recent studies have shown that a conserved RNA-binding protein, LIN28, which is expressed in mouse spermatogonia [28, 29], binds to stem loops of Mirlet7 precursors and inhibits maturation of the Mirlet7 family by blocking both DROSHA- and DICER-mediated processing of the Mirlet7 precursors and accelerating degradation of the Mirlet7 precursors [30–32]. We therefore examined whether repression of Lin28 by RA signaling mediated the induction of Mirlet7 family miRNAs. To test this possibility, we used qPCR and Western blot analysis to examine the mRNA and protein levels of Lin28 in both in vitro and in vivo experiments with or without RA treatment. Although Lin28 expression was unchanged in the in vitro experiments 8 h after RA treatment, RA signaling significantly reduced the mRNA levels of Lin28 in both experimental systems within 24 h of RA treatment (Fig. 3, c and d). As shown in Figure 3, e and f, the protein levels of LIN28 in both RA-treated spermatogonia and VAD testes were significantly decreased compared with that of non-RA-treated spermatogonia and VAD testes (average densitometry ratio of LIN28 and β-ACTIN [mean ± SD] for nontreated spermatogonial cells: 0.175 ± 0.044, n = 4; for RA-treated spermatogonial cells: 0.114 ± 0.008, n = 4; P < 0.05 [Fig. 3e]; and average densitometry ratio of LIN28 and β-ACTIN (mean ± SD) for nontreated testes: 0.207 ± 0.015, n = 11; for RA-treated testes: 0.145 ± 0.023, n = 11; P < 0.01 [Fig. 3f]). These results suggest that RA could induce the expression of Mirlet7 family members by repression of Lin28. In addition, the level of Lin28 in P19 cells was significantly decreased by RA treatment, whereas there was no significant difference in the expression of pri-Mirlet7d/a/f and pri-Mirlet7g between ethanol and RA treatment (Supplemental Fig. S1, b and c).
FIG. 3.
Posttranscriptional induction of Mirlet7 expression by RA signaling correlates with Lin28 repression. a and b) Quantitative RT-PCR analysis of the abundance of pri-Mirlet7s in both the in vitro model (a) and in vivo model (b) with ethanol vehicle or RA treatment. For the in vitro model, THY+ spermatogonia from 8 to 10 mice in each experiment were pooled and were treated with ethanol or RA (0.7 μM) for 24 h in vitro. For the in vivo model, VAD male mice were treated with sesame oil or RA for 24 h. Total RNA was subjected qRT-PCR, and Rps2 was used for normalization between samples. The levels of pri-Mirlet7d/a/f and pri-Mirlet7g were not significantly different between vehicle and RA treatment in both in vitro (P > 0.5 for both, n = 3; Student t-test) and in vivo (P > 0.5 for both, n = 16; Student t-test) model systems. c and d) Quantitative RT-PCR analysis of the Lin28 expression in both in vitro (c) and in vivo (d) models treated with vehicle or RA. Expression of Lin28 was significantly decreased in both in vitro (*P < 0.001, n = 3; Student t-test) and in vivo (*P < 0.001, n = 16; Student t-test) model systems within 24 h of RA treatment, whereas Lin28 expression was not significantly changed in the in vitro (P > 0.5, n = 5; Student t-test) model 8 h after RA treatment. e and f) Western blot analysis of the LIN28 protein in both in vitro (e) and in vivo (f) models treated with vehicle or RA. The level of LIN28 was decreased in both in vitro and in vivo model systems within 24 h of RA treatment. For the in vitro model, the result shown is representative of two independent experiments. For the in vivo model, the result shown is representative of three independent experiments. Equal protein loading was confirmed by β-ACTIN levels.
Retinoic Acid Receptors (RARs) Associated with Conserved Regions Upstream of Lin28
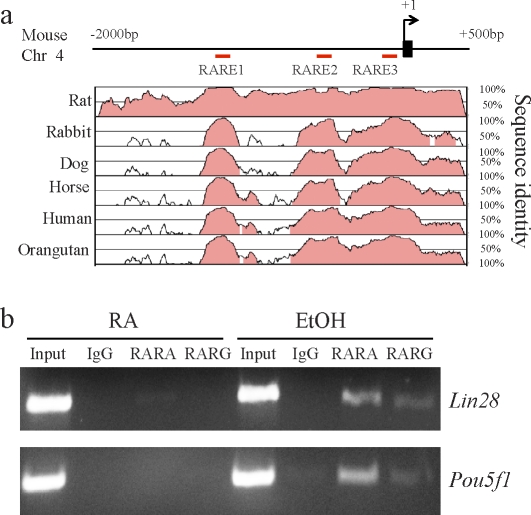
The action of RA on gene expression is mediated by two families of nuclear hormone receptors, the RARs (isoforms α, β, and γ) and the retinoid X receptors (RXRs; isoforms α, β, and γ), which work as RAR/RXR heterodimers. The RAR/RXR heterodimers bind to RA response elements (RAREs), typically composed of two direct repeats of a core hexameric motif, PuG(G/T)TCA, separated by a 5-bp spacer sequence (referred to as DR5) [33]. Promoters of many RA-regulated genes carry RAREs. To examine whether Lin28 is directly regulated by RA signaling, we performed ChIPs to determine whether RARs associate with a conserved locus upstream of Lin28. Inspection of ∼2-kb sequences upstream of the transcription initiation site of Lin28 by TESS (www.cbil.upenn.edu/cgi-bin/tess/tess) [34] revealed the presence of three putative RAREs (designated as RARE1, RARE2, and RARE3; Fig. 4a). We used Vista software [35, 36] to identify several conserved regions ∼2 kb upstream of Lin28 and RAREs with sites showing high conservation among mammals (Fig. 4a). RARE2 (AGGTCAGCGCCA), which exhibits a conserved homology to RARE sequences [37, 38], is located between −647 and −658 relative to the first transcription start site of Lin28 and is also situated 3′ to an Sp1 consensus binding site. Interestingly, the RARE located closely downstream of the Sp1 binding site in the Pou5f1 promoter is believed to mediate the RA-induced repression of Pou5f1 in P19 cells [39, 40]. We therefore designed PCR amplicons within the RARE2 to evaluate RAR binding in ChIP samples. As a positive control, an amplicon was designed within the promoter region of Pou5f1. Compared with ChIP samples generated with an IgG, a strong signal of the RARE2 region amplicon in both RARA and RARG ChIP samples was observed in RA-untreated P19 cells, whereas the signal in RARA ChIP samples was greatly reduced by RA treatment, and no signal in RARG ChIP samples was obtained in RA-treated P19 cells (Fig. 4b). Thus, RARs are associated with the conserved RARE2 region just upstream of Lin28 gene, suggesting that RA signaling directly regulates the expression of Lin28. We found that both RARA and RARG interact with the Pou5f1 promoter in untreated P19 cells, but not in RA-treated P19 cells. This result is consistent with the previous observation that in P19 cells, the Pou5f1 promoter was occupied by RAR/RXR heterodimers before RA treatment, but this occupancy was lost following the treatment [41, 42], demonstrating the specificity of our findings.
FIG. 4.
The RARs associated with the conserved region upstream of Lin28. a) Vista analysis of phylogenetic conservation encompassing upstream of Lin28. Vista was used to generate pairwise alignments between the genomic sequence from mouse and that from the indicated species. Red bars indicate locations of the RAREs, which are present in the conserved regions of Lin28. b) Chromatin immunoprecipitation analysis showing interaction of both RARA and RARG with the RARE2 (shown in a) of Lin28. The ChIP DNA samples obtained with RARs antibody and IgG control were subjected to PCR with specific primers encompassing RARE2 upstream of Lin28 (upper) or the Pou5f1 promoter as positive controls (lower). The RARs bind to both RARE2 and the Pou5f1 promoter in RA-untreated P19 cells, whereas only a very weak signal for RARA binding to RARE2 was detected in RA-treated P19 cells. No PCR product was detected in ChIP samples generated with an IgG control.
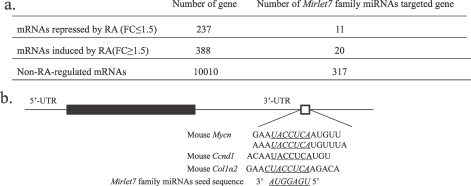
Putative Targets of the Mirlet7 Family miRNAs During RA-Induced Spermatogonia Differentiation
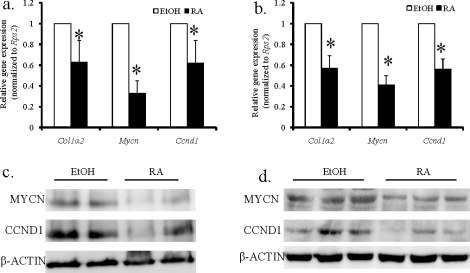
We used bioinformatics and mRNA microarray analysis to identify Mirlet7 target genes during RA-induced spermatogonial differentiation. We first investigated whether any genes expressed in mouse spermatogonia are putative targets of Mirlet7 family miRNAs. Based on computationally predicted target genes found in the database TargetScan (www.targetscan.org) [9, 43], Mirlet7 family miRNAs are predicted to target 325 mRNAs in mouse spermatogonia (www.wsu.edu/∼griswold/microarray.html) [44]. We next determined whether any RA-regulated mRNAs were the targets of Mirlet7 family miRNAs. Results from a microarray analysis of RA-untreated and 24-h RA-treated VAD mouse testes were used for this purpose (www.wsu.edu/∼griswold/microarray.html) [44]. From these data, we found that Mirlet7 miRNAs potentially target 11 RA-repressed, 20 inducible, and 317 non-RA-regulated genes (Fig. 5a and Supplemental Table S2). Some of the mRNAs (11) in the array that were decreased in RA-treated VAD testis showed conserved Mirlet7 family miRNA-binding sites in their 3′ UTRs with 7-mer or 8-mer seeds (Fig. 5b). Among these genes, Mycn, Ccnd1, and Col1a2 are known to be involved in spermatogonial development [45–48]. We further confirmed by qRT-PCR and Western blot analysis that expression of Ccnd1, Col1a2, and Mycn was significantly downregulated in both RA-treated spermatogonia (Fig. 6, a and c; Fig 6c, average densitometry ratio of N-MYC and β-ACTIN [mean ± SD) for non-RA-treated cells: 0.503 ± 0.055, n = 4; for RA-treated cells: 0.264 ± 0.053, n = 4; P < 0.05; average densitometry ratio of CCND1 and β-ACTIN [mean ± SD]: for non-RA-treated cells: 0.130 ± 0.009, n = 4; for RA-treated cells: 0.098 ± 0.007, n = 4; P < 0.05) and VAD testes (Fig. 6, b and d; Fig. 6d, average densitometry ratio of N-MYC and β-ACTIN [mean ± SD] for non-RA-treated testes: 0.376 ± 0.018, n = 11; for RA-treated testes: 0.305 ± 0.007, n = 11; P < 0.05; average densitometry ratio of CCND1 and β-ACTIN [mean ± SD] for non-RA-treated testes: 0.197 ± 0.01, n = 11; for RA-treated testes: 0.171 ± 0.006, n = 11; P < 0.05). It is plausible that RA may increase the expression of Mirlet7 which, in turn, downregulates proliferation-relevant gene expression and promotes spermatogonial differentiation.
FIG. 5.
The RA-regulated genes potentially targeted by Mirlet7 family miRNAs. a) Number of genes potentially targeted by Mirlet7 family members in RA-treated VAD testes. The mRNAs are subclassified into RA-repressed, RA-induced, and nonregulated by RA but expressed in spermatogonia based on microarray analysis of THY+ spermatogonia, vehicle, or RA-treated VAD testes (www.wsu.edu/∼griswold/microarray.html). FC, fold change. b) Mirlet7 miRNAs target the Ccnd1, Col1a2, and Mycn 3′ UTR. Sequence alignment shows a putative Mirlet7 complementary site in the Col1a2 and Ccnd1 3′ UTR and two putative Mirlet7 complementary sites in the Mycn 3′ UTR.
FIG. 6.
Quantitative RT-PCR (a and b) and Western blot analysis (c and d) of Mycn, Ccnd1, and Col1a2 in both in vitro (a and c) and in vivo (b and d) models treated with vehicle or RA. Data from qRT-PCR analyses are expressed as fold differences compared with vehicle-treated samples (mean ± SD, *P < 0.01, n = 3 for in vitro model, n = 14 for in vivo model; Student t-test). Rps2 was used for normalization between samples. Compared with non-RA-treated THY+ spermatogonia (EtOH; c) and VAD testes (EtOH; d), RA-treated THY+ spermatogonia (RA; c) and VAD testes (RA; d) had reduced expression of MYCN and CCND1. For the in vitro model, THY+ spermatogonia isolated from 8 to 10 pups were pooled in each group, and the result shown is representative of two independent experiments. For the in vivo model, protein was extracted from VAD testis, and the result shown is representative of three independent experiments. Equal protein loading was confirmed by β-ACTIN levels.
DISCUSSION
The role of RA in the initiation of spermatogonial differentiation has been well described [3, 4, 6, 7, 49, 50]. However, the mechanisms underlying spermatogonial differentiation remain to be explored. We proposed in this study that the posttranscriptional regulation of genes by miRNAs may be a molecular mechanism contributing to spermatogonial differentiation. For this study, we focused on determining the role of Mirlet7 family miRNAs in RA-induced spermatogonial differentiation. We chose this family because: 1) mature Mirlet7 is highly conserved across metazoans [51]; 2) the Mirlet7 family processing regulator LIN28 is expressed in mouse undifferentiated spermatogonia [29]; and 3) its role in multiple differentiation events in various tissues has been well established [25, 52].
The testis has a complex miRNA signature, and its role in testis function is just beginning to be examined. We found in this study that the Mirlet7 family miRNAs are expressed in spermatogonia and spermatocytes. We have presented evidence here that RA-induced spermatogonial differentiation is accompanied by an increase of six members of mature Mirlet7 miRNAs. The fact that the levels of Mirlet7 primary transcripts were not controlled by RA was striking and suggested that a common pathway was implicated in the processing of Mirlet7 family miRNAs. In embryonic stem cells and primordial germ cells, the pluripotency factor LIN28 has been shown to act as a specific inhibitor of all Mirlet7 family miRNA processing [30, 32, 53]. Lin28 is expressed in undifferentiated spermatogonia [28, 29], and our microarray experiments and qPCR analysis demonstrate that Lin28 is repressed within 24 h of RA treatment in spermatogonia. Chromatin immunoprecipitation analyses have further shown that RARs associate directly with a conserved RARE upstream of Lin28. These results indicate that inhibition of Lin28 could account for the RA-driven induction of the Mirlet7 miRNAs in spermatogonia.
There is extensive evidence to document a potent differentiation and antiproliferation activity of the Mirlet7 family. The Mirlet7 miRNAs are associated with cellular differentiation, and enforced expression of Mirlet7 inhibits self-renewal/proliferation and promotes differentiation [54–56]. These functions are attributable to the capacity of Mirlet7 to silence the self-renewal program by suppressing many downstream targets, including Ras, Hmga2, Myc, Mycn, Ccnd1, and even Lin28 itself [54, 56–60]. To assess the contribution of Mirlet7 miRNAs to the regulation of spermatogonial differentiation, we determined whether the expression of their potential targets can be changed as a result of RA in this process. Identification of an miRNA target by using an mRNA transcriptome database is relatively complex because most miRNAs regulate gene expression through translational repression rather than mRNA degradation in animals [61]. To overcome this obstacle, we first focused only on the genes with expression levels that showed a significant inverse correlation with Mirlet7 miRNAs. By analyzing the expression profile from VAD mice treated with RA, we found a downregulation of 11 genes within 24 h of RA treatment. Of the 11 gene candidates, we chose Mycn, Ccnd1, and Col1a2 for further investigation. These genes were selected because 1) Ccnd1, a key cell cycle regulator, has been implicated in spermatogonial proliferation, particularly during the G1/S transition [47, 48]; 2) Mycn, which is expressed in undifferentiated spermatogonia, has been shown to be involved in glial cell line-derived neurotrophic factor (GDNF)-induced SSC self-renewal/proliferation [46, 62, 63]; and 3) Col1a2 is associated with type A spermatogonia [45]. Using a bioinformatics approach and experimental validation, we identified Mycn, Ccnd1 and Col1a2 as targets of Mirlet7 in spermatogonia. We found that the expression of Mycn, Ccnd1, and Col1a2 is significantly reduced, whereas the level of Mirlet7 family miRNAs is dramatically increased upon RA-induced spermatogonial differentiation. Moreover, the direct downregulation of Mycn, Ccnd1, and Col1a2 by Mirlet7 miRNAs has been experimentally validated in other systems [52, 54, 64]. These studies suggest that Mirlet7 miRNAs play a critical role in spermatogonial proliferation and differentiation through targeting key genes.
It is interesting to note that Lin28 is itself a target gene for Mirlet7, suggesting that the regulatory interaction between Mirlet7 and Lin28 functions as a feed-forward loop. A possible mechanism could be that RA induces spermatogonial differentiation, resulting in the inhibition of Lin28 expression. With the decrease of Lin28, Mirlet7 levels rapidly increase. This increase in Mirlet7 levels could then in turn suppress its negative regulator Lin28 in spermatogonia. Moreover, repression of Mycn and Ccnd1 by Mirlet7 miRNAs blocks spermatogonial proliferation and promotes differentiation. Furthermore, Myc and Mycn have been shown to be a global regulator of miRNA expression and can activate expression of stem cell-associated miRNAs (such as the Mir-17-92 cluster and Mir-290 cluster) to stimulate cell proliferation/self-renewal [65]. Like Mycn, Myc is also a direct target for Mirlet7 [57]. Interestingly, Myc is highly expressed in undifferentiated spermatogonia [63]. The downregulation of Mycn and even Myc by Mirlet7 could prevent the expression of stem cell-associated miRNAs in spermatogonia. Therefore, our studies suggest an RA signal-Lin28-Mirlet7-Mycn regulatory loop that may contribute to RA-induced spermatogonial differentiation.
Last, our results provide new insight into the role of an RA-regulated miRNA, Mirlet7, in spermatogonial differentiation. However, this study does not exclude the possibility that other RA-regulated miRNAs may also play a role in spermatogonial differentiation. Future experiments are necessary to uncover additional Mirlet7 targets and other RA-regulated miRNAs, and to clarify the role of miRNAs in the regulation of spermatogonial differentiation.
ACKNOWLEDGMENTS
We thank Dr. Cécile Rochette-Egly for providing antibodies. We thank Dr. Cathryn Hogarth for critical reading of this manuscript.
Footnotes
Supported by National Institutes of Health grant HD10808.
REFERENCES
- Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol 2008; 24: 263 286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction 2001; 121: 347 354 [DOI] [PubMed] [Google Scholar]
- Griswold MD, Bishop PD, Kim KH, Ping R, Siiteri JE, Morales C. Function of vitamin A in normal and synchronized seminiferous tubules. Ann N Y Acad Sci 1989; 564: 154 172 [DOI] [PubMed] [Google Scholar]
- Morales C, Griswold MD. Retinol-induced stage synchronization in seminiferous tubules of the rat. Endocrinology 1987; 121: 432 434 [DOI] [PubMed] [Google Scholar]
- Doyle TJ, Oudes AJ, Kim KH. Temporal profiling of rat transcriptomes in retinol-replenished vitamin A-deficient testis. Syst Biol Reprod Med 2009; 55: 145 163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod 2008; 78: 537 545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod 2008; 79: 35 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350 355 [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19: 92 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister SC, Tizard ML, Doran TJ, Sinclair AH, Smith CA. Sexually dimorphic microRNA expression during chicken embryonic gonadal development. Biol Reprod 2009; 81: 165 176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E, Babak T, Chua G, Hughes T. Moens PB. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res 2008; 16: 243 260 [DOI] [PubMed] [Google Scholar]
- Michalak P, Malone JH. Testis-derived microRNA profiles of African clawed frogs (Xenopus) and their sterile hybrids. Genomics 2008; 91: 158 164 [DOI] [PubMed] [Google Scholar]
- Mishima T, Takizawa T, Luo SS, Ishibashi O, Kawahigashi Y, Mizuguchi Y, Ishikawa T, Mori M, Kanda T, Goto T, Takizawa T. MicroRNA (miRNA) cloning analysis reveals sex differences in miRNA expression profiles between adult mouse testis and ovary. Reproduction 2008; 136: 811 822 [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed microRNAs. Dev Biol 2007; 311: 592 602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Lu Y, Sun H, Qiu W, Tao D, Liu Y, Chen H, Yang Y, Zhang S, Li X, Ma Y. Microarray profiling of microRNAs expressed in testis tissues of developing primates. J Assist Reprod Genet 2009; 26: 179 186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Lu Y, Sun H, Tao D, Zhang S, Liu W, Ma Y. A microarray for microRNA profiling in mouse testis tissues. Reproduction 2007; 134: 73 79 [DOI] [PubMed] [Google Scholar]
- Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH, Durand P, Samarut J, Pain B, Rouault JP. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA 2010; 16: 720 731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, Tang F, Hajkova P, Lao K, O'Carroll D, Das PP, Tarakhovsky A, Miska EA, Surani MA. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One 2008; 3: e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007; 131: 1273 1286 [DOI] [PubMed] [Google Scholar]
- Tamminga J, Kathiria P, Koturbash I, Kovalchuk O. DNA damage-induced upregulation of miR-709 in the germline downregulates BORIS to counteract aberrant DNA hypomethylation. Cell Cycle 2008; 7: 3731 3736 [DOI] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol 2009; 19: 630 639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Hecht NB. The DNA/RNA-binding protein, translin, binds microRNA122a and increases its in vivo stability. J Androl 2008; 29: 572 579 [DOI] [PubMed] [Google Scholar]
- Yu Z, Raabe T, Hecht NB. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol Reprod 2005; 73: 427 433 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403: 901 906 [DOI] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer; 17: F19 F36 [DOI] [PubMed] [Google Scholar]
- Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc 2009; 4: 107 115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402 408 [DOI] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 2001; 27: 422 426 [DOI] [PubMed] [Google Scholar]
- Zheng K, Wu X, Kaestner KH, Wang PJ. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol 2009; 9: 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 2008; 32: 276 284 [DOI] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 2008; 283: 21310 21314 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science 2008; 320: 97 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 2004; 328: 1 16 [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics 2008; 21: 2.6.1 2.6.15 [DOI] [PubMed] [Google Scholar]
- Bray N, Dubchak I, Pachter L. AVID: a global alignment program. Genome Res 2003; 13: 97 102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and tools for whole-genome alignments. Genome Res 2003; 13: 73 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa R, Yu VC, Naar A, Kyakumoto S, Han Z, Silverman S, Rosenfeld MG, Glass CK. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev 1993; 7: 1423 1435 [DOI] [PubMed] [Google Scholar]
- Lee CH, Wei LN. Characterization of an inverted repeat with a zero spacer (IR0)-type retinoic acid response element from the mouse nuclear orphan receptor TR2-11 gene. Biochemistry 1999; 38: 8820 8825 [DOI] [PubMed] [Google Scholar]
- Pikarsky E, Sharir H, Ben-Shushan E, Bergman Y. Retinoic acid represses Oct-3/4 gene expression through several retinoic acid-responsive elements located in the promoter-enhancer region. Mol Cell Biol 1994; 14: 1026 1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester I, Scholer HR. Regulation of the Oct-4 gene by nuclear receptors. Nucleic Acids Res 1994; 22: 901 911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shushan E, Sharir H, Pikarsky E, Bergman Y. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol Cell Biol 1995; 15: 1034 1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ozato K. Genomic footprinting of retinoic acid regulated promoters in embryonal carcinoma cells. Methods 1997; 11: 197 204 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15 20 [DOI] [PubMed] [Google Scholar]
- Griswold Lab Microarray Data. Affymetrix GeneChip Data, Washington State University; Pullman, WA.: 2007. World Wide Web (URL: http://www.wsu.edu/∼griswold/microarray/). (January 12, 2007) [Google Scholar]
- He Z, Feng L, Zhang X, Geng Y, Parodi DA, Suarez-Quian C, Dym M. Expression of Col1a1, Col1a2 and procollagen I in germ cells of immature and adult mouse testis. Reproduction 2005; 130: 333 341 [DOI] [PubMed] [Google Scholar]
- Braydich-Stolle L, Kostereva N, Dym M, Hofmann MC. Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Dev Biol 2007; 304: 34 45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Jiang J, Kokkinaki M, Dym M. Nodal signaling via an autocrine pathway promotes proliferation of mouse spermatogonial stem/progenitor cells through Smad2/3 and Oct-4 activation. Stem Cells 2009; 27: 2580 2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer TL, Roepers-Gajadien HL, Gademan IS, Kal HB, de Rooij DG. Involvement of the D-type cyclins in germ cell proliferation and differentiation in the mouse. Biol Reprod 2000; 63: 1893 1898 [DOI] [PubMed] [Google Scholar]
- Li H, Clagett-Dame M. Vitamin A deficiency blocks the initiation of meiosis of germ cells in the developing rat ovary in vivo. Biol Reprod 2009; 81: 996 1001 [DOI] [PubMed] [Google Scholar]
- Li H, Palczewski K, Baehr W, Clagett-Dame M. Vitamin A deficiency results in meiotic failure and accumulation of undifferentiated spermatogonia in prepubertal mouse testis. Biol Reprod 2011; 84: 336 341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000; 408: 86 89 [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A; 107: 1876 1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 2009; 460: 909 913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature; 463: 621 626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol 2010; 12: 1101 1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 2008; 10: 987 993 [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007; 67: 9762 9770 [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell 2005; 120: 635 647 [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007; 21: 1025 1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007; 315: 1576 1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature 2008; 455: 64 71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Lee J, Inoue K, Ogonuki N, Miki H, Toyokuni S, Ikawa M, Nakamura T, Ogura A, Shinohara T. Pluripotency of a single spermatogonial stem cell in mice. Biol Reprod 2008; 78: 681 687 [DOI] [PubMed] [Google Scholar]
- Koji T, Izumi S, Tanno M, Moriuchi T, Nakane PK. Localization in situ of c-myc mRNA and c-myc protein in adult mouse testis. Histochem J 1988; 20: 551 557 [DOI] [PubMed] [Google Scholar]
- Ji J, Zhao L, Budhu A, Forgues M, Jia HL, Qin LX, Ye QH, Yu J, Shi X, Tang ZY, Wang XW. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol; 52: 690 697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV. Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005; 435: 839 843 [DOI] [PubMed] [Google Scholar]