Abstract
Plants provide unique opportunities to study the mechanistic basis and evolutionary processes of adaptation to diverse environmental conditions. Complementary laboratory and field experiments are important for testing hypothesis reflecting long term ecological and evolutionary history. For example, these approaches can infer whether local adaptation results from genetic tradeoffs (antagonistic pleiotropy), where native alleles are best adapted to local conditions, or if local adaptation is caused by conditional neutrality at many loci, where alleles show fitness differences in one environment, but not in the contrasting environment. Ecological genetics in natural populations of perennial or outcrossing plants also may differ substantially from model systems. In this review of the evolutionary genetics of plant adaptation, we emphasize the importance of field studies for understanding the evolutionary dynamics of model and non-model systems, highlight a key life history trait (flowering time), and discuss emerging conservation issues.
Keywords: Evolutionary genetics, Evolutionary and ecological functional genomics, local adaptation, antagonistic pleiotropy, genotype by environment interaction, drought tolerance
Linking genotype to phenotype in the field and the laboratory
Advances in genomics and DNA sequencing technology are revolutionizing our understanding of natural genetic variation. Sequence signatures of natural selection can be detected in patterns of nucleotide polymorphism within and among populations. These approaches have identified strong selective sweeps in humans [e.g., 1], characterized genome- wide rates of positive selection in Drosophila [2], and detected diverse modes of natural selection in plants [3,4]. Furthermore, recent statistical improvements provide more robust inference of genomic regions that have been influenced by local adaptation [5,6], including the identification of correlations between environmental factors and allele frequencies that are likely adaptive [7].
Nevertheless, massive data sets on nucleotide polymorphism and divergence often fail to identify signatures of recent, strong selection on individual loci within or among populations. For example, resequencing of replicate Drosophila melanogaster populations subject to 600 generations of divergent phenotypic selection found no evidence for fixation of new, unconditionally advantageous mutations [8], perhaps because evolutionary changes were based on standing genetic variation in linkage equilibrium with nearby polymorphisms. Similar results were found in a whole genome analysis of 179 human genomes, which showed little evidence for classic selective sweeps [9]. Such findings are not inconsistent with recent adaptive evolution, however, since clear signatures of selection are not expected when adaptive evolutionary change employs standing genetic variation rather than new mutations, or when polygenic complex traits evolve by subtle changes in allele frequency at many loci. Even when population genomic studies succeed in identifying a set of genes or broader genomic regions that are likely to have been recent targets of selection, it is difficult to infer the phenotypic traits that were affected by the changes in genome sequence, and the ecological circumstances that imposed the changes in natural selection. For these reasons, analyses of adaptive evolution will benefit greatly from a combination of approaches that link genomic studies with manipulative experiments on individual phenotypes and environmental variables [10].
Among the experimental systems in biology, plants provide excellent opportunities to study the interaction between genetic and environmental variation, which produces the complex traits observed in nature. Manipulative experiments such as reciprocal transplants can test for local adaptation to relevant natural environments. Forward genetic approaches have provided key insights into the genes that underlie plant adaptations in model systems under controlled laboratory conditions. In this review, we emphasize the need to complement laboratory genomic analyses with field studies of natural populations to understand evolutionary processes in model and non-model systems. Evolutionary studies in plants may be more difficult to conduct in long-lived and outcrossing species [11], yet ecological genetics in natural populations of perennial or outcrossing plants may differ substantially from annual, naturally inbred species that are widely used in plant biology. Here, we: (a) discuss the merits of studying emerging plant model systems and briefly review ecogenomic techniques that can be employed in these systems; (b) review the genetic basis of flowering time, an example of a key phenotypic trait that has been extensively characterized in Arabidopsis thaliana, and highlight the ways in which emerging model systems can enhance our understanding of the genetics of adaptation in this trait; and (c) discuss the importance of studying plant adaptations in a world that is rapidly changing.
Emerging model systems are focused on phenotypic traits in environments that are ecologically and evolutionarily relevant
Early studies of evolutionary genetics of plant adaptation focused on Arabidopsis thaliana in the laboratory [12], but more recently, evolutionary genomic resources are being developed for many other plant species, and Arabidopsis thaliana studies are being conducted in the wild. (For a synopsis of model and emerging model species, resources available for each species, and life history characteristics, see Table 1.) Investigations of ecological model organisms, including perennials and outcrossers, will elucidate the breadth of evolutionary processes that maintain phenotypic and genetic variation in natural populations [13]. For example, is most variation actively maintained by balancing, frequency-dependent or divergent selection? Or, is variation due to mutation-selection balance, or immigration of maladapted alleles? Do species with different life history strategies and mating systems use similar genetic pathways to produce convergent phenotypes? Studies of ecological model species have substantially advanced our understanding of the genetic basis of phenotypic traits (e.g., flower color, Box 1).
Table 1.
Characteristics of genomically-enabled emerging model systems for evolutionary ecology.
| Plant Ecological Model Systems |
Transformation | Genome sequence2 | QTL mapping | RILs or IILs | Positional cloning | Interspecific mapping3 |
Diversity collection4 | Genome Size (Mb) | Life History | Breeding system |
|---|---|---|---|---|---|---|---|---|---|---|
| Aquilegia species | −1 | + | + | − | − | + | + | 301 | P | O |
| Arabidopsis lyrata | + | + | + | − | − | H | + | 207 | P | I, O |
| Arabidopsis thaliana | + | + | + | + | + | − | + | 125 | A | I |
| Arabis alpina | + | S | + | − | − | ? | + | 380 | P | I |
| Boechera species | + | S | + | + | + | + | + | 200 | P | I, D |
| Brachypodium distachyon | + | + | + | − | + | − | + | 270 | A | I |
| Brassica & Raphanus | + | S | + | + | + | + | + | 550 | B | I, O |
| Capsella rubella & grandiflora | − | S | − | − | − | + | + | 202 | A | I, O |
| Helianthus species | + | S | + | + | − | + | + | 3500 | B | O |
| Mimulus species | + | + | + | + | + | + | + | 500 | B | I, O |
| Nicotiana attenuata | + | S | − | − | − | − | − | 2300 | A | I |
| Oryza species | + | S | + | + | + | + | − | 357 | B | I, O |
| Panicum capillare & hallii | − | S | − | − | − | ? | − | 300 | B | I |
| Zea species, teosinte | − | + | + | − | + | + | + | 2500 | B | O |
In all cases: + means yes, − means no, and ? means unknown.
Genome sizes indicate the smallest known values within each group. Genome sizes for Boechera and Capsella were calculated from relative genome sizes [99] scaled to Arabidopsis lyrata = 207 Mb [100].
Interspecific mapping refers to homoploid, fertile F1s providing fertile F2 progeny.
Diversity collection signifies the existence of publically available intra- or -interspecific accessions.
A = annual; B = both annual and perennial genotypes exist; D = diplosporous apomixes; H = hybrid breakdown exhibited by interspecific crosses; I = predominantly inbreeding; IILs = inbred introgression lines; O = predominantly outcrossing; P = Perennial; RILs = recombinant inbred lines; S = sequencing in progress;
Box 1. Biosynthetic pathways and ecological consequences of flower color.
Flower color presents a robust example of a trait with a well-defined, phylogenetically- conserved, biosynthetic pathway [86]. Anthocyanin pigments are the most common, widely distributed, floral pigment in angiosperms [86]. Flux down the three main branches of the anthocyanin biosynthetic pathway determines the production of three major classes of anthocyanins, and, therefore, flower color: pelargonidin (red or orange flowers), cyanidin (blue or magenta flowers), and delphinidin (dark blue to purple flowers) [86]. Many macroevolutionary transitions in flower color result from changes in the class of anthocyanin produced [86]. Studies of model (Petunia spp., Antirrhinum majus, and Ipomoea spp.) and non- model organisms have highlighted the importance of anthocyanins both in flower color and visitation by pollinators and have linked phenotype to genotype within the appropriate ecological context [32,86,87].
Flower color is an ideal trait to address evolutionary questions about gene duplication, pleiotropy, and the types of genes (e.g., structural or regulatory) that influence adaptation [32,86,88,89]. Anthocyanin pigments are produced in floral tissue, where they determine flower color, as well as vegetative and other tissues, where they influence herbivore resistance, desiccation resistance and other fitness-related traits [88]. Mutations in the genes that regulate enzymes in the anthocyanin pathway can influence floral color without impairing anthocyanin production in vegetative tissue; however, mutations to structural genes that influence flower color could have adverse pleiotropic effects on other physiological or morphological traits [32,86,88]. Consequently, several key questions about flower color evolution remain unresolved, such as whether flower color polymorphism influences fitness directly, or if fitness variation results from selection on correlated vegetative traits [86].
Floral traits are particularly amenable to dissection at the genetic level, as closely-related lineages can often be crossed under controlled conditions [87]. For example, a recent elegant study of intraspecific flower color variation in Phlox drummondii demonstrated that cis- regulatory mutations to two genes in the anthocyanin biosynthetic pathway affected flower color, pollinator behavior, as well as pre- and post-zygotic isolation [90]. Additionally, flower color can be experimentally altered by painting flowers, which permits researchers to study natural selection on flower color without disrupting genetic correlations among floral and vegetative traits and to disentangle the effects of pollinators and other environmental factors as agents of selection [86]. Ecological model species provide many opportunities to investigate flower color evolution.
Complex trait variation in natural populations differs from domesticated crops and introduced species in several important respects. First, native species can be studied in the environments where they originally evolved, influenced by ecological adaptation to biotic [e.g., 14] and abiotic [e.g., 15] environments over long periods of time. Alternatively, crop populations and introduced species occupy ephemeral habitats that may be very different from the habitats where they originated. Secondly, native species in undisturbed habitats may retain heritable variation influenced by mutation, genetic drift, demography, and adaptation over thousands of generations. In contrast, agricultural and introduced species have experienced domestication or migration bottlenecks that eliminate rare alleles from populations, as well as possible admixture among divergent founder genotypes. For these reasons, studies in natural populations are especially important for testing hypothesis that reflect long term ecological and evolutionary history. Furthermore, it is important that field studies expose experimental individuals to naturally-recruiting vegetation, since competitors can influence the expression of quantitative traits and the evolutionary dynamics of plant species [16,17]. Field studies of native species can also explicitly test for adaptation to the biotic community by experimentally manipulating the density or species composition of competitors, herbivores, mutualists or pathogens [16,17].
Experimental approaches
A growing body of literature from trees suggests that the genetic basis of adaptation can be addressed even in long-lived perennial and woody species [e.g., 18]. When nonmodel species are closely related to model organisms (e.g., in the family Brassicaceae), molecular resources developed for the model can be exploited in evolutionary studies of the non-model species [4]. In plant with short generation times, experimental mapping populations such as recombinant inbred lines (RILs) and near-isogenic lines (NILs) can be generated to detect and fine map Quantitative Trait Loci (QTL) [19,20]. Pedigreed lines cannot, however, be easily produced for most long-lived perennial species, but other ecogenomics approaches can be used to investigate the genetic basis of adaptation in nonmodel organisms [21–25]. Briefly, QTL mapping approaches can be applied to natural populations when relatedness of individuals is unknown in methods such as genome wide association studies (GWAS) [26]. GWAS explicitly tests for the association between phenotypic traits and allelic variation at many loci, and has a number of advantages over traditional QTL mapping of pedigreed lines [27]. For one, natural populations generally have experienced more recombination than experimental populations and GWAS can, therefore, map QTL with more precision [28]. Additionally, GWAS incorporate greater allelic diversity than family-based QTL methods, which rely on phenotypic and genetic differences between the parental lines from controlled crosses [28]. However, population-based methods like GWAS need to control for population structure, which can confound the detection of significant QTL (Box 2).
Box 2. Complications of association mapping techniques.
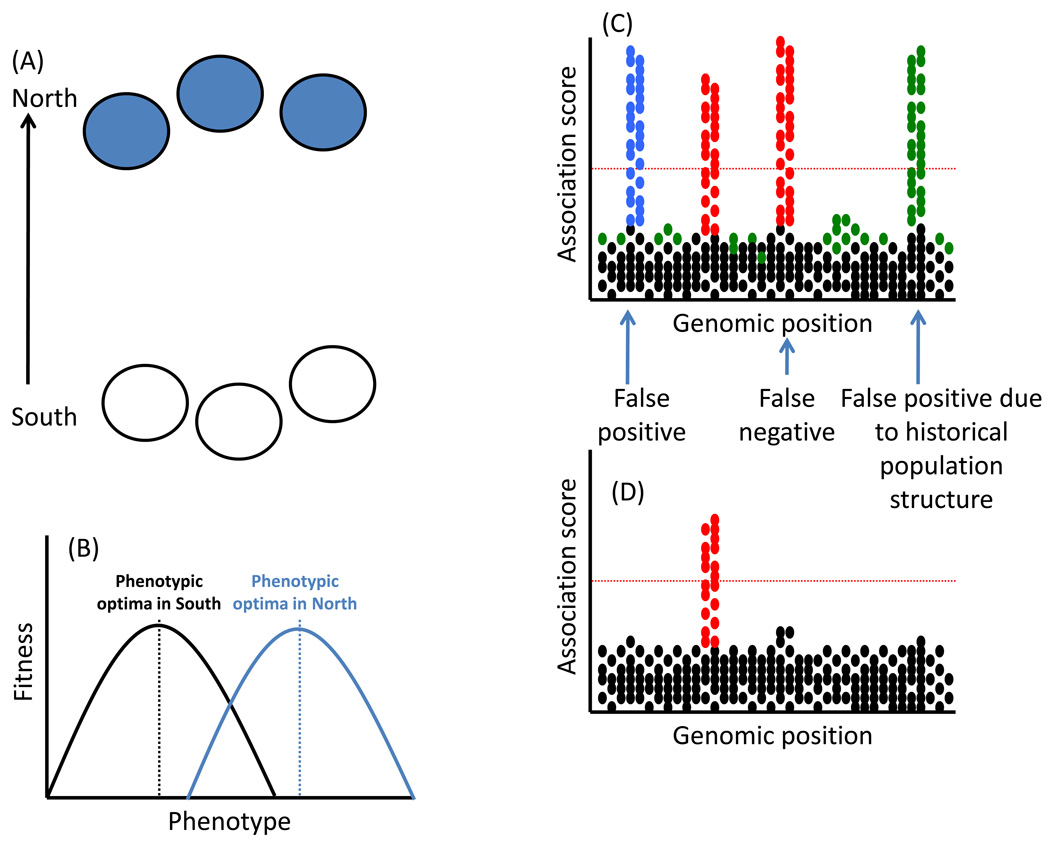
Population demography, genetic drift, and historical events such as population bottlenecks or founder events can confound the discovery of genes that encode adaptive traits [27,91]. These evolutionary forces may be particularly pervasive in non-model systems under field conditions. For example, genetic drift, limited gene flow, and divergent natural selection can simultaneously drive population differentiation across heterogeneous landscapes. Divergent selection can generate allelic variation at loci that influence ecologically-relevant phenotypic traits, but genetic drift operates across the genome affecting variation at neutral and non-neutral loci. When populations are subdivided because of both divergent selection and genetic drift, phenotypic variation caused by adaptive loci may be spuriously correlated with allelic variation at neutral loci (Figure I). In such cases, population-based approaches such as genome wide association studies (GWAS) have difficulty identifying adaptive loci that underlie phenotypic variation [25,26]. Statistical tools to control for population structure can eliminate false positives, i.e., marker loci that appear to be associated with phenotypic variation; however, statistical corrections can also discard adaptive loci that truly influence ecologically-relevant traits (false negatives) [61]. As population structure cannot always be controlled statistically, GWAS may best be applied within (not among) populations [25,26].
GWAS results also depend on the frequency of alleles in natural populations; an allele that has a large phenotypic effect is unlikely to be detected in GWAS if it occurs at low frequency [28]. The statistical power to detect a QTL with a phenotypic effect increases substantially with the frequency of functional alleles [28]. Furthermore, the precise genetic architecture of adaptation could be difficult to resolve with GWAS. If phenotypic variation is the result of many QTLs of small effect, those small effect QTLs could be difficult to detect. Complementing GWAS with other experimental approaches can highlight genomic regions that have important phenotypic effects. Independent evidence derived from traditional linkage mapping of lineages derived from experimental crosses, candidate gene approaches, and transgenics (when possible) can be used to corroborate and test the results of GWAS and elucidate the genetic basis of adaptation in nonmodel systems [28,61,91,92]. All QTL mapping approaches require fine-mapping and ultimate cloning of the QTL to identify causal genes.
In contrast with QTL mapping, population genomics techniques identify regions of the genome that exhibit patterns of polymorphism or divergence that implicate natural selection rather than neutral processes [29]. Population genomics approaches can detect the signature of selection at specific loci [29], but are conducted in the absence of phenotypic data [23] and, therefore, cannot elucidate the genetic basis of phenotypes. Additional techniques such as transcription profiling can be used to link phenotype with genotype, and will be more important as sequencing costs decline [21,23]. Finally, humans share key traits with some nonmodel organisms, such as long generation time and complex population history, and investigations of humans suffer from the inability to create pedigreed lines. Evolutionary biologists interested in perennial and annual species could adopt techniques developed for studies of humans, which can complement manipulative QTL studies [30].
Reciprocal transplant experiments with mapping populations such as RILs or NILs are necessary to study the genetic basis of local adaptation
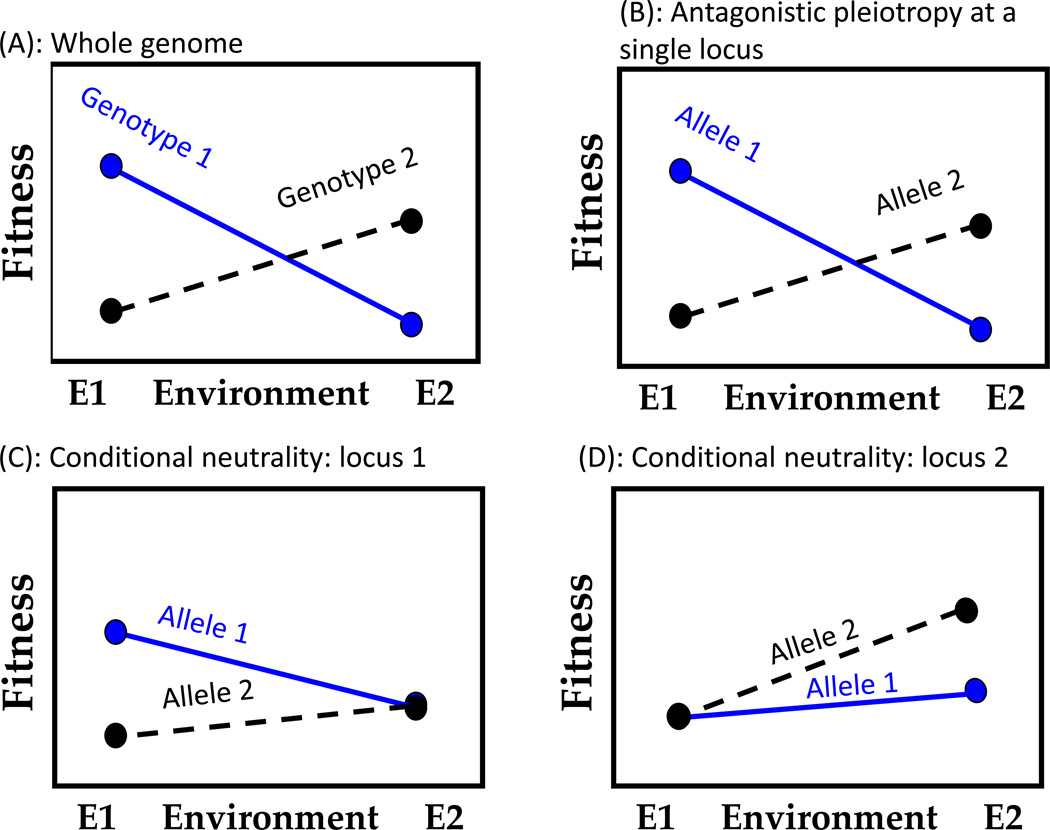
Due to their sedentary nature, plants may be strongly influenced by locally variable selection, which often results in local adaptation [31]. Divergent selection can drive reproductive isolation and ecological speciation [32]; however, the genetic basis of local adaptation remains unresolved. Local adaptation could result from tradeoffs at key loci where native alleles show a fitness advantage relative to foreign alleles (antagonistic pleiotropy) [13,33]. Alternatively, multiple independent loci could interact to produce local adaptation at the organismal level, if alleles at some loci are beneficial in only one environment, but neutral in the contrasting environment, and alleles at other loci show the opposite pattern (conditional neutrality) [13,33]. Both conditional neutrality and antagonistic pleiotropy can be inferred from QTL by environment interactions when genotypes are exposed to two or more environments and fitness-related QTLs are mapped. Distinguishing between these alternative hypotheses, however, requires explicit consideration of the fitness effects of alleles in their native habitats or simulated environmental conditions (Figure 1). Theoretically, homogenizing gene flow would spread conditionally beneficial alleles throughout metapopulations, thereby reducing the influence of conditional neutrality on local adaptation [33]. Nonetheless, to-date, the literature presents more examples of conditional neutrality [34–37] than antagonistic pleiotropy [38], although several studies reveal both conditional neutrality and antagonistic pleiotropy for different fitness components, sets of environments, or loci [33,39]. Additionally, chromosomal inversions can contribute to local adaptation and reproductive isolation, especially when inversions capture loci that display antagonistic pleiotropy [40, and references therein].
Figure 1.
In spatially heterogeneous landscapes, divergent selection in contrasting environments can result in the evolution of local adaptation [31]. Owing to their sedentary nature, many plant species are adapted to local conditions [31], which can be graphically represented as elevated fitness in their home environment and depressed fitness in an alternative environment (A). In all panels of this figure, the genotype or allele is labeled with its environment of origin (1 or 2). The genetic basis of local adaptation has only rarely been investigated in plants. Local adaptation at the organismal level could be due to antagonistic pleiotropy at the single locus or QTL level (B), where native alleles have a fitness advantage relative to foreign alleles. Alternatively, an allele could be beneficial in its native environment, but have no fitness costs in the contrasting environment (C and D). This pattern of conditional neutrality can contribute to local adaptation at the whole-genome level when several independent loci interact to influence fitness, and alleles derived from contrasting environments are favored at different loci. For example, at one locus, an allele derived from environment 1 is conditionally advantageous (C), whereas an allele from environment 2 is conditionally advantageous at a second locus (D). Investigating local adaptation requires reciprocal transplant or common garden experiments in the native habitats.
Demonstration of antagonistic pleiotropy requires higher statistical power, as the fitness advantage of the local allele must be significant in two contrasting environments. This results in an ascertainment bias against detecting antagonistic pleiotropy. Although QTL mapping provides little evidence that antagonistic pleiotropy maintains genetic variation among populations, detecting this historical mechanism of balancing selection requires fitness measurements of native alleles or QTLs in undisturbed populations where they originated, which has rarely been attempted. Clearly, discerning the genetic mechanism for local adaptation (and phenotypic plasticity, Box 3) requires additional field and laboratory studies of non-model species with different mating systems and life history strategies.
Box 3. The genetic basis of phenotypic plasticity.
Extensive interhabitat gene flow, and temporally variable environments where conditions change within the lifetime of an individual, can favor genotypes that shift their phenotype based on the environment they encounter, i.e., phenotypically plastic forms [93,94]. Phenotypic plasticity is an important strategy to maximize fitness in temporally and spatially heterogeneous landscapes, and QTL by environment interactions for phenotypic traits can reveal the genetic basis of plasticity [95]. For adaptive phenotypic plasticity to evolve, individuals must be capable of responding to predictable environmental signals, and plastic genotypes must have a fitness advantage over genotypes that are incapable of altering their phenotypes [93,94].
Plasticity in phenotypic traits can be heritable [96] and subject to natural selection [17,94]. Nevertheless, little is known about regulation and genetic control of phenotypic plasticity, although QTL can be mapped directly for plasticity, when plasticity is defined as a change in phenotype over environments (e.g., the slope of the reaction norm) or phenotypic variance across environments [95]. Phenotypic plasticity could result from heritable epigenetic effects that influence gene expression at different developmental stages or in different environments [97,98]. More evolutionary studies of phenotypic plasticity are needed to: (a) quantify heritability of phenotypic plasticity, (b) evaluate selection on plasticity in temporally and spatially variable habitats, and (c) dissect the genetic basis of plasticity. Indeed flowering phenology, one of the best known plant phenotypes (see main text), is a highly plastic trait, as the timing of flowering varies in response to seasonal cues; heritability of flowering time, as well as selection on this trait and causal genes underlying flowering time are often known.
Genetic basis of natural variation in flowering time, a key phenotypic trait: Translating from model organisms to non-model species in natural environments
The initiation of reproduction is a critical life history transition for all species. In plants, the exact timing of reproduction has clear fitness consequences; flowering too early or too late can reduce the number of potential mates [41], increase floral damage due to adverse conditions like frosts [42], and risk incomplete seed development prior to the onset of harsh winter or drought seasons. Selection can optimize flowering to coincide with favorable environmental conditions, thus contributing to local adaptation in heterogeneous landscapes [34,43–45]. Not surprisingly, plant species have evolved to flower in response to reliable environmental cues, such as the duration of winter (vernalization), snowmelt date, photoperiod, temperature, nutrient levels and precipitation [20,42,46–48].
Flowering time has been investigated extensively at the molecular and developmental level in the model species Arabidopsis thaliana [reviewed in 49,50,51] and some crop species [52]. In Arabidopsis, the flowering time gene network consists of more than 60 genes [49], which are regulated by four pathways: photoperiod, autonomous, vernalization, and gibberellin [49,50]. These four pathways integrate extrinsic and intrinsic signals to promote flowering at appropriate times during the growing season. A recent review provides an excellent synopsis of the flowering time gene network [50].
Orthologs of Arabidopsis flowering time genes influence the transition to reproduction in many monocotyledonous and dicotyledonous species [50,53]. For example, the floral integrator FLOWERING LOCUS T (FT), has conserved effects on flowering time in Arabidopsis and a diverse array of plant species [49,53–59]. Furthermore, duplication of FT homologs has been implicated in sunflower domestication [59] and an ortholog of FT co-localizes with a QTL that affects the timing of flowering under controlled conditions and the probability of flowering in field gardens in the mustard Boechera stricta [20]. Recent work suggests that the protein encoded by FT is an important component of “florigen,” a transmissible signal that travels from leaf tissue to meristems where it initiates flowering [49,51]. Grasses, which are only distantly related to Arabidopsis, rely on orthologs of Arabidopsis flowering time genes to flower in response to environmental signals (vernalization in temperate lineages, photoperiod and temperature); however, flowering in these monocotyledonous species is also regulated by genes that have no known Arabidopsis homologs or have somewhat different functions than their Arabidopsis counterparts [50].
QTL mapping approaches have detected loci that underlie natural variation in flowering time in Arabidopsis, and co-localize with genes from the flowering time gene network, which were primarily described using induced mutations [49]. However, these studies have been largely conducted under controlled laboratory conditions, and some variation in flowering time in Arabidopsis accessions remains unexplained [49]. Additionally, QTLs for flowering time in Arabidopsis detected in field studies may not correspond with QTLs mapped in the laboratory and/or do not co-localize with genes known to be from the flowering time gene network [60,61]. Future research should complement laboratory with field studies to investigate the genetic basis of this key life history trait under realistic natural conditions, which can vary spatially and temporally. The lack of congruence between field and laboratory studies in Arabidopsis [60,61] and the closely-related crucifer, Boechera stricta [20], suggest that additional abiotic or biotic signals may control flowering time in nature, but these signals have not been modeled effectively under laboratory conditions. For example, precipitation influences flowering phenology, and flowering at specific times can enable escape from drought [48,62]. Nevertheless, we know little about genes that promote or inhibit flowering in response to rainfall or soil water content [63]. Additionally, tropical cereals such as rice and maize do not require vernalization to flower [50]; other agronomic and natural species that inhabit tropical regions could also rely on somewhat different environmental signals than Arabidopsis to promote flowering. Experimental manipulations in the field are crucial to investigate the evolutionary dynamics of flowering phenology and detect QTL that differentially influence the transition to flowering in response to biotic factors or abiotic stresses like drought in tropical and temperate systems. Research in nonmodel systems under natural conditions could illuminate additional, as yet undescribed, floral regulatory pathways and relationships among genes in different pathways.
Little is known about the genetic basis of flowering time in perennial species [20,55]. In the iteroparous perennial crucifer, Arabis alpina, PERPETUAL FLOWERING 1 (PEP1), an ortholog of the Arabidopsis FLOWERING LOCUS C (FLC) gene, underlies the perennial response to vernalization [46]. In the annual Arabidopsis, vernalization elicits chromatin modifications that repress FLC and promote flowering; in contrast PEP1 is only transiently repressed by vernalization in A. alpina, which permits this perennial species to flower at the appropriate time of the growing season over many years [46]. Unlike annuals, perennial species can delay reproduction if conditions for flowering are suboptimal. Thus, annuals and perennials may differ fundamentally in flowering time response to environmental conditions and the genetic architecture of flowering phenology. What extrinsic and intrinsic cues trigger key life history transitions in perennial species? Are these cues similar to those that initiate transitions in annual species? Future studies of the environmental signals and genetic basis of flowering time in perennials will reveal evolutionarily important differences among species with different life history strategies.
Importance of plant adaptation for emerging global problems
Drought tolerance
Water availability is a fundamental determinant of plant performance in natural and agriculture populations. Strategies for coping with drought stress include drought escape (often by reproducing before the onset of water limitation) [48], or dehydration avoidance (perhaps by growth of deep roots to exploit subterranean water supplies)[64]. Although traits such as these are amenable to genomic analyses of complex trait variation, attempts to understand the interactions between plant phenotypes and water-limited environments have been among the most difficult problems in plant biology [65]. These challenges reflect the diversity of environmental conditions that can impose drought stress in the field, uncertainty of how component traits can be integrated into whole plant phenotypes to deal effectively with water stress, and the need for interdisciplinary experiments combining molecular biology, quantitative genetics, and physiological ecology [66].
Genetic studies of natural variation in drought tolerance have examined plant performance under field conditions [67,68], or have used genomic approaches to investigate plant responses to water limitation in the laboratory [69]. However, few studies have combined both field phenotypes and lab genomics [70], especially for ecological model systems [15]. Because drought tolerance is very important for crop productivity and human welfare, large research investments have been made to elucidate mechanisms of water use efficiency. Consequently, there is enormous potential for evolutionary studies to apply these results for hypothesis testing in ecological genetics.
Conservation genetics
Habitats in contemporary landscapes are highly fragmented, and species are being exposed to novel environmental stresses due to climate change, the expanding range of invasive species, and habitat degradation [48,71–74]. Evolutionary biologists who explicitly design experimental studies with these conservation priorities in mind can simultaneously test hypotheses derived from theory, and generate results that can be applied to emerging global problems.
Invasive species can have devastating effects on ecosystem processes, individual-level fitness of native species, community dynamics, and species diversity [75]. Evolutionary studies of invasive species have addressed a number of fundamental questions, such as: Does phenotypic plasticity facilitate invasion [76]? What are the evolutionary dynamics of invasive species in their novel ranges [72,77]? Is clinal variation in traits due to adaptation in the novel range, or multiple invasions from the native range [78]? Invasive species are well-suited to studies of rapid evolution to novel conditions, natural selection in the wild, and the effects of population bottlenecks and founder effects on additive genetic variation [72,77]. In contrast with native species, invasive species may experience reduced selection for tolerance and resistance to specialized pests, and self-fertilization may be advantageous as a mechanism of reproductive assurance [72]. Many more empirical studies are needed to understand natural selection on phenotypic traits in invasive species and the evolutionary biology of invaders [72].
Habitat fragmentation reduces the number of populations as well as average population size and increases the extent of geographic isolation [73]. Population bottlenecks, genetic drift, inbreeding and reduced gene flow can erode genetic diversity in small fragmented populations, resulting in increased homozygosity, depressed fitness, and population (and species) extinction [73,74]. Even long-lived wind-pollinated tree species can show reduced diversity, altered breeding systems, and increased inbreeding due to fragmentation [79]. Limited genetic variation can constrain adaptive evolution, especially under novel environmental conditions; thus, habitat fragmentation might impede the ability of species to adapt to altered climatic regimes [73,74,80]. Empirical studies to-date have largely ignored the implications of fragmentation for the evolutionary potential of species, especially in the context of global climate change.
Anthropogenic climate change has already caused species extinctions, altered the abundance of species, disturbed interspecific interactions, resulted in altitudinal and latitudinal shifts in geographic ranges of species, and altered phenology [81,82]. Contemporary climate change is occurring significantly more rapidly than geological climate change; species may not have the dispersal abilities needed to track preferred conditions, given the extent of fragmentation [83]. Climate change will likely impose considerable selection for stress tolerance. Nevertheless, few studies have explicitly tested whether species are adapting to novel conditions [reviewed in 80]. Given their shorter generation times, annuals and short-lived perennials may adapt more readily to a changing climate than long-lived herbaceous and woody perennials. Additional studies are needed to determine whether plant species with different life history strategies will be capable of adapting to novel conditions. Plant species are flowering significantly earlier than ever before [81], flowering time is highly responsive to environmental conditions [20], and much is known about the genetic basis of flowering time in model species [50]. Therefore, this life history trait could provide valuable insights into evolutionary response to climate change.
Population-level studies of habitat fragmentation focus on genetic diversity and inbreeding [73,79]. Similarly, studies of global climate change test the ecological and evolutionary responses of species to predicted conditions [81]. We know almost nothing about how habitat fragmentation influences adaptive evolution under projected climate change. Nevertheless, environmental stresses (e.g., climate change) can exacerbate inbreeding depression [84]; thus, inbred fragmented populations could show a reduced capacity to adapt to contemporary and changing conditions relative to large unfragmented populations. Fragmented populations with reduced genetic diversity might lack variation for key ecological traits such as drought-tolerance. Assisted migration to suitable habitats, in conjunction with conservation of habitat corridors, could be necessary to prevent dramatic declines in species and genetic diversity [85].
Concluding remarks
Plant evolutionary biologists owe a great deal to model organisms like Arabidopsis thaliana. Molecular and developmental studies of Arabidopsis have greatly advanced our understanding of the evolutionary genetics of critical phenotypic traits under controlled laboratory conditions, and increasingly under field conditions. However, simplified laboratory environments often do not reflect the complex suite of abiotic and biotic conditions that organisms experience in nature. This discrepancy limits the generalities that can be drawn from studies done entirely in controlled settings [20,60,61]. For example, QTLs identified in field studies are not always detected under laboratory conditions (and vice versa) [20,60,61]. Additionally, heritability values calculated in laboratory studies overestimate heritabilities measured in the field and may, therefore, alter our perspective on the capacity of species to respond to selection [11]. Emerging and ecological model systems will provide insight into the evolutionary forces that shape phenotypes in undisturbed natural populations, and will illuminate the evolutionary genetics of species that differ in life histories (e.g., perennial vs. annual), mating systems (e.g., selfing vs. outcrossing), growth strategies (e.g., herbaceous vs. tree), and other important characteristics. Future efforts need to complement field and laboratory studies to dissect the genetic basis of ecologically-relevant phenotypes, and understand the processes that promote and constrain adaptive evolution.
Box 2, Figure I.
(A) A set of 6 hypothetical populations subdivided into northern and southern groups, which have experienced divergent natural selection and genetic drift. (B) Divergent selection favors different phenotypic optima in the North and South. (C) Uncorrected genome wide association study reveals several genomic regions significantly associated with phenotype (the dotted red line indicates significance threshold). Regions with high association scores (metric of the relationship between genotype and phenotype) result from natural selection causing local adaptation (red), as well as historical population structure (green) and random false positives (blue). (D) Statistical controls for population structure eliminate false positives (unselected loci) and cause false negatives (adaptive loci). False negatives could potentially be detected via family-based QTL mapping of North by South controlled cross.
Acknowledgements
T.M.-O. and J.A. were supported by grants from the NIH (R01 GM086496 to T.M.-O.) and NSF (EF-0723447 to T.M.-O.). J.W. was supported by grants from the NIH (GM073990) and NSF (IOS-1024966 and EF-0723814). We thank R. Colautti, C. Olson-Manning and T. Pendergast for valuable discussions, numerous colleagues for sharing information on ecological model systems for Table 1, and editor Joël Savard and two anonymous reviewers for critiques of a previous draft.
Glossary terms
- Antagonistic pleiotropy
Genetic tradeoffs that occur at the level of a single locus. Antagonistic pleiotropy can maintain genetic variation across the landscape, if alleles at a locus underlying a critical fitness component show clear home-site advantage (elevated fitness in home environment, depressed fitness in contrasting environment)
- Conditional neutrality
At a single locus, or QTL, an allele may show a fitness advantage in its home environment, but no fitness cost in the contrasting environment. Conditional neutrality can contribute to local adaptation at the organismal level when multiple loci interact to determine fitness, and alleles from different environments are conditionally favored at different loci
- Forward genetic approaches
These approaches investigate the genetic basis of phenotypes and contrast with reverse genetic approaches (e.g., gene silencing), which identify the phenotypic effects of known genes
- Iteroparous vs. semelparous, and annual vs. perennial
Iteroparous species mate during multiple reproductive cycles over their lives, whereas semelparous species experience only one mating season prior to senescence. Annual species generally complete their entire life cycles within one year, and are therefore semelparous. Perennial species require multiple years to complete their life cycles. However, not all perennial species are iteroparous, rather some very long-lived species mate only once after many years of vegetative growth, and are therefore semelparous
- Near-isogenic Lines (NILs)
Inbred lines that segregate only at a QTL of interest, against an identical genomic background, can be used to fine-map ecologically-relevant QTL. Near-isogenic lines (NILs) can be produced by crossing two inbred lines to create heterozygous F1 progeny creating F2 lines through self-pollination and single seed descent, then backcrossing the progeny to the parental lines, and selecting appropriate lines for further study based on trait expression
- Recombinant Inbred Lines (RILs)
For species tolerant of inbreeding, two inbred parental lines can be crossed to generate heterozygous F1 hybrids. Through 6–8 generations of self-pollination and single seed descent, recombinant inbred lines (RILs) can be produced. RILs are primarily homozygous within a line (expected homozygosity of F6 RILs: 96.9%), which allows researchers to expose genetically nearly identical individuals to multiple environments to study the genetic basis of local adaptation and phenotypic plasticity. Additionally, recombination of the parental genomes increases genetic variation and facilitates QTL detection
- Selective sweep
The rapid rise to fixation (allele frequency = 1) of a beneficial allele reduces genetic variation at linked loci.
- Vernalization
Exposure of juvenile plants (not seeds) to non-freezing winter temperatures can promote flowering in some temperate species. Vernalization specifically refers to the length of winter. Vernalization is often an absolute requirement for flowering, but species can vary in the duration of vernalization they need to experience
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Reviews Genetics. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sattath S, et al. Pervasive Adaptive Protein Evolution Apparent in Diversity Patterns around Amino Acid Substitutions in Drosophila simulans. Public Library of Science Genetics. 2011;7:e1001302. doi: 10.1371/journal.pgen.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakker E, et al. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner TL, et al. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nature Genetics. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- 5.Excoffier L, et al. Detecting loci under selection in a hierarchically structured population. Heredity. 2009;103:285–298. doi: 10.1038/hdy.2009.74. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, et al. Population differentiation as a test for selective sweeps. Genome Research. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coop G, et al. Using Environmental Correlations to Identify Loci Underlying Local Adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke MK, et al. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature. 2010;467:587–590. doi: 10.1038/nature09352. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez RD, et al. Classic Selective Sweeps Were Rare in Recent Human Evolution. Science. 2011;331:920–924. doi: 10.1126/science.1198878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pritchard J, Di Rienzo A. Not by sweeps alone. Nature Reviews Genetics. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geber M, Griffen LR. Inheritance and natural selection on functional traits. International Journal of Plant Science. 2003;164:S21–S42. [Google Scholar]
- 12.Metcalf CJE, Mitchell-Olds T. Life history in a model system: opening the black box with Arabidopsis thaliana. Ecology Letters. 2009;12:593–600. doi: 10.1111/j.1461-0248.2009.01320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell-Olds T, et al. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nature Reviews Genetics. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- 14.Parachnowitsch AL, Caruso CM. Predispersal seed herbivores, not pollinators, exert selection on floral traits via female fitness. Ecology. 2008;89:1802–1810. doi: 10.1890/07-0555.1. [DOI] [PubMed] [Google Scholar]
- 15.Knight CA, et al. Expression profiling and local adaptation of Boechera holboellii populations for water use efficiency across a naturally occurring water stress gradient. Molecular Ecology. 2006;15:1229–1237. doi: 10.1111/j.1365-294X.2006.02818.x. [DOI] [PubMed] [Google Scholar]
- 16.Dechaine JM, et al. Constraints on the evolution of adaptive plasticity: Costs of plasticity to density are expressed in segregating progenies. New Phytologist. 2007;176:874–882. doi: 10.1111/j.1469-8137.2007.02210.x. [DOI] [PubMed] [Google Scholar]
- 17.Donohue K, et al. Evidence of adaptive divergence in plasticity: density- and site-dependent selection on shade-avoidance responses in Impatiens capensis. Evolution. 2000;54:1956–1968. doi: 10.1111/j.0014-3820.2000.tb01240.x. [DOI] [PubMed] [Google Scholar]
- 18.Neale DB, Ingvarsson PK. Population, quantitative and comparative genomics of adaptation in forest trees. Current Opinion in Plant Biology. 2008;11:149–155. doi: 10.1016/j.pbi.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Schranz ME, et al. Ecological genomics of Boechera stricta: identification of a QTL controlling the allocation of methionine- vs branched-chain amino acid-derived glucosinolates and levels of insect herbivory. Heredity. 2009;102:465–474. doi: 10.1038/hdy.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson JT, et al. Life-history QTLs and natural selection on flowering time in Boechera stricta, a perennial relative of Arabidopsis. Evolution. 2011;65:771–787. doi: 10.1111/j.1558-5646.2010.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson JT, Mitchell-Olds T. Ecological genetics and genomics of plant defenses: evidence and approaches. Functional Ecology. 2011;25:312–324. doi: 10.1111/j.1365-2435.2010.01785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrera CM, Bazaga P. Quantifying the genetic component of phenotypic variation in unpedigreed wild plants: tailoring genomic scan for within-population use. Molecular Ecology. 2009;18:2602–2614. doi: 10.1111/j.1365-294X.2009.04229.x. [DOI] [PubMed] [Google Scholar]
- 23.Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- 24.Manel S, et al. Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Molecular Ecology. 2010;19:3760–3772. doi: 10.1111/j.1365-294X.2010.04717.x. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell-Olds T. Complex-trait analysis in plants. Genome Biology. 2010;11:113. doi: 10.1186/gb-2010-11-4-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atwell S, et al. Genome-wide association study of 107 phenotypes in a common set of Arabidopsis thaliana inbred lines. Nature. 2010 doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingvarsson PK, Street NR. Association genetics of complex traits in plants. New Phytologist. 2011;189:909–922. doi: 10.1111/j.1469-8137.2010.03593.x. [DOI] [PubMed] [Google Scholar]
- 28.Myles S, et al. Association Mapping: Critical considerations shift from genotyping to experimental design. The Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohenlohe PA, et al. Using population genomics to detect selection in natural populations: Key concepts and methodological considerations. International Journal of Plant Science. 2010;171:1059–1071. doi: 10.1086/656306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritchard J, et al. The genetics of human adaptation: Hard sweeps, Soft sweeps, and polygenic adaptation. Current Biology. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leimu R, Fischer M. A meta-analysis of local adaptation in plants. Public Library of Science One. 2008;3:e4010. doi: 10.1371/journal.pone.0004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bomblies K. Doomed lovers: Mechanisms of isolation and incompatibility in plants. Annual Review of Plant Biology. 2010;61:109–124. doi: 10.1146/annurev-arplant-042809-112146. [DOI] [PubMed] [Google Scholar]
- 33.Hall MC, et al. Is local adaptation in Mimulus guttatus caused by trade-offs at individual loci? Molecular Ecology. 2010;19:2739–2753. doi: 10.1111/j.1365-294X.2010.04680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verhoeven KJF, et al. Habitat-specific natural selection at a flowering-time QTL is a main driver of local adaptation in two wild barley populations. Molecular Ecology. 2008;17:3416–3424. doi: 10.1111/j.1365-294X.2008.03847.x. [DOI] [PubMed] [Google Scholar]
- 35.Verhoeven KJF, et al. The genetic basis of adaptive population differentiation: a quantitative trait locus analysis of fitness traits in two wild barley populations from contrasting habitats. Evolution. 2004;58:270–283. [PubMed] [Google Scholar]
- 36.Gardner KM, Latta RG. Identifying loci under selection across contrasting environments in Avena barbata using quantitative trait locus mapping. Molecular Ecology. 2006;15:1321–1333. doi: 10.1111/j.1365-294X.2005.02835.x. [DOI] [PubMed] [Google Scholar]
- 37.Lowry DB, et al. Genetic and physiological basis of adaptive salt tolerance divergence between coastal and inland Mimulus guttatus. New Phytologist. 2009;183:776–788. doi: 10.1111/j.1469-8137.2009.02901.x. [DOI] [PubMed] [Google Scholar]
- 38.Li ZK, et al. QTL X environment interactions in rice. I. Heading date and plant height. Theoretical and Applied Genetics. 2003;108:141–153. doi: 10.1007/s00122-003-1401-2. [DOI] [PubMed] [Google Scholar]
- 39.Weinig C, et al. Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics. 2003;165:321–329. doi: 10.1093/genetics/165.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowry DB, Willis JH. A Widespread Chromosomal Inversion Polymorphism Contributes to a Major Life-History Transition, Local Adaptation, and Reproductive Isolation. Public Library of Science Biology. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elzinga JA, et al. Time after time: flowering phenology and biotic interactions. Trends in Ecology & Evolution. 2007;22:432–439. doi: 10.1016/j.tree.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Inouye DW. Effects of climate change on phenology, frost damage, and floral abundance of montane wildflowers. Ecology. 2008;89:353–362. doi: 10.1890/06-2128.1. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, et al. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21199–21204. doi: 10.1073/pnas.1007431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall MC, Willis JH. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution. 2006;60:2466–2477. [PubMed] [Google Scholar]
- 45.Sandring S, Ågren J. Pollinator-mediated selection on floral display and flowering time in the perennial herb Arabidopsis lyrata. Evolution. 2009;63:1292–1300. doi: 10.1111/j.1558-5646.2009.00624.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang R, et al. PEP1 regulates perennial flowering in Arabis alpina. Nature. 2009;459:423–427. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- 47.Kim DH, et al. Vernalization: Winter and the Timing of Flowering in Plants. Annual Review of Cell and Developmental Biology. 2009;25:277–299. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- 48.Franks SJ, et al. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehrenreich IM, et al. Candidate Gene Association Mapping of Arabidopsis Flowering Time. Genetics. 2009;183:325–335. doi: 10.1534/genetics.109.105189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. Trends in Genetics. 2010;26:519–527. doi: 10.1016/j.tig.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Turck F, et al. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 52.Jung C, Muller AE. Flowering time control and applications in plant breeding. Trends in Plant Science. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz C, et al. Cis-regulatory Changes at FLOWERING LOCUS T Mediate Natural Variation in Flowering Responses of Arabidopsis thaliana. Genetics. 2009;183:723–732. doi: 10.1534/genetics.109.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skøt L, et al. Allelic variation in the perennial ryegrass Flowering Locus T gene is associated with changes in flowering time across a range of populations. Plant Physiology. 2011;155:1013–1022. doi: 10.1104/pp.110.169870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayama R, et al. A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. The Plant Cell. 2007;19:2988–3000. doi: 10.1105/tpc.107.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin M-K, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. The Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shalit A, et al. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8392–8397. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blackman BK, et al. The role of recently derived FT paralogs in sunflower domestication. Current Biology. 2010;20:1–7. doi: 10.1016/j.cub.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinig C, et al. Novel loci control variation in reproductive timing in Arabidopsis thaliana in natural environments. Genetics. 2002;162:1875–1884. doi: 10.1093/genetics/162.4.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brachi B, et al. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. Public Library of Science Genetics. 2010;6:e1000940. doi: 10.1371/journal.pgen.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu CA, et al. Natural variation for drought-response traits in the Mimulus guttatus species complex. Oecologia. 2010;162:23–33. doi: 10.1007/s00442-009-1448-0. [DOI] [PubMed] [Google Scholar]
- 63.Hausmann N, et al. Quantitative trail loci affecting delta-13C and response to differential water availability in Arabidopsis thaliana. Evolution. 2005;59:81–96. [PubMed] [Google Scholar]
- 64.Bernier J, et al. The large-effect drought-resistance QTL qtl12.1 increases water uptake in upland rice. Field Crops Research. 2009;110:139–146. [Google Scholar]
- 65.Reynolds M, Tuberosa R. Translational research impacting on crop productivity in drought-prone environments. Current Opinion in Plant Biology. 2008;11:171–179. doi: 10.1016/j.pbi.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 66.Tardieu F, Tuberosa R. Dissection and modelling of abiotic stress tolerance in plants. Current Opinion in Plant Biology. 2010;13:206–212. doi: 10.1016/j.pbi.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Maherali H, et al. Adaptive Value and Costs of Physiological Plasticity to Soil Moisture Limitation in Recombinant Inbred Lines of Avena barbata. American Naturalist. 2010;175:211–224. doi: 10.1086/649598. [DOI] [PubMed] [Google Scholar]
- 68.Sherrard M, et al. Water stress alters the genetic architecture of functional traits associated with drought adaptation in Avena barbata. Evolution. 2009;63:702–715. doi: 10.1111/j.1558-5646.2008.00580.x. [DOI] [PubMed] [Google Scholar]
- 69.Juenger T, et al. Exploring genetic and expression differences between physiologically extreme ecotypes: comparative genomic hybridization and gene expression studies of Kas-1 and Tsu-1 accessions of Arabidopsis thaliana. Plant, Cell and Environment. 2010;33:1268–1284. doi: 10.1111/j.1365-3040.2010.02146.x. [DOI] [PubMed] [Google Scholar]
- 70.Oh S-J, et al. Overexpression of the Transcription Factor AP37 in Rice Improves Grain Yield under Drought Conditions. Plant Physiology. 2009;150:1368–1379. doi: 10.1104/pp.109.137554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ackerly DD, et al. The geography of climate change: implications for conservation biogeography. Diversity and Distributions. 2010;16:476–487. [Google Scholar]
- 72.Colautti RI, Barrett SCH. Natural selection and genetic constraints on flowering phenology in an invasive plant. International Journal of Plant Science. 2010;171:960–971. [Google Scholar]
- 73.Ouborg NJ, et al. The rough edges of the conservation genetics paradigm for plants. Journal of Ecology. 2006;94:1233–1248. [Google Scholar]
- 74.Willi Y, et al. Limits to the adaptive potential of small populations. Annual Review of Ecology Evolution and Systematics. 2006;37:433–458. [Google Scholar]
- 75.Hoffmeister TS, et al. Ecological and evolutionary consequences of biological invasion and habitat fragmentation. Ecosystems. 2005;8:657–667. [Google Scholar]
- 76.Parker IM, et al. An evolutionary approach to understanding the biology of invasions: local adaptation and general purpose genotype in the weed Verbascum thapsus. Conservation Biology. 2003;17:59–72. [Google Scholar]
- 77.Lankau RA, et al. Evolutionary limits ameliorate the negative impact of an invasive plant. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15362–15367. doi: 10.1073/pnas.0905446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keller SR, et al. Adaptation and colonization history affect the evolution of clines in two introduced species. New Phytologist. 2009;183:678–690. doi: 10.1111/j.1469-8137.2009.02892.x. [DOI] [PubMed] [Google Scholar]
- 79.Jump AS, Peñuelas J. Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8096–8100. doi: 10.1073/pnas.0510127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoffmann AA, Willi Y. Detecting genetic responses to environmental change. Nature Reviews Genetics. 2008;9:421–432. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- 81.Parmesan C. Ecological and evolutionary responses to recent climate change. Annual Review of Ecology Evolution and Systematics. 2006;37:637–669. [Google Scholar]
- 82.Lenoir J, et al. A significant upward shift in plant species optimum evolution during the 20th Century. Science. 2008;320:1768–1771. doi: 10.1126/science.1156831. [DOI] [PubMed] [Google Scholar]
- 83.Massot M, et al. Climate warming, dispersal inhibition and extinction risk. Global Change Biology. 2008;14:461–469. [Google Scholar]
- 84.Fox CW, Reed DH. Inbreeding depression increases with environmental stress: An experimental study and meta-analysis. Evolution. 2010;65:246–258. doi: 10.1111/j.1558-5646.2010.01108.x. [DOI] [PubMed] [Google Scholar]
- 85.Damschen EI, et al. Corridors increase plant species richness at large scales. Science. 2006;313:1284–1286. doi: 10.1126/science.1130098. [DOI] [PubMed] [Google Scholar]
- 86.Rausher MD. Evolutionary transitions in floral color. International Journal of Plant Science. 2008;169:7–21. [Google Scholar]
- 87.Bradshaw HD, Schemske DW. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426:176–178. doi: 10.1038/nature02106. [DOI] [PubMed] [Google Scholar]
- 88.Streisfeld MA, Rausher MD. Population genetics, pleiotropy, and the preferential fixation of mutations during adaptive evolution. Evolution. 2011 doi: 10.1111/j.1558-5646.2010.01165.x. [DOI] [PubMed] [Google Scholar]
- 89.Cooley A, et al. Gene duplication in Mimulus underlies parallel floral evolution via independent trans-regulatory changes. Current Biology. 2011 doi: 10.1016/j.cub.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 90.Hopkins R, Rausher MD. Identification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature. 2011;469:411–414. doi: 10.1038/nature09641. [DOI] [PubMed] [Google Scholar]
- 91.Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nature Reviews Genetics. 2010;11:867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- 92.Hamblin MT, et al. Population genetics of genomics-based crop improvement methods. Trends in Genetics. 2011;27:98–106. doi: 10.1016/j.tig.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 93.Anderson JT, et al. Phenotypic plasticity despite source-sink population dynamics in a long-lived perennial plant. New Phytologist. 2010;188:856–867. doi: 10.1111/j.1469-8137.2010.03404.x. [DOI] [PubMed] [Google Scholar]
- 94.Baythavong BS, Stanton ML. Characterizing selection on phenotypic plasticity in response to natural environmental heterogeneity. Evolution. 2010;64:2904–2920. doi: 10.1111/j.1558-5646.2010.01057.x. [DOI] [PubMed] [Google Scholar]
- 95.Lacaze X, et al. Genetics of phenotypic plasticity: QTL analysis in barley, Hordeum vulgare. Heredity. 2009;102:163–173. doi: 10.1038/hdy.2008.76. [DOI] [PubMed] [Google Scholar]
- 96.Steinger T, et al. Evolution in stressful environments II: Adaptive value and costs of plasticity in response to low light in Sinapis arvensis. Journal of Evolutionary Biology. 2003;16:313–323. doi: 10.1046/j.1420-9101.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- 97.Richards CL, et al. What role does heritable epigenetic variation play in phenotypic evolution? Bioscience. 2010;60:232–237. [Google Scholar]
- 98.Feng S, et al. Epigenetic Reprogramming in Plant and Animal Development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oyama RK, et al. The shrunken genome of Arabidopsis thaliana. Plant Systematics and Evolution. 2008;273:257–271. [Google Scholar]
- 100.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics. doi: 10.1038/ng.807. (forthcoming) [DOI] [PMC free article] [PubMed] [Google Scholar]