Abstract
Escherichia coli topoisomerase I (TopA) cleaves and rejoins one strand of double-stranded DNA to relax the negatively supercoiled DNA. Structurally, TopA contains an N-terminal catalytic fragment and a C-terminal zinc-binding region that is required for relaxation of the negatively supercoiled DNA. Here we report that E. coli TopA is an iron and zinc binding protein. The UV–Vis absorption measurements and metal content analyses reveal that TopA purified from E. coli cells grown in the rich LB medium contains both iron and zinc. However, TopA purified from E. coli cells grown in the M9 minimal medium has negligible amounts of zinc or iron and no topoisomerase activity. Nevertheless, supplement of exogenous zinc or iron in E. coli cells grown in the M9 minimal medium produces the zinc- or iron-bound TopA, respectively. Whereas the zinc-bound TopA is fully active to relax the negatively super-coiled DNA, the iron-bound TopA has little or no enzyme activity. Furthermore, excess iron in the M9 minimal medium is able to compete with the zinc binding in TopA in E. coli cells and attenuate the topoisomerase activity, suggesting that E. coli TopA may be modulated by iron and zinc binding in vivo.
Keywords: Topoisomerase I, Zinc, Iron, Metalloprotein
Introduction
DNA topoisomerases are widely distributed in organisms ranging from bacteria to humans (Forterre and Gadelle 2009), and are essential for DNA replication, transcription, recombination, and chromatin remodeling (Champoux 2001; Schoeffler and Berger 2008). Topoisomerases have two subfamilies: type I enzymes that cleave and rejoin one strand of double-stranded DNA to relax the supercoiled DNA and type II enzymes that cleave and rejoin both strands of double-stranded DNA to allow pass of a region of duplex from the same or a different molecule (Champoux 2001). Escherichia coli topoisomerase I (TopA) belongs to type I subfamily (Wang 2002; Tse-Dinh 2009). When gene topA encoding topoisomerase I is deleted, the mutant E. coli cells are not viable at low temperature (<30°C) (Stupina and Wang 2005), and are hypersensitive to high temperature (>50°C) or oxidative stresses (Qi et al. 1999; Tse-Dinh 2000). Structurally, E. coli TopA has an N-terminal catalytic fragment (67 kDa) and a C-terminal zinc-binding region (30 kDa). While the N-terminal catalytic fragment is sufficient for cleaving the single-stranded DNA, the C-terminal zinc-binding region is required for relaxing the negatively supercoiled DNA (Tse-Dinh and Beran-Steed 1988; Tse-Dinh 1991; Ahumada and Tse-Dinh 2002) and for interacting with RNA polymerase (Cheng et al. 2003). The crystal structure of the E. coli TopA N-terminal fragment revealed that the catalytic core has a toroidal fold enclosing a central hole to accommodate substrate DNA (Lima et al. 1994, Baker et al. 2009). However, the crystal structure of full-length E. coli TopA containing both the N-terminal fragment and the C-terminal zinc-binding region is still not available.
Zinc is an essential trace metal that facilitates correct folding of proteins, stabilizes the domain structure, and plays important catalytic roles in enzymes (Berg and Shi 1996). In E. coli, zinc accumulates to similar levels as calcium and iron (~0.2 mM) under normal growth conditions (Outten and O’Halloran 2001). Depletion of zinc in growth medium results in slow-growth phenotype and activation of zinc uptake systems (Graham et al. 2009), indicating that zinc has crucial roles in E. coli cells. It has been shown that zinc binding in E. coli TopA is essential for relaxing the negatively supercoiled DNA (Tse-Dinh and Beran-Steed 1988). Here, we find that E. coli TopA is able to bind not only zinc, but also iron in E. coli cells, depending on the iron and zinc contents in the growth medium. Whereas the zinc-bound TopA is active to relax the negatively supercoiled DNA, the iron-bound TopA has very little or no such enzyme activity. Furthermore, excess iron in the M9 minimal medium is able to compete with the zinc binding in TopA in E. coli cells and attenuate the TopA topoisomerase activity. The results suggest that TopA topoisomerase activity may be regulated by the iron and zinc binding in E. coli cells.
Materials and methods
Protein purification
The DNA fragment encoding topoisomerase I (TopA) was amplified from the wild-type E. coli genomic DNA with PCR using two primers: TopA-1 (5′-AGGTGCATGGGTAAAGCTCTTGTC-3′) and TopA-2 (5′-GAATTAAAGCTTATTTTTTTCCTTCAACCC-3′). For the TopA N-terminal fragment (containing amino acid 597 residues), two primers: TopA-1 and TopAN-2 (5′-ACAAAAGCTTCAGTCAATGCTGG TCAGAAC-3′), were used for PCR. For the TopA C-terminal fragment (containing 268 amino acid residues), two primers: TopAC-1 (5′-CAGGAGCTCCTGCCCGACTTGTGGTCGC-3′) and TopA-2, were used for PCR. Each PCR product was digested with restriction enzymes NdeI and HindIII, and ligated to an expression vector pET28b+ (Novagen co). The cloned DNA fragments were confirmed by direct sequencing using T7 primer (Genomic Facility, LSU). Recombinant TopA and the N-terminal and C-terminal fragments were expressed in E. coli BL21 strain in either rich LB (Luria–Bertani) medium (Affymetrix/USB) or the M9 minimal medium supplemented with glucose (0.2%), thiamin (5 μg/ml) and 20 amino acids (each at 10 μg/ml). The growth media and all chemicals were prepared with double-distilled de-ionized water. The proteins were purified following the procedure described in (Duan et al. 2009), and the purity of purified protein was over 95%, judging from the SDS/PAGE followed by the Coomassie blue staining. The protein concentration of purified TopA was measured at 280 nm using an extinction coefficient of 95.7 cm−1 mM−1. The UV–Vis absorption spectra of protein samples were measured in a Beckman DU640 UV–Vis spectrometer equipped with a temperature control.
Topoisomerase activity assay
The negatively supercoiled DNA relaxation assay was carried out following the procedure described in (Tse-Dinh and Beran-Steed 1988; Tse-Dinh 1991; Ahumada and Tse-Dinh 2002). Briefly, purified TopA was incubated in buffer (10 μl) containing Tris (50 mM, pH 8.0) and MgSO4 (3 mM) at 37°C for 10 min before the negatively supercoiled plasmid pUC18 DNA (250 ng) was added to reaction solutions. After incubation at 37°C for 30 min, reactions were stopped by adding 2.5 μl stop solution containing 5% SDS and 15% glycerol. The reaction solution was then loaded onto a 1% agarose gel in TEA buffer. The agarose gel was run at 2 V cm−1 for 16 h, stained with ethidium bromide, and photographed.
Metal content analyses
Total iron content in protein samples was determined using an iron indicator FerroZine following the procedures described in (Cowart et al. 1993). Total zinc content in protein samples was determined using a zinc indicator PAR (4-(2-pyridylazo)-resorcinol) following the procedures described in Bae et al. (2004). The zinc and iron contents in purified proteins were also analyzed by the Inductively Coupled Plasma-Emission Spectrometry (ICP-ES) (Chemical Analysis Laboratory, University of Georgia). Both metal content analyses produced similar results.
Results
E. coli topoisomerase I (TopA) is an iron and zinc binding protein
When recombinant E. coli topoisomerase I (TopA) is expressed in E. coli cells grown in the rich LB medium, purified TopA is fully active to relax the negatively supercoiled DNA as reported previously (Tse-Dinh and Beran-Steed 1988). However, we noticed that purified TopA has a distinct reddish color (Fig. 1 inset). The UV–Vis absorption measurements showed that purified TopA has two major absorption peaks at 482 and 563 nm (Fig. 1), indicative of possible iron binding in the protein. Both absorption peaks are completely eliminated when the protein is reduced with sodium dithionite (Fig. 1) and re-appear after the reduced TopA is exposed to air for 30 min (data not shown), indicating that the iron center in TopA is redox active. The total metal content analyses of purified TopA revealed that there are 0.51 ± 0.12 iron atoms and 1.82 ± 0.41 zinc atoms per TopA monomer (n = 3), confirming that purified TopA contains both iron and zinc.
Fig. 1.
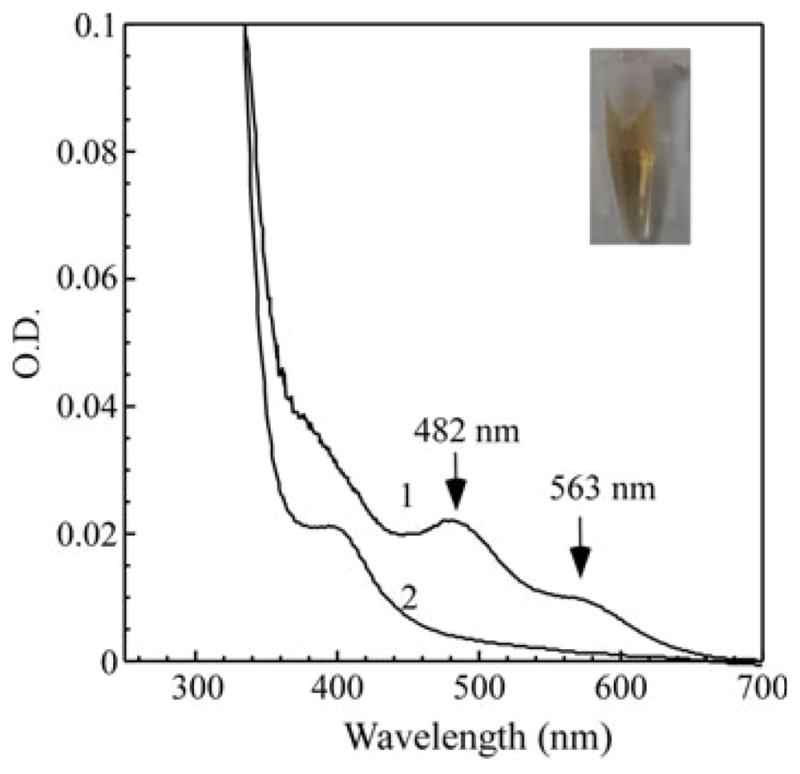
E. coli topoisomerase I (TopA) is an iron and zinc binding protein. Recombinant TopA was expressed in E. coli cells grown in the rich LB medium under aerobic growth conditions. Purified TopA (35 μM) was dissolved in buffer containing Tris (20 mM, pH 8.0) and NaCl (200 mM) before (spectrum 1) and after (spectrum 2) addition of sodium dithionite (2 mM). Inset is a photograph of purified TopA (700 μM)
To determine whether the iron binding in TopA is associated with the C-terminal zinc-binding region, we expressed the N-terminal catalytic fragment and the C-terminal zinc-binding region of TopA in E. coli cells grown in the rich LB medium. Figure 2 shows that while purified C-terminal fragment (30 kDa) (TopA-C) has the same absorption peaks at 482 and 563 nm as TopA, purified N-terminal fragment (67 kDa) (TopA-N) does not have such absorption peaks, suggesting that the C-terminal zinc-binding region, but not the N-terminal fragment, of TopA, is involved in the iron binding in E. coli cells.
Fig. 2.
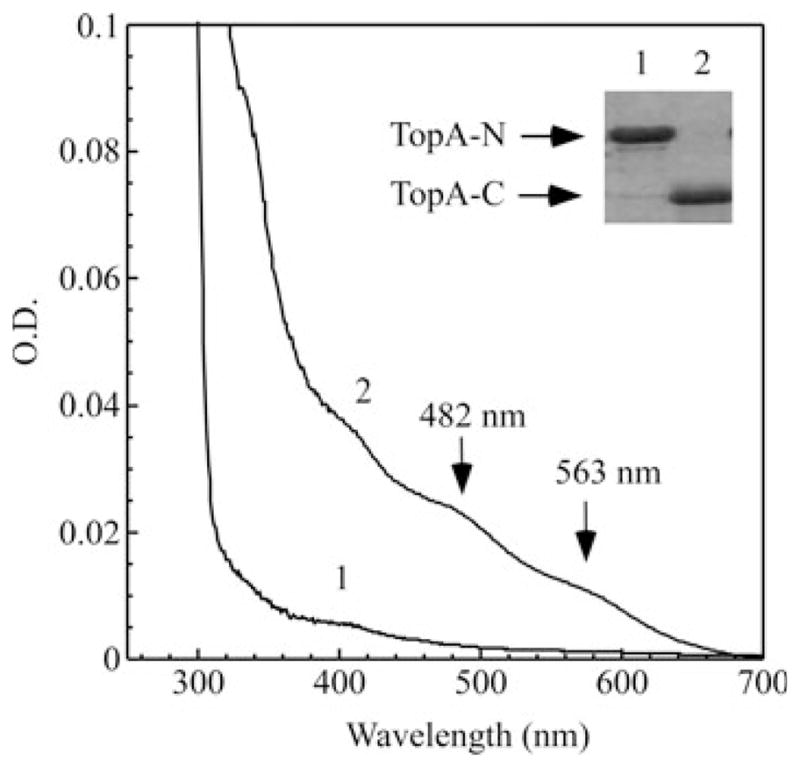
The iron binding activity of the N-terminal and C-terminal fragments of TopA in vivo. The N-terminal and C-terminal fragments of TopA were cloned and expressed in E. coli cells grown in the rich LB medium. Spectrum 1, purified N-terminal fragment of TopA. Spectrum 2, purified C-terminal fragment of TopA. The concentration of purified proteins was about 30 μM. Insert is a photograph of the SDS/PAGE gel of purified N-terminal fragment (TopA-N, 70 kDa) (1) and C-terminal fragment (TopA-C, 30 kDa) (2) purified from E. coli cells
The iron and zinc binding activity of TopA in E. coli cells grown in the M9 minimal medium
To further explore the zinc and iron binding activity of TopA, we expressed the protein in E. coli cells grown in the M9 minimal medium for better control of metal contents in the growth medium. Figure 3a shows that TopA purified from E. coli cells grown in the M9 minimal medium has no absorption peaks at 482 and 563 nm. The total metal content analyses revealed that purified TopA has 0.13 ± 0.02 iron atoms and 0.20 ± 0.04 zinc atoms per TopA monomer (n = 3). However, when the M9 minimal medium is supplemented with exogenous iron (20 μM), TopA purified from E. coli cells has the absorption peaks at 482 and 563 nm, indicating the iron binding in TopA (Fig. 3a). The metal content analyses confirmed that purified TopA contains 1.15 ± 0.12 iron atoms and 0.14 ± 0.02 zinc atoms per TopA monomer (n = 3). In parallel, when the M9 minimal medium is supplemented with exogenous zinc (20 μM), purified TopA has no indication of iron binding (Fig. 3a) and contains 0.02 ± 0.01 iron atoms and 2.44 ± 0.31 zinc atoms per TopA monomer (n = 3). Thus, the iron and zinc binding in TopA expressed in E. coli cells can be modulated by the iron and zinc content in the M9 minimal medium.
Fig. 3.
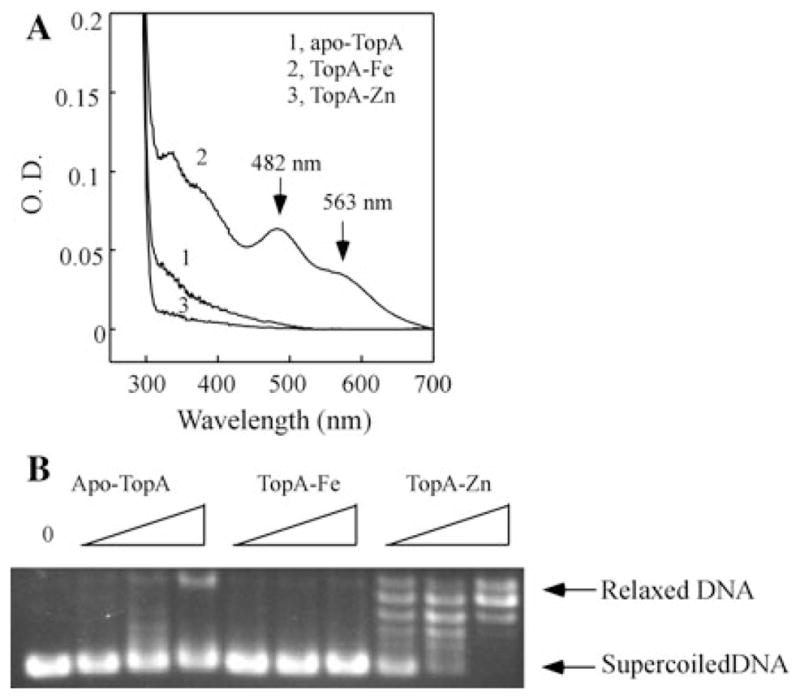
Iron and zinc binding in TopA in E. coli cells grown in the M9 minimal medium. a UV–Vis absorption spectra of TopA purified from E. coli cells grown in the M9 minimal medium supplemented with nothing (spectrum 1), Fe(NH4)2SO4 (20 μM) (spectrum 2), or ZnSO4 (20 μM) (spectrum 3). The protein concentration of purified TopA was 10 μM. b The enzyme activity of TopA purified from E. coli cells grown in the M9 minimal medium supplemented with nothing (apoTopA), Fe(NH4)2SO4 (20 μM) (Fe-TopA), or ZnSO4 (20 μM) (Zn-TopA). The negatively supercoiled pUC18 plasmid (250 ng) was incubated with purified TopA (50, 100, and 200 nM) at 37°C for 30 min. The reaction products were resolved on 1% agarose gel
Apo-TopA, the iron-bound TopA and the zinc-bound TopA as prepared in Fig. 3a are then subject to the analyses of topoisomerase activity. Figure 3b shows that while the zinc-bound TopA is fully active to relax the negatively supercoiled DNA, similar to the TopA purified from E. coli cells grown in the rich LB medium (Tse-Dinh and Beran-Steed 1988), apo-TopA or the iron-bound TopA has very little or no enzyme activity under the experimental conditions.
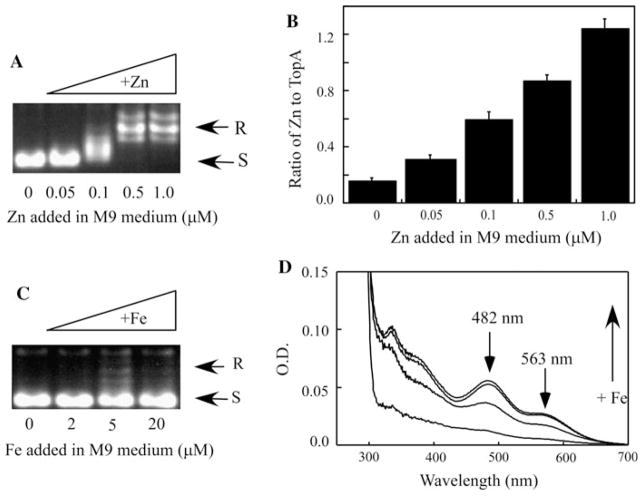
We then attempted to compare the relative binding activity of TopA for zinc and iron in vivo. Recombinant TopA was expressed in E. coli cells grown in the M9 minimal medium supplemented with increasing concentrations of exogenous zinc or iron. To our surprise, supplement of 0.5 μM exogenous zinc in the M9 minimal medium is sufficient to produce an active TopA expressed in E. coli cells (Fig. 4a). As the concentration of exogenous zinc in the M9 minimal medium is increased from 0 to 1 μM, the zinc binding in TopA is progressively increased (Fig. 4b). In parallel, when the M9 minimal medium is supplemented with increased concentrations of iron (0–20 μM), the iron binding in purified TopA is also gradually increased (Fig. 4d). However, TopA remains essentially inactive regardless of iron binding in the protein (Fig. 4c). Thus, TopA appears to have a relatively higher binding affinity for zinc than for iron in E. coli cells, and that zinc binding, but not iron binding, in TopA can restore its topoisomerase activity.
Fig. 4.
Relative iron and zinc binding activity of TopA in E. coli cells. a The enzyme activity of TopA purified from E. coli cells grown in the M9 minimal medium supplemented with 0, 0.05, 0.1, 0.5, and 1.0 μM ZnSO4. The concentration of purified TopA used for the enzyme activity assay was 100 nM. b The zinc content of TopA purified from E. coli cells grown in the M9 minimal medium supplemented with 0, 0.05, 0.1, 0.5, and 1.0 μM ZnSO4. c The enzyme activity of TopA purified from E. coli cells grown in the M9 minimal medium supplemented with 0, 2.0, 5.0, and 20 μM Fe(NH4)2SO4. The concentration of purified TopA used for the enzyme activity assay was 100 nM. d UV–Vis absorption spectra of TopA purified from E. coli cells grown in the M9 minimal medium supplemented with 0, 2.0, 5.0, and 20 μM Fe(NH4)2SO4. The protein concentration of purified TopA was 10 μM
Modulation of TopA activity by iron and zinc binding in E. coli cells
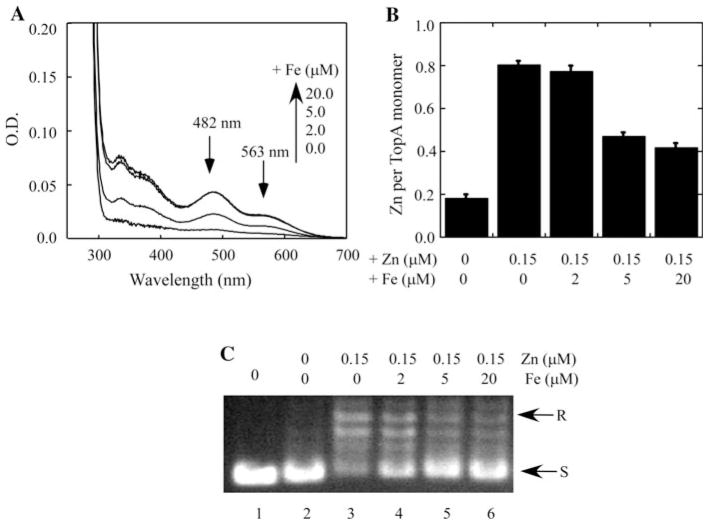
Because TopA can bind both iron and zinc in E. coli cells, and the iron-bound TopA is inactive to relax the negatively supercoiled DNA, we reasoned that excess iron may compete for the zinc-binding in TopA to attenuate the enzyme activity. To test this hypothesis, TopA is expressed in E. coli cells grown in the M9 minimal medium supplemented with a fixed concentration of zinc (0.15 μM) and increasing concentrations of iron (0–20 μM). A low concentration of zinc (0.15 μM) was chosen, because higher concentration of zinc (>0.5 μM) in the M9 minimal medium precluded any competition of iron binding in recombinant TopA in E. coli cells (data not shown), further suggesting that TopA has a relatively higher binding affinity for zinc than iron in E. coli cells.
Nevertheless, as the concentration of exogenous iron in M9 minimal medium (containing 0.15 μM zinc) is increased, the iron binding in TopA is gradually increased (Fig. 5a) with a concomitant decrease of the zinc binding in the protein (Fig. 5b). The parallel topoisomerase activity analyses indicated that the enzyme activity is significantly decreased as the iron binding in TopA is increased (panel C), indicating that excess iron in the M9 minimal medium can effectively compete with the zinc binding in TopA in E. coli cells to modulate the enzyme activity.
Fig. 5.
Competition of iron and zinc binding in TopA in E. coli cells. Recombinant TopA was purified from E. coli cells grown in the M9 minimal medium supplemented with 0.15 μM ZnSO4 and 0, 2.0, 5.0, and 20 μM Fe(NH4)2SO4. a The UV–Vis absorption spectra of purified TopA. The protein concentration was 20 μM. b Zinc content analyses of purified TopA. The zinc content in purified TopA was represented as the ratio of zinc to TopA monomer. c The enzyme activity of TopA purified from the E. coli cells grown in M9 minimal medium with 0.15 μM ZnSO4 and 0, 2.0, 5.0, and 20 μM Fe(NH4)2SO4 (lane 3–6), respectively. Lane 1 no enzyme added. Lane 2 apo-TopA. The concentration of purified TopA used for the enzyme activity assay was 100 nM. The data are representatives from three independent experiments
Discussion
In search for unknown iron binding proteins in E. coli, we unexpectedly found that YrdD, a C-terminal homolog of the E. coli topoisomerase I (TopA), is a novel iron/zinc binding protein (unpublished data). The finding led us to inquire whether the E. coli TopA is also an iron/zinc binding protein. Here, we report that when E. coli TopA is expressed in E. coli cells grown in the rich LB medium, purified TopA, like YrdD, contains both iron and zinc. The results are in contrast with the previous report that recombinant E. coli TopA contains only zinc atoms (Tse-Dinh and Beran-Steed 1988). We attributed this discrepancy to the higher concentrations of TopA (30–100 μM) that were used for the metal content analysis in this study (Fig. 1) in contrast to 1–2 μM TopA used in the previous study (Tse-Dinh and Beran-Steed 1988). When recombinant TopA is expressed in E. coli cells grown in the M9 minimal medium, purified TopA has negligible amounts of iron or zinc. However, supplement of exogenous iron or zinc in the M9 minimal medium produces the iron or zinc-bound TopA in E. coli cells, respectively (Fig. 3a). The negatively supercoiled DNA relaxation analyses reveal that while the zinc-bound TopA is active, both apo-TopA and the iron-bound TopA have very little or no enzyme activity under the experimental conditions (Fig. 3b), suggesting that although TopA is able to bind both iron and zinc, iron cannot substitute for zinc in restoring the TopA enzyme activity. Finally, we find that excess iron in the M9 minimal medium is able to compete with the zinc binding in TopA in E. coli cells, resulting in attenuation of the topoisomerase activity (Fig. 5).
It has been reported that iron and zinc have a similar ligand binding coordination in proteins (Dauter et al. 1996). For example, in the estrogen receptor zinc finger protein, the in vitro substitution of zinc with iron does not significantly affect its target DNA binding activity (Conte et al. 1996). In the tristetraprolin-2D, a protein involved in regulating inflammatory response, binding of zinc or iron has the same RNA binding activity (diTargiani et al. 2006). In the neural zinc finger factor 1, binding of zinc or iron also shows the similar DNA binding activity (Besold et al. 2010). In the GATA-1 zinc finger protein, the iron-bound form has an even higher binding affinity for DNA than the zinc-bound form (Omichinski et al. 1993). Thus, at least in these cases, zinc and iron could mutually interchange for the protein function. On the other hand, in the classical zinc finger protein TFIIIA, the iron-bound form can no longer recognize the target DNA (Hanas et al. 1983). Our finding that substitution of zinc with iron in E. coli TopA fails to restore the enzyme activity (Fig. 4) represents another example that iron and zinc are functionally not interchangeable in proteins. It may be envisioned that binding of iron or zinc in TopA will lead to subtle structural change in such that only zinc binding in the specific zinc-binding site(s) in TopA will allow TopA to relax the negatively supercoiled DNA.
Topoisomerases have been proposed as the prominent targets of antibacterial and anticancer therapeutic agents (Nitiss 2002; Mitscher 2005; Pommier 2006; Tse-Dinh 2009). Understanding the potential regulation of TopA by iron and zinc binding may provide new insights for developing therapeutic drugs targeting topoisomerases. Nevertheless, a number of questions remain to be further investigated on the iron and zinc binding in TopA. First, since TopA has three zinc binding sites in its C-terminal region (Tse-Dinh and Beran-Steed 1988; Ahumada and Tse-Dinh 2002), it is pertinent to determine whether zinc binding in all three zinc-binding sites is required for the full enzyme activity. Second, if TopA is capable of binding both iron and zinc in vivo, it would be interesting to demonstrate whether TopA with different combinations of iron and zinc binding is catalytically active. TopA purified from E. coli cells grown in the rich LB medium is fully active even though the protein contains 0.51 ± 0.12 iron atoms and 1.82 ± 0.41 zinc atoms per TopA monomer (Fig. 1), indicating that partial iron binding in TopA does not significantly affect its enzyme activity. However, if excess iron is included in the M9 minimal medium, the enzyme activity of TopA is considerably attenuated (Fig. 5). While detailed structural and functional change of TopA upon binding of iron or zinc remains to be further investigated, we propose that modulation of the topoisomerase activity of TopA by iron and zinc binding could be physiologically relevant.
Unlike zinc which is redox inert, iron center in proteins can often promote production of hydroxyl free radicals via Fenton reaction under aerobic conditions (Conte et al. 1996). Thus, binding of iron instead of zinc in TopA would increase the risk of promoting cellular oxidative damages. Interestingly, a recent study showed that addition of an iron chelator 2,2′-dipyridyl to E. coli cell culture can alleviate the TopA cleavage complex-mediated cell killing by preventing production of hydroxyl free radicals (Liu et al. 2009). The finding that TopA contains a redox active iron center in E. coli cells may provide a direct biochemical link between the TopA cleavage complex-mediated cell killing and the iron-mediated cellular oxidative damage.
Acknowledgments
The authors would like to thank Dr. Anne Grove (LSU) for the advice on the topoisomerase activity assay. This study was supported by the National Institutes of Health grant (CA107494); the China National Natural Science Foundation grant (30770448); and Natural Science Foundation of Zhejiang Province grants (Y2081075 and Y507233). A.P.L. was supported by the Howard Hughes Medical Institute Undergraduate Research Program at LSU.
Abbreviations
- TopA
E. coli topoisomerase I
- EPR
Electron paramagnetic resonance
Contributor Information
Jianxin Lu, Laboratory of Molecular Medicine, Wenzhou Medical College, Wenzhou 325035, Zhejiang, People’s Republic of China.
Wu Wang, Laboratory of Molecular Medicine, Wenzhou Medical College, Wenzhou 325035, Zhejiang, People’s Republic of China.
Guoqiang Tan, Laboratory of Molecular Medicine, Wenzhou Medical College, Wenzhou 325035, Zhejiang, People’s Republic of China.
Aaron P. Landry, Department of Biological Sciences, Louisiana State University, 202 Life Sciences Building, Baton Rouge, LA 70803, USA
Peng Yi, Laboratory of Molecular Medicine, Wenzhou Medical College, Wenzhou 325035, Zhejiang, People’s Republic of China.
Fan Si, Laboratory of Molecular Medicine, Wenzhou Medical College, Wenzhou 325035, Zhejiang, People’s Republic of China.
Yaguang Ren, Laboratory of Molecular Medicine, Wenzhou Medical College, Wenzhou 325035, Zhejiang, People’s Republic of China.
Huangen Ding, Email: hding@lsu.edu, Laboratory of Molecular Medicine, Wenzhou Medical College, Wenzhou 325035, Zhejiang, People’s Republic of China. Department of Biological Sciences, Louisiana State University, 202 Life Sciences Building, Baton Rouge, LA 70803, USA.
References
- Ahumada A, Tse-Dinh YC. The role of the Zn(II) binding domain in the mechanism of E. coli DNA topoisomerase I. BMC Biochem. 2002;3:13. doi: 10.1186/1471-2091-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JB, Park JH, Hahn MY, Kim MS, Roe JH. Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J Mol Biol. 2004;335:425–435. doi: 10.1016/j.jmb.2003.10.065. [DOI] [PubMed] [Google Scholar]
- Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- Besold AN, Lee SJ, Michel SL, Sue NL, Cymet HJ. Functional characterization of iron-substituted neural zinc finger factor 1: metal and DNA binding. J Biol Inorg Chem. 2010;15:583–590. doi: 10.1007/s00775-010-0626-1. [DOI] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Cheng B, Zhu CX, Ji C, Ahumada A, Tse-Dinh YC. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J Biol Chem. 2003;278:30705–30710. doi: 10.1074/jbc.M303403200. [DOI] [PubMed] [Google Scholar]
- Conte D, Narindrasorasak S, Sarkar B. In vivo and in vitro iron-replaced zinc finger generates free radicals and causes DNA damage. J Biol Chem. 1996;271:5125–5130. doi: 10.1074/jbc.271.9.5125. [DOI] [PubMed] [Google Scholar]
- Cowart RE, Singleton FL, Hind JS. A comparison of bathophenanthrolinedisulfonic acid and ferrozine as chelators of iron(II) in reduction reactions. Anal Biochem. 1993;211:151–155. doi: 10.1006/abio.1993.1246. [DOI] [PubMed] [Google Scholar]
- Dauter Z, Wilson KS, Sieker LC, Moulis JM, Meyer J. Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein. Proc Natl Acad Sci USA. 1996;93:8836–8840. doi: 10.1073/pnas.93.17.8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- diTargiani RC, Lee SJ, Wassink S, Michel SL. Functional characterization of iron-substituted tristetraprolin-2D (TTP-2D, NUP475–2D): RNA binding affinity and selectivity. Biochemistry. 2006;45:13641–13649. doi: 10.1021/bi060747n. [DOI] [PubMed] [Google Scholar]
- Duan X, Yang J, Ren B, Tan G, Ding H. Reactivity of nitric oxide with the [4Fe-4S] cluster of dihydroxyacid dehydratase from Escherichia coli. Biochem J. 2009;417:783–789. doi: 10.1042/BJ20081423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, McLeod CW, Poole RK. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, Zinc transport and zinc-independent proteins. J Biol Chem. 2009;284:18377–18389. doi: 10.1074/jbc.M109.001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas JS, Hazuda DJ, Bogenhagen DF, Wu FY, Wu CW. Xenopus transcription factor A requires zinc for binding to the 5S RNA gene. J Biol Chem. 1983;258:14120–14125. [PubMed] [Google Scholar]
- Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67 K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- Liu IF, Annamalai T, Sutherland JH, Tse-Dinh YC. Hydroxyl radicals are involved in cell killing by bacterial topoisomerase I cleavage complex. J Bacteriol. 2009;191:5315–5319. doi: 10.1128/JB.00559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitscher LA. Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial agents. Chem Rev. 2005;105:559–592. doi: 10.1021/cr030101q. [DOI] [PubMed] [Google Scholar]
- Nitiss JL. DNA topoisomerases in cancer chemotherapy: using enzymes to generate selective DNA damage. Curr Opin Investig Drugs. 2002;3:1512–1516. [PubMed] [Google Scholar]
- Omichinski JG, Trainor C, Evans T, Gronenborn AM, Clore GM, Felsenfeld G. A small single-“finger” peptide from the erythroid transcription factor GATA-1 binds specifically to DNA as a zinc or iron complex. Proc Natl Acad Sci USA. 1993;90:1676–1680. doi: 10.1073/pnas.90.5.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- Qi H, Menzel R, Tse-Dinh YC. Increased thermosensitivity associated with topoisomerase I deletion and promoter mutations in Escherichia coli. FEMS Microbiol Lett. 1999;178:141–146. doi: 10.1111/j.1574-6968.1999.tb13770.x. [DOI] [PubMed] [Google Scholar]
- Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- Stupina VA, Wang JC. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J Biol Chem. 2005;280:355–360. doi: 10.1074/jbc.M411924200. [DOI] [PubMed] [Google Scholar]
- Tse-Dinh YC. Zinc (II) coordination in Escherichia coli DNA topoisomerase I is required for cleavable complex formation with DNA. J Biol Chem. 1991;266:14317–14320. [PubMed] [Google Scholar]
- Tse-Dinh YC. Increased sensitivity to oxidative challenges associated with topA deletion in Escherichia coli. J Bacteriol. 2000;182:829–832. doi: 10.1128/jb.182.3.829-832.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse-Dinh YC. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 2009;37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse-Dinh YC, Beran-Steed RK. Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repetitive zinc-binding domains. J Biol Chem. 1988;263:15857–15859. [PubMed] [Google Scholar]
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]