Abstract
Several studies have shown that promoters of protein-coding genes are origins of pervasive non-coding RNA transcription and can initiate transcription in both directions. However, only recently have researchers begun to elucidate the functional implications of this bidirectionality and noncoding RNA production. Increasing evidence indicates that non-coding transcription at promoters influences the expression of protein-coding genes, revealing a new layer of transcriptional regulation. This regulation acts at multiple levels, from modifying local chromatin to enabling regional signal spreading and more distal regulation. Moreover, the bidirectional activity of a promoter is regulated at multiple points during transcription, giving rise to diverse types of transcripts.
Bidirectionality is an inherent feature of promoters
In the past decade, genomic research has focused on protein-coding genes. However, recent studies have revealed a myriad of non-coding transcripts in different organisms [1, 2]. While protein-coding transcription represents the output of only less than 2% of the human genome, more than 70% of the genome is transcribed [3]. The unexpected level of transcriptome complexity has led to the suggestion that non-coding RNAs (ncRNAs) comprise a previously hidden layer of genomic programming and that ncRNA-directed regulatory circuits underpin complex genetic phenomena in eukaryotes [4]. A large portion of reported ncRNA transcription occurs in the proximity of protein-coding genes, particularly at promoters but also at ends of coding regions and intragenically. In this review, we focus on the generation and functional consequences of non-coding transcription initiating from bidirectional promoters.
Divergent, bidirectional transcription of protein-coding genes has been recognized for many years [5], and the more recent sequencing and annotation of whole genomes has revealed that bidirectional organization of coding genes is common [6, 7]. Further technological advancements over the last few years, however, have revealed an abundance of previously unobserved non-coding transcripts near the promoters of protein-coding genes in different organisms, including bacteria [8–11], yeast [12–15], fruit fly [16], mouse [17, 18], human [19] and plants [20].
To understand the diversity of ncRNAs found near promoters, one must consider the technologies used to detect them as they bias our accounts of ncRNAs. The study of transcripts at promoters in turn provides information about the frequency of bidirectional transcription, the control of directionality in bidirectional promoters, and the importance of transcriptional regulation mediated by promoter-associated transcription. Here we comment on the main features of promoter-associated transcripts and technologies for detecting them (detailed further in Table 1 and Box 1, respectively).
Table 1.
Examples of non-coding transcripts associated with protein-coding genes detected in different studies (See Figure 1). Note that different technologies measure different aspects of transcription; thus, the transcripts listed here may include some redundancy.
| Abbreviation | Full name | Transcript length (nt) | Transcript stability | Organism | Technology | Measured molecule | Reference |
|---|---|---|---|---|---|---|---|
| GRO-seq read | Global run-on sequencing read | - | - | Human | GRO-seq | RNA polymerase II activity | [25] |
| NET-seq read | Native elongating transcript sequencing read | - | - | Yeast | NET-seq | Nascent RNA | [26] |
| PASR | Promoter associated short RNA | 22–200 | Stable | Human | Tiling array | Short RNA | [21, 22] |
| TASR | Termination associated short RNA | 22–200 | Stable | Human | Tiling array | Short RNA | [21] |
| PALRs | Promoter associated long RNA | hundreds – >1000 | Stable | Human | Tiling array | RNA | [21] |
| sRNA | Short RNA | <200 | Stable | Human | Tiling array | Short RNA | [21] |
| lRNA | Long RNA | >200 | Stable | Human | Tiling array | RNA | [21] |
| Short RNA (sRNA) | Short RNA | <200 | Stable | Human | RNA sequencing | Short RNA | [22] |
| 50–200 | Stable | Human | Tiling array | Short RNA | [17] | ||
| tiRNA | Transcription initiation RNA | ~18 | Stable | Human, chicken, Drosophila | RNA sequencing | Short RNA | [23] |
| 12–27 | Stable | Human, chicken | RNA sequencing | Short RNA | [23] | ||
| TSSa-RNA | Transcription start site associated RNA | 20–90 | Stable | Mouse | RNA sequencing | Short RNA | [18] |
| SUT | Stable unannotated transcript | ~760 | Stable | Yeast | Tiling array | RNA | [13] |
| PROMPT | Promoter upstream transcript | - | Unstable | Human | RNA sequencing | RNA | [24] |
| CUT | Cryptic unstable transcript | ~200–600 | Unstable | Yeast | Tiling array | RNA | [13, 14] |
Box 1. Different technologies used to study bidirectional transcription.
Enabled by the development of new genomic technologies, an abundance of new transcripts has been described (reviewed in [1] and [2]). These technologies interrogate gene expression at multiple steps, from the initiation of transcription to the final transcripts produced.
Detection of transcriptional activity. Nuclear run-ons measure the density of transcribing RNA polymerases over a genomic region. This is done by allowing transcriptionally engaged polymerases to resume elongation in the presence of labeled nucleotides while preventing new transcription initiation events from occurring. The labeled RNAs produced during this run-on reaction are thus indicative of the elongation activity of the polymerases. The genome-wide implementation of nuclear run-on, the global run-on (GRO-seq), yields a snapshot of the occupancy of active, engaged polymerases in a strand-specific manner independent of transcript length or stability [25].
Detection of nascent RNAs. Immunoprecipitation of RNAs bound to RNA polymerase II followed by sequencing (NET-seq) allows strand-specific measurements of engaged RNA polymerase II [26]. This provides an RNA polymerase II occupancy profile, independent of the polymerase’s ability to elongate. Thus, nascent transcripts attached to paused or backtracked polymerases are also measured.
Whole transcriptome profiling. Analyzing total RNA, polyA-enriched RNAs or rRNA-depleted RNA by strand-specific tiling arrays or RNA-seq is the most widely used method to study transcriptomes [3, 13, 15].
Detection of short RNAs. Purification of short (18 to 200 nt) RNAs enables their profiling independently of full-length RNAs. This subpopulation of transcripts has varying origins, from short products of transcription to fragments produced post-transcriptionally from longer molecules. This approach has been carried out using both tiling arrays [21] and sequencing [18, 22, 23].
TSS sequencing. Sequencing of the 5’ terminal part of capped mRNA transcripts has been used to detect active promoters [29, 30]. The sequencing of capped transcripts permits genome-wide mapping of TSSs and thus identification of bidirectional promoters. This can be combined with the identification of transcription termination sites [97–99] to define both transcript boundaries [100].
Profiling of RNA degradation mutants. Knocking down components of the RNA degradation machinery has been applied to enrich samples for short-lived RNA molecules [13, 14, 24]. These RNAs could, in some cases, be precursors of the short RNA molecules detected with other techniques.
Different technologies demonstrate the bidirectionality of promoters
The extent of ncRNA transcription has been investigated using different approaches, including measurements of steady-state RNA levels [13, 14, 17, 18, 21–24], RNA polymerase activity [25], and nascent RNAs associated with RNA polymerase II [26]. These studies have resulted in a catalog of non-coding transcripts near protein-coding genes and have revealed both protein/non-coding and non-coding/non-coding bidirectional transcription (Figure 1).
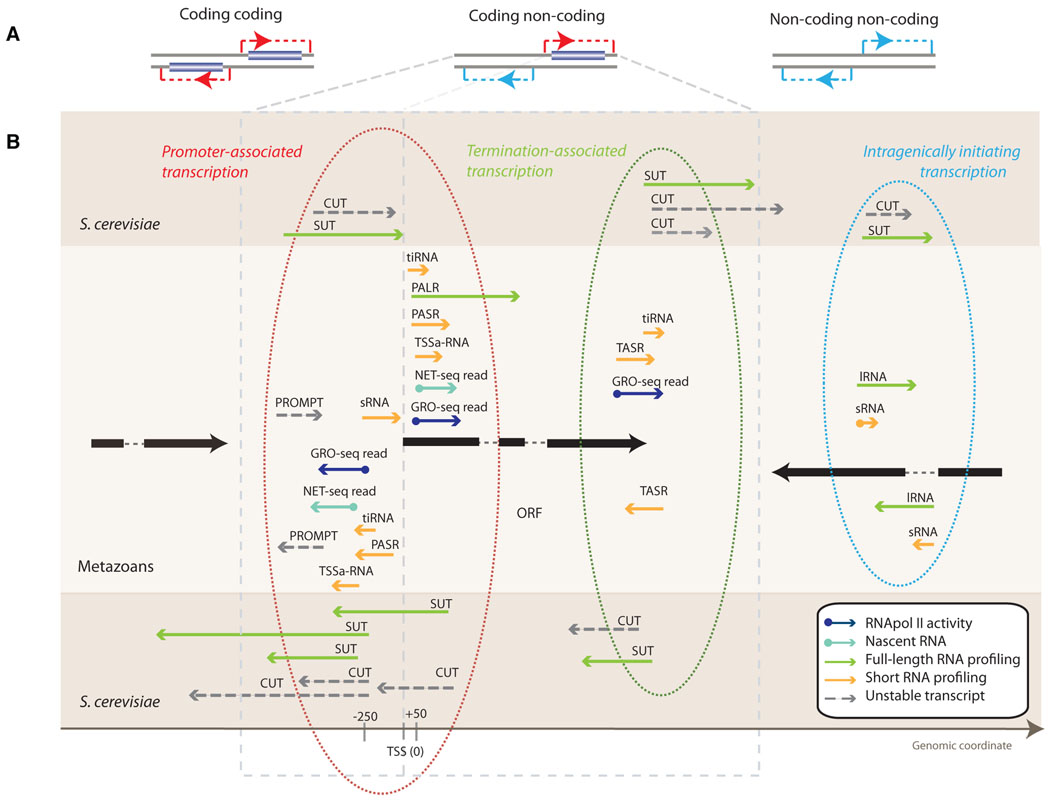
Figure 1. Examples of pervasive bidirectional transcription.
A) Transcription at a bidirectional promoter involves pairs of protein-coding transcripts, pairs of non-coding transcripts, or coding and non-coding transcripts. The dark blue boxes represent protein-coding genes. B) Schematic examples of non-coding RNAs in proximity to protein-coding genes reported in diverse studies and organisms. The black bars represent proteincoding transcripts and the arrowheads the direction of transcription. The narrow arrows represent long stable transcripts (green), short stable transcripts (yellow), unstable transcripts (dashed gray), nascent RNAs (turquoise) and actively transcribed RNAs (blue). As indicated by arrow directions, non-coding RNAs can be generated from either strand relative to the coding transcript. Different types of ncRNA transcription are circled and shown on different protein-coding transcripts. The background shading indicates species where transcripts were defined. See Table 1 for details. Transcripts are represented as described in different studies; note that in reality, some may be the same non-coding transcript but profiled with a different technique (see text for discussion). Note also that the lengths of noncoding RNAs are often not precisely known and are not represented here in accurate proportion to protein-coding gene lengths.
A large body of evidence for promoter bidirectionality derives from studies that have used expression profiling of steady-state RNA levels, targeting either all transcripts or specific subpopulations of different lengths and stabilities. A widely used approach in higher eukaryotes is to specifically profile short RNAs that are otherwise difficult to distinguish from overlapping long transcripts. This has revealed short transcripts of varying sizes located on both strands near promoters [17, 18, 21–23] (Figure 1, Table 1). Transcripts also differ in their stability. As transcript abundance in a cell is dependent on the rates of both synthesis and decay [27, 28], some transcripts are not detected by RNA profiling of wild-type cells due to their short lifetimes. Manipulation of the RNA degradation pathways has been used to uncover these so-called cryptic unstable transcripts (CUTs) that usually initiate from shared promoters [13, 14, 24]. In addition to profiling transcripts with specific size and stability, bidirectional transcription has been studied using protocols that capture 5’ termini of transcripts and reveal transcription start sites (TSSs) [29, 30].
Evidence for pervasive bidirectional initiation from promoters also derives from studies that directly measure transcription. These studies have been performed either by detecting all nascent transcripts (that is, those attached to RNA polymerase) or by targeting only transcripts that are being elongated. Global run-on sequencing (GRO-seq), a method that measures actively elongating RNAs (Box 1), has revealed RNA polymerase-mediated transcription in two directions at 55% of human promoters, with only a small bias in orientation [25]. Promoter bidirectionality has also been shown in yeast by sequencing nascent transcripts that co-purify with RNA polymerase II (NET-seq, Box 1) [26].
A systematic application of these diverse technologies will provide the most comprehensive information necessary to characterize non-coding transcription at promoters in terms of transcript type and number. Clearly, some of the non-coding transcripts detected reflect distinct biological entities (e.g. transcripts with characteristic biogenesis and degradation mechanisms (CUTs [13, 14], PROMPTs [24]). Other transcripts may not represent independent transcription events but rather post-transcriptional processing products of longer ncRNAs generated from the same genomic locus [22]. Furthermore, some of the observed diversity is likely due to differences in detection techniques. Therefore, the capacity of each technology should be taken into account when comparing results. For example, NET-seq cannot identify nascent transcripts close to the TSS as they are too short to map uniquely to the genome, while in GRO-seq these transcripts are elongated during the run-on reaction, allowing them to be mapped. In another example, when profiling steady state transcript abundance for specific sizes, one should note that the different transcript lengths detected could be arbitrary subpopulations from a continuum of sizes. Nevertheless, the diversity of the applied approaches shows that transcriptional initiation at most, if not all, promoters occurs in a bidirectional manner, even if the final transcription products are mainly observed in one orientation.
By studying different steps of the transcription process, and measuring specific subpopulations of transcripts, one can obtain complementary insights into the activity and regulation of bidirectional transcription. Thus, recent genomic developments substantiate the pervasiveness and illustrate the complexity of bidirectional transcription. These observations coupled with the anticipated functional implications of ncRNAs on genome regulation display the need for further investigation of transcriptional initiation at promoters.
Architecture of bidirectional promoters
A promoter can be defined as a region of DNA that directs the transcription of a downstream unit [31, 32]. To understand how bidirectional transcription works, it is important to study the role that chromatin structure plays in the assembly of the transcription machinery and how chromatin is dynamically regulated [33–36].
Nucleosome-depleted regions are hallmarks of promoters
Active promoters generally contain an 80 to 300 bp nucleosome-depleted region (NDR) [34, 37, 38] flanked by two well-positioned nucleosomes (Figure 2). These nucleosomes often contain the histone variant H2A.Z [39–41]. The association between NDRs and TSSs suggests that assembly of the transcription machinery occurs in NDRs [37, 38]; and further studies have shown that transcripts often originate bidirectionally from these regions [13, 14]. The regular positioning of nucleosomes with respect to promoters is conserved from yeast to humans [34], excluding minor differences. In yeast, the location of the TSS is just inside the +1 nucleosome, whereas in metazoans this nucleosome is positioned further downstream, leaving the TSS accessible [42]. This could allow different ways of transcriptional regulation. In yeast, the presence of the +1 nucleosome at the TSS could play a role in transcription initiation [37], while in metazoans the +1 nucleosome may contribute to transcription elongation by helping to pause RNA polymerase II [34]. There are also gene-specific differences in the degree of nucleosome depletion at promoters [33, 36], which allow variations in gene regulation and transcriptional plasticity. NDRs are not only associated with the 5’ ends of genes, but are also found at their 3’ ends, where they may be involved in transcription termination [43], antisense transcription [13, 43], or gene looping, where start and termination sites interact during transcription [43, 44].
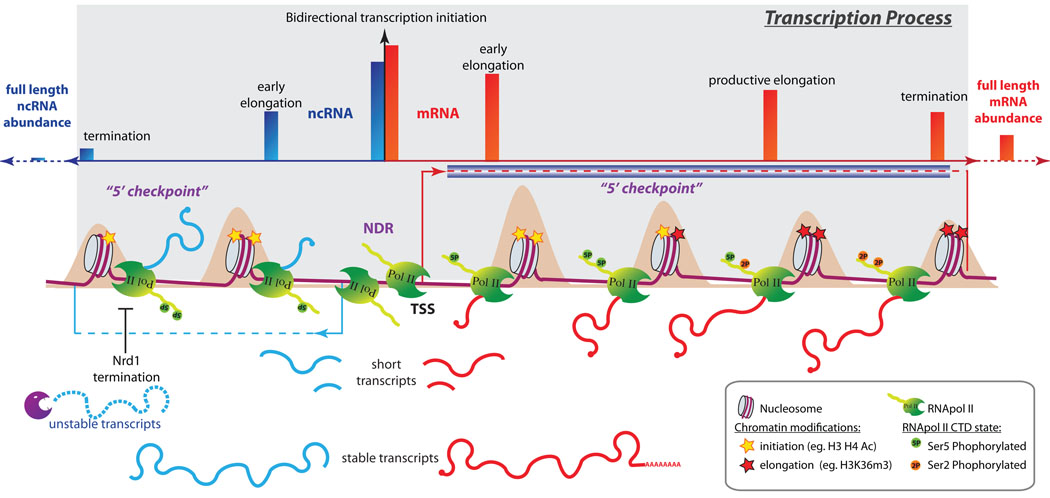
Figure 2. Model for bidirectional transcription.
Bidirectional transcription of an mRNA (on the right in red) and a ncRNA (on the left in blue) is depicted as an example. The dark blue box represents the protein-coding region. Transcription starts bidirectionally from the nucleosome-depleted region (NDR). Nucleosome density is represented by a beige pattern behind the genomic regions. Only a portion of the RNA polymerase II that assembles at the promoter will enter into early elongation. This entry is characterized by Ser5 phosphorylation of the polymerase CTD and chromatin modifications specific to transcription initiation and early elongation. Before transcribing further, the polymerase passes a “5’ checkpoint” where it pauses, terminates, or commits to productive elongation. If the polymerase does not proceed through this checkpoint, transcription will be terminated through the Nrd1 pathway, producing an unstable transcript. If the polymerase proceeds through the checkpoint, it will enter into productive elongation that is associated with characteristic chromatin modifications and CTD phosphorylation. Once RNA polymerase II has terminated transcription, final transcript abundance is determined by transcript stability. Bidirectional transcription is regulated at multiple steps following transcription initiation. The number of RNA polymerase II molecules that pass each of these steps, represented by red (sense) and blue (antisense) bars in the upper panel, decreases as they move further away from the promoter. All of these regulatory steps are differentially controlled for both orientations.
As the presence of an NDR is important for transcription initiation, the precise organization of chromatin is critical for dictating where transcription starts. Spurious transcription is suppressed via several mechanisms [45], of which one of the most intensively studied is histone modification [35, 46]. A well-known example is the suppression of intragenic cryptic transcription by histone deacetylation. This is catalyzed by the Rpd3S complex that recognizes H3K36me produced by Set2 during RNA polymerase II elongation [35, 47, 48, Churchman, 2011 #72]. Another mechanism involves limiting access of the transcription machinery by remodeling nucleosome distributions [49–51]. Chromatin remodeling plays an important role in promoter regulation by influencing the positioning of nucleosomes on the promoter [33, 36]. In these cases, regulation is affected by displacing the nucleosomes from 5’ and 3’ NDRs [52] or by modifying the size of the NDR [53]. Chromatin remodeling also acts to repress spurious transcription at intragenic locations where transcription should not occur [33, 36, 54].
In summary, a NDR is a site for transcription initiation where the transcriptional machinery assembles, potentially in both orientations. Further regulatory steps define how far the polymerase can continue when transcribing in a bidirectional manner.
Direction is regulated throughout the transcription process
If evidence of pervasive transcription at promoters indicates that promoters are generally capable of initiating transcription in two directions, why do we detect productive elongation mostly in one orientation? Once the transcription machinery has assembled onto DNA, it has the option to start transcription in both directions. Data collected by measuring transcriptional activity at the start of transcription show that RNA polymerase assembly and initiation occur in almost equal proportions in both orientations [25]. However, nascent sense transcripts are at least eight times more abundant than divergent transcripts at more than half of yeast promoters [26]. This shows that transcription in the divergent orientation decreases even during the transcription process as RNA polymerase II moves further away from the promoter, which is probably due to post-recruitment regulation. Indeed, a large proportion of initiation events do not produce a final stable transcript [55, 56], reflecting possible regulation after RNA polymerase II recruitment. Although at any given moment 60% of RNA polymerase II present in a cell has initiated transcription, less than 10% will produce a stable sense transcript [56]. Therefore, the relative amount of transcripts produced from a bidirectional promoter is controlled via several regulatory steps during the transcription cycle (initiation, elongation and termination).
The progression of RNA polymerase II along a transcription unit is accompanied by a series of chromatin modifications [35, 46] and RNA polymerase II C-terminal domain (CTD) phosphorylation [57–59]. RNA polymerase II is assembled onto the preinitation complex (PIC) with its CTD hypophosphorylated. During early elongation the CTD becomes rapidly phosphorylated on Ser5 (Figure 2). In yeast, this phosphorylation helps to recruit machinery such as mRNA-capping enzymes and H3K4 methyltransferase, as well as the early termination complex (Nrd1-Nab3) for promoting termination of short transcripts [60]. During the early elongation step, a 5' checkpoint has been hypothesized, at which the polymerase chooses between pause, termination, or commitment to productive elongation [57, 61] (Figure 2). This checkpoint could enable the regulation of promoter directionality [62] by committing the already initiated transcription to termination. One factor that could be involved in this checkpoint is the prolyl isomerase Ess1. Ess1 is recruited by the CTD phosphorylated on Ser5 and enhances the dephosphorylation of Ser5P by Ssu72. This leads to the dissociation of the polymerase from the Nrd1-Nab3 termination complex and hence productive elongation [63]. This checkpoint model is also in agreement with current data showing that promoters contain chromatin modifications indicative of bidirectional initiation, while marks of productive elongation are only seen in one orientation [18, 46, 64]. The regulation of transcription after RNA polymerase II recruitment is a well-characterized process, both in metazoans [64, 65] and in yeast [66, 67]. This regulation could be one of the sources of short RNAs detected on both sides of promoters [18, 22, 23], whose length may thus reflect the point at which non-productive transcription was terminated (Figure 2).
If RNA polymerase II passes the initial 5’ checkpoint and enters productive elongation, the level of Ser5P decreases, whereas the level of Ser2P increases. Ser2P is required to recruit machinery that cotranscriptionally modifies chromatin, polyadenylates, and terminates transcription [58] (Figure 2). Finally, the amount of divergent transcripts is also controlled at the post-transcriptional level by regulating transcript stability [13, 24]. Hence, the extent of bidirectional transcription from a promoter is regulated at several steps, allowing varying levels of divergent transcription. Transcription of two protein-coding genes from a bidirectional promoter demonstrates that production of full-length transcripts in both orientations is possible, even though the majority of promoters appear to display orientation preference. Mechanisms for the establishment and maintenance of this preference remain speculative (Box 2).
Box 2. Potential mechanisms for directional regulation of transcription.
Although promoters are generally capable of actively initiating transcription in two directions, in most cases productive elongation is seen primarily in one orientation. Thus, a mechanism must exist to regulate a putative 5’ checkpoint and thus dictate this asymmetry after bidirectional initiation. This could include:
Specific sequence signals present in the promoter or coding region. It has been proposed that the nucleotide composition around the promoter affects its bidirectionality [86].
Chromatin modifications. Previous rounds of transcription could mark the orientation favored in subsequent rounds. One example of such epigenetic memory is the co-transcriptional trimethylation of H3K36 that recruits the deacetylase Rpd3S [47]. This deacetylase has been shown to repress spurious transcription within the coding region [35, 48], but it has also been shown to decrease the bidirectionality of a downstream promoter [26]. Another example could be ncRNA transcription in the proximity of the promoter. This could influence the direction of transcription initiation by inducing chromatin remodeling favoring one orientation.
3D structure of transcription. Transcriptional memory could be maintained by using spatial mechanisms such as DNA looping linking the promoter with the favored 3’ end [101, 102].
Functional consequences of pervasive transcription at bidirectional promoters
Given that several transcripts are produced from the same promoter, the promoter must act as a regulatory unit to couple their transcription. The effect of this transcription on gene activity (local or distal) could be mediated by either the transcription process itself or by the produced transcripts. The consequences of ncRNA transcription from a bidirectional promoter thus depend on transcript length, sequence and stability. Although there are an increasing number of case studies demonstrating that ncRNAs generated from bidirectional promoters have functional roles (see below), it is not clear which proportion is functional. Here we classify instances of bidirectional transcription into those affecting the bidirectional promoter, neighboring protein-coding genes, or more distal genes. To illustrate their potential functions, we utilize examples, where informative, of other promoter-associated transcripts with characterized effects.
Bidirectional promoters couple two divergent protein-coding genes
Divergent organization of two protein-coding genes has been widely reported in different organisms [6, 68–71]. A study in humans indicated that the number of divergent gene pairs does not correlate with gene density [72], suggesting that maintaining their coupling by a shared promoter is beneficial. This is also supported by partial conservation of such pairs between species [6]. Divergent pairs are mainly co-expressed, but some display opposite regulation [6, 70]. The anti-correlated regulation could be due to competition for the same pool of polymerases and associated factors, or the recruitment of chromatin modifiers (see mechanism for coding/non-coding pairs in Figure 3A and B). Bidirectional promoters often couple protein-coding genes involved in the same process [6, 70, 72], allowing for coordinated temporal (e.g. [73]) and environmental (e.g. [74]) responses.
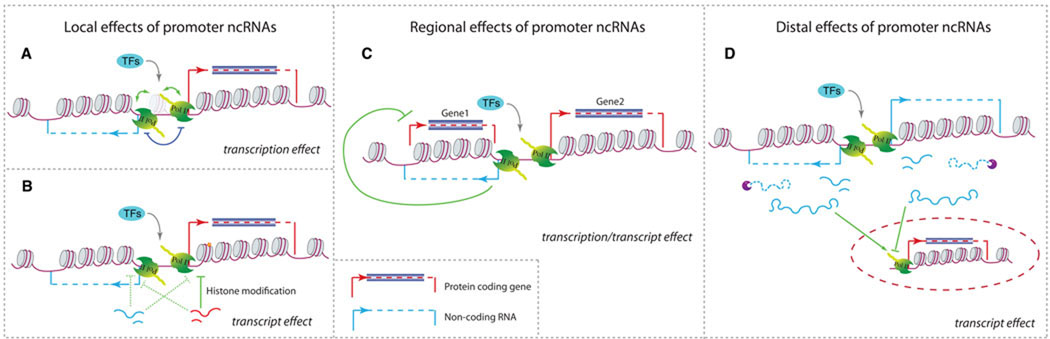
Figure 3. Functional consequences of bidirectional promoters.
Non-coding RNA transcription from bidirectional promoters couples the expression of protein-coding genes through local effects (A and B), regional signal spreading (C), or distal effects (D). A) Transcription of ncRNAs reorganizes (green arrows) the chromatin in bidirectional regions or competes (dark blue line) with the protein-coding gene for the same pool of polymerases and accessory factors. Transcription of ncRNAs can modify (left green arrow) the local chromatin structure, for example by displacing positioned nucleosomes (transparent one), therefore facilitating (right green arrow) the transcription of the protein-coding gene. B) ncRNA transcripts affect histone modifications of nearby chromatin. The solid green line shows the repression effect published in case studies [17, 77]; dashed green lines show possible effects suggested in this review. C) Transcription factors co-regulate the expression of a ncRNA (blue arrow) and Gene 2. The ncRNA then spreads the signal to the overlapping Gene 1 (green line) and represses its expression (red arrows) as demonstrated recently [78]. D) ncRNAs affect activation or repression (green arrows) of distal genes (gene in dashed oval). The bidirectional promoter can generate only ncRNAs or a protein-coding gene and a ncRNA (not shown). The direction of transcription is represented for non-coding transcription (blue arrow) and protein-coding transcription (red arrow). Protein-coding regions are represented by dark blue boxes.
ncRNA transcription influences local chromatin
Nucleosomes are a significant barrier for transcription [75]; hence, their exclusion permits local transcription. Transcription of a ncRNA from a shared promoter could activate protein-coding gene expression by displacing positioned nucleosomes, even when the RNAs are short [23] (Figure 3A). A study in Schizosaccharomyces pombe has shown that upstream transcription from the same strand of the fbp1+ locus induces local remodeling of chromatin and is a prerequisite for the activation of the downstream gene [76]. Although in this case, transcription arises from the same strand as that of the protein-coding gene, divergent transcription could modify local chromatin in a similar manner.
Pervasive transcription around a promoter region could also act to create a reservoir of RNA polymerase, which would facilitate rapid activation of the protein-coding gene when required [24]. In addition, transcription could promote upstream initiation through negative supercoiling of DNA [18], serve as a barrier to block the spread of repressive chromatin [62], or keep the promoter open to decrease stochastic variation of gene expression (minimizing its intrinsic transcriptional noise) [36].
Non-coding transcription can also be repressive, for instance by removing bound transcription factors from the protein-coding gene promoter or by competing for the same pool of general transcription factors (Figure 3 A). In the case of the TPI1 gene, the unstable ncRNA and TPI1 mRNA appear to originate from different pre-initiation complexes co-regulated by the same transcription activators. These transcripts would therefore compete for the same polymerases and accessory factors [14].
ncRNA transcripts recruit modifiers to reorganize local chromatin
Non-coding transcripts themselves can also act to influence protein-coding gene expression by modifying the surrounding chromatin (Figure 3B). In these cases, ncRNA transcripts cause either activation or repression of the bidirectionally-oriented gene. For example, same-strand short RNA transcription takes place at promoters of non-transcribing genes targeted by the Polycomb repressive complex-2 (PRC2), a complex that catalyzes histone trimethylation (H3K27me3) [17]. The ncRNAs recruit PRC2 to downstream genes, leading to a block in RNA polymerase II elongation [17]. It has also been shown that a ncRNA transcribed from the promoter of the human cyclin D1 (CCND1) gene represses its expression. This is accomplished by the recruitment of an RNA-binding protein that represses the activating effect of two histone acetyltransferases on CCND1 [77].
Divergent transcription spreads regulatory signals to neighboring genes
Divergent ncRNAs generated from a shared bidirectional promoter can extend to overlap an upstream gene, especially in compact genomes. For example, in yeast, 55% of stable ncRNAs initiating from a bidirectional promoter overlap an upstream ORF on the opposite strand, forming antisense transcripts [78] (Figure 3C). Antisense expression has been shown to repress the expression of sense protein-coding genes, for instance through transcriptional interference or histone modifications [79–83]. Possible regulatory mechanisms of overlapping antisense transcription have been extensively reviewed [2, 84, 85]. Here, we will focus on the consequences of antisense repression on protein-coding genes in the context of their initiation from bidirectional promoters.
One important consequence of bidirectional promoters is that the regulation of a single gene could spread to neighboring genes through the divergently transcribed ncRNA (Figure 3C). An example of a ncRNA that couples two tandem protein-coding genes is the transcript SUT719 in yeast, which initiates from a bidirectional promoter shared with GAL80 and extends to overlap the upstream SUR7 gene promoter [78]. SUT719 is co-regulated with GAL80 via a Gal4 binding site in the bidirectional promoter. SUT719 expression inhibits SUR7 expression. Thus, the activation signal for GAL80 directly represses SUR7 expression through the production of an antisense transcript from the bidirectional promoter. This coupling of expression through local spreading of regulatory signals by ncRNAs from bidirectional promoters could be a general feature of the transcriptome, especially in species with a compact genome. In yeast, the ncRNA-mediated coupling of two tandem genes (such as SUR7 and GAL80) has been shown to decrease their co-expression genome-wide [78]. Furthermore, regional signal spreading could extend to influence more distal locations. In fact, multiple examples in mammalian genomes have been described where at least three transcription units are connected to each other by overlapping transcripts or divergent promoters [86]. Such ‘chaining’ of events could result in linking of up to 11 transcription units [86]. Hence, the local organization of transcription units could have a significant impact on genome-wide transcriptional regulation, complexity and evolution.
ncRNAs influence the expression of distal protein-coding genes
While local regulatory effects can occur independently of transcript stability, distal effects are typically thought to be more dependent on stable transcript species. However, it has been observed that distal genomic regions physically interact in 3D nuclear space [44, 87, 88]. This creates local environments where distal genomic regions are subject to similar influences, such as chromatin modifiers, transcription factors, or transcripts. In these environments, the act of ncRNA transcription may also affect genes distally located on the same or different chromosomes.
ncRNAs generated by bidirectional promoters could have enhancer-like functions to regulate distal genes (Figure 3D). A study of mouse cortical neurons discovered a novel class of bidirectional ncRNAs (enhancer RNAs) that are transcribed from enhancer domains [89]. These bidirectionally transcribed ncRNAs are generally short and non-polyadenylated, and it has been suggested that their synthesis is important for activation of the corresponding ORFs [89]. Moreover, it has been shown that the depletion of long intergenic non-coding RNAs (lincRNAs) initiating from a bidirectional promoter in humans decreases the expression of distal protein-coding genes, along with other lincRNAs [90].
Further evidence exists for long-range regulation of protein-coding genes by ncRNAs. Separate transfection of sense and antisense short RNAs transcribed from both strands either upstream or overlapping the first exon of c-MYC results in reduced expression of c-MYC mRNA in HeLa cells [22]. In yeast, ectopical expression of antisense ncRNAs represses the expression of the Ty1 retrotransposons [80] and the phosphate transporter PHO84 [91]. Although there is no direct evidence for bidirectionality of its promoter, HOTAIR, a 2.2kb ncRNA transcribed in antisense orientation to the HOXC gene in humans, represses the distal HOXD gene cluster through an interaction with chromatin modifiers [92].
In summary, the consequences of transcription at bidirectional promoters range from local effects enabled by the transcription process itself, to distal effects mediated by the transcripts generated. Coupled transcription from bidirectional promoters is therefore used by the cell as an additional mechanism for the regulation of gene expression.
Concluding remarks and future perspectives
Much of the increased phenotypic complexity of higher eukaryotes is thought to arise from gene regulation rather than an increase in protein-coding gene numbers [93–95]. In addition to regulation mediated by distantly acting regulatory regions, expression of ncRNAs close to gene promoter regions provides a convenient means to locally regulate and fine-tune gene expression levels by exploiting shared chromatin, shared sequence, or sequence complementarity. While some of the non-coding transcription generated at protein-coding gene promoters may simply be transcriptional noise, there is mounting evidence that some of it is involved in gene regulation.
The interleaved, overlapping organization of transcripts has considerable consequences for research practices and genetics studies in general: mutating a region could affect not only the gene of interest, but also its associated non-coding transcripts and thus also nearby genes (e.g. [96]). The complex regulatory architecture also means that, when mapping a phenotype to a certain genomic locus, it is necessary to consider ncRNAs and bidirectional promoters as causes for phenotypic variability.
Several outstanding questions remain regarding the scope and functional consequences of promoter-associated non-coding transcription (Box 3). Further discovery and classification of these transcripts based on ncRNA features, subcellular localization, and correlation with protein-coding gene expression levels, promise to provide further insight into their functional consequences on cellular and organismal phenotypes.
Box 3. Outstanding questions.
How many different ncRNA species are transcribed from promoters? Is the diversity of measured ncRNAs a reflection of different RNA biogenesis pathways or differences in detection methodologies?
How many divergent ncRNAs are functional as transcripts or through the act of their transcription?
What are the molecular mechanisms for the regulation of bidirectional transcription?
What are the dynamics of transcription and the life cycle of transcripts (from initiation to termination)?
Is there cell-to-cell variation in divergent transcription? Does it affect the regulation of protein-coding genes?
What are the roles of divergent transcription in evolution, genome organization, and genetic networks?
Acknowledgements
We thank Raeka Aiyar, Wolfgang Huber, Julien Gagneur, Zhenyu Xu and Joël Savard for critical comments on the manuscript. This work was supported by grants to L.M.S. from the National Institutes of Health and the Deutsche Forschungsgemeinschaft. V.P. is supported by an EMBO postdoctoral Fellowship.
Glossary
- Backtracked polymerase
RNA polymerase II molecules that have moved backwards on the DNA template, leaving a misaligned 3’ end of the RNA that requires the aid of accessory factors to resume elongation.
- Bidirectional promoter
A genomic region of DNA that initiates transcription in both orientations. Different definitions of bidirectional promoters have been applied in diverse studies. For example, in yeast, transcripts were considered to originate from a bidirectional promoter when their start sites were within the same nucleosome-depleted region [13]. In metazoans, a distance cutoff of less than 1000 bp has been frequently used to define bidirectional promoters [6, 86].
- Bidirectional transcription
In this review, bidirectional transcription refers to transcription from a bidirectional promoter. In general, however, the term has also been used to describe overlapping transcription from both strands of a genomic region.
- Chromatin
Combination of DNA, nucleosomes, and other proteins to pack and organize the eukaryotic genome. Here, chromatin structure refers to the wrapping of genomic DNA around nucleosomes.
- Cryptic transcription
Production of non-canonical transcripts of unknown function that arises especially when there is interference with the chromatin modification or remodeling machinery.
- CTD
C-terminal domain of RNA polymerase II consisting of multiple repeats of the heptapeptide YSPTSPS that is used as a binding platform for other factors during the transcription cycle.
- Histone modifications
Post-translational modifications on histones that alter interactions with DNA and other proteins. The common nomenclature is the name of the histone followed by the single letter amino acid abbreviation, the amino acid position in the protein and the type of modification (e.g. H3K36me).
- Nascent RNA
Newly produced RNA that is physically associated with the RNA polymerase during the transcription process.
- Nucleosome-depleted region (NDR)
Region of DNA with very low nucleosome occupancy that is flanked by well-ordered nucleosomes at both sides. Thus it is also referred to as a nucleosome-free region (NFR).
- Pervasive transcription
Transcription that is not solely restricted to well-defined functional features, such as protein-coding gene regions, but that takes place non-randomly throughout the genome.
- Post-recruitment regulation
Regulation of the transcription process following the recruitment of RNA polymerase II to the promoter.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carninci P. RNA dust: where are the genes? DNA Res. 2010;17:51–59. doi: 10.1093/dnares/dsq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 3.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000459. e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck CF, Warren RA. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988;52:318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trinklein ND, et al. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermsen R, et al. Chance and necessity in chromosomal gene distributions. Trends Genet. 2008;24:216–219. doi: 10.1016/j.tig.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Guell M, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. doi: 10.1126/science.1176951. [DOI] [PubMed] [Google Scholar]
- 9.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 10.Wade JT, et al. Reply to "Concerns about Recently Identified Widespread Antisense Transcription in Escherichia coli". MBio 1. 2010 doi: 10.1128/mBio.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins TT, et al. A strand-specific RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000569. e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neil H, et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 15.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2010 doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, et al. Genome-wide transcription analyses in rice using tiling microarrays. Nat Genet. 2006;38:124–129. doi: 10.1038/ng1704. [DOI] [PubMed] [Google Scholar]
- 21.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 22.Fejes-Toth KSV, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, Gingeras TR. Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taft RJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 24.Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 25.Core LJ, et al. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller C, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Martinez J, et al. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 30.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 31.Juven-Gershon T, et al. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandelin A, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 33.Rach EA, et al. Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1001274. e1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, et al. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 38.Raisner RM, et al. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Tolstorukov MY, et al. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009;19:967–977. doi: 10.1101/gr.084830.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Sullivan JM, et al. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 45.Cheung V, et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Lickwar CR, et al. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One. 2009;4:e4886. doi: 10.1371/journal.pone.0004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends Genet. 2010;26:476–483. doi: 10.1016/j.tig.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 52.Whitehouse I, et al. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 53.Yadon AN, et al. Chromatin remodeling around nucleosome-free regions leads to repression of noncoding RNA transcription. Mol Cell Biol. 2010;30:5110–5122. doi: 10.1128/MCB.00602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tirosh I, et al. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol. 2010;11:R49. doi: 10.1186/gb-2010-11-5-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 56.Pelechano V, et al. A complete set of nascent transcription rates for yeast genes. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015442. e15442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mayer A, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 59.Kim H, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vasiljeva L, et al. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Margaritis T, Holstege FC. Poised RNA polymerase II gives pause for thought. Cell. 2008;133:581–584. doi: 10.1016/j.cell.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 62.Seila AC, et al. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 63.Singh N, et al. The Ess1 prolyl isomerase is required for transcription termination of small noncoding RNAs via the Nrd1 pathway. Molecular cell. 2009;36:255–266. doi: 10.1016/j.molcel.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pelechano V, et al. Regulon-specific control of transcription elongation across the yeast genome. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000614. e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, et al. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol. 2008;28:1393–1403. doi: 10.1128/MCB.01733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen BA, et al. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- 69.Johnson ZI, Chisholm SW. Properties of overlapping genes are conserved across microbial genomes. Genome Res. 2004;14:2268–2272. doi: 10.1101/gr.2433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin JM, et al. Transcription factor binding and modified histones in human bidirectional promoters. Genome Res. 2007;17:818–827. doi: 10.1101/gr.5623407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dhadi SR, et al. Genome-wide comparative analysis of putative bidirectional promoters from rice, Arabidopsis and Populus. Gene. 2009;429:65–73. doi: 10.1016/j.gene.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 72.Adachi N, Lieber MR. Bidirectional gene organization: a common architectural feature of the human genome. Cell. 2002;109:807–809. doi: 10.1016/s0092-8674(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 73.Guarguaglini G, et al. Expression of the murine RanBP1 and Htf9-c genes is regulated from a shared bidirectional promoter during cell cycle progression. Biochem J. 1997;325(Pt 1):277–286. doi: 10.1042/bj3250277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hansen JJ, et al. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum Genet. 2003;112:71–77. doi: 10.1007/s00439-002-0837-9. [DOI] [PubMed] [Google Scholar]
- 75.Kulaeva OI, et al. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat Res. 2007;618:116–129. doi: 10.1016/j.mrfmmm.2006.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirota K, et al. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 77.Wang X, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu Z, et al. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol. 2011;7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hongay CF, et al. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 80.Berretta J, et al. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Camblong J, et al. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 82.Houseley J, et al. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pinskaya M, et al. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilusz JE, et al. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Engstrom PG, et al. Complex Loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lanctot C, et al. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 88.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 89.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Camblong J, et al. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes & development. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 94.Pennisi E. Deciphering the genetics of evolution. Science. 2008;321:760–763. doi: 10.1126/science.321.5890.760. [DOI] [PubMed] [Google Scholar]
- 95.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 96.Tufarelli C, et al. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 97.Yoon OK, Brem RB. Noncanonical transcript forms in yeast and their regulation during environmental stress. Rna. 2010;16:1256–1267. doi: 10.1261/rna.2038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beck AH, et al. 3'-end sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS One. 2010;5:e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ng P, et al. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- 101.Tan-Wong SM, et al. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes & development. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Laine JP, et al. A physiological role for gene loops in yeast. Genes & development. 2009;23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]