Abstract
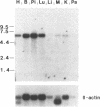
We have cloned a series of overlapping cDNA clones encoding a 5194 bp transcript for human DNA methyltransferase (DNA MTase). This sequence potentially codes for a protein of 1495 amino acids with a predicted molecular weight of 169 kDa. The human DNA MTase cDNA has eighty percent homology at the nucleotide level, and the predicted protein has seventy-four percent identity at the amino acid level, to the DNA MTase cDNA cloned from mouse cells. Like the murine DNA MTase, the amino terminal two-thirds of the human protein contains a cysteine-rich region suggestive of a metal-binding domain. The carboxy terminal one-third of the protein shows considerable similarity to prokaryotic (cytosine-5)-methyltransferases. The arrangement of multiple motifs conserved in the prokaryotic genes is preserved in the human DNA MTase, including the relative position of a proline-cysteine dipeptide thought to be an essential catalytic site in all (cytosine-5)-methyltransferases. A single 5.2 kb transcript was detected in all human tissues tested, with the highest levels of expression observed in RNA from placenta, brain, heart and lung. DNA MTase cDNA clones were used to screen a chromosome 19 genomic cosmid library. The DNA MTase-positive cosmids which are estimated to span a genomic distance of 93 kb have been localized to 19p13.2-p13.3 by fluorescence in situ hybridization. Isolation of the cDNA for human DNA MTase will allow further study of the regulation of DNA MTase expression, and of the role of this enzyme in establishing DNA methylation patterns in both normal and neoplastic cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L. DNA methylation. The effect of minor bases on DNA-protein interactions. Biochem J. 1990 Jan 15;265(2):309–320. doi: 10.1042/bj2650309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S. B., Makos M., Wu J. J., Yen R. W., de Bustros A., Vertino P., Nelkin B. D. Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer Cells. 1991 Oct;3(10):383–390. [PubMed] [Google Scholar]
- Bestor T. H. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988 Oct 20;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Carrano A. V., Lamerdin J., Ashworth L. K., Watkins B., Branscomb E., Slezak T., Raff M., de Jong P. J., Keith D., McBride L. A high-resolution, fluorescence-based, semiautomated method for DNA fingerprinting. Genomics. 1989 Feb;4(2):129–136. doi: 10.1016/0888-7543(89)90291-7. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983 Jan 6;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Buckley J. D. The role of DNA methylation in cancer. Adv Cancer Res. 1990;54:1–23. doi: 10.1016/s0065-230x(08)60806-4. [DOI] [PubMed] [Google Scholar]
- Mabry M., Nakagawa T., Baylin S., Pettengill O., Sorenson G., Nelkin B. Insertion of the v-Ha-ras oncogene induces differentiation of calcitonin-producing human small cell lung cancer. J Clin Invest. 1989 Jul;84(1):194–199. doi: 10.1172/JCI114140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos T., Pfeifer G. P., Hoppe F., Drahovsky D., Müller-Hermelink H. K. Proliferation-associated expression of DNA methyltransferase in human embryonic lung cells. Virchows Arch B Cell Pathol Incl Mol Pathol. 1989;56(6):371–375. doi: 10.1007/BF02890039. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Drahovsky D. DNA methyltransferase polypeptides in mouse and human cells. Biochim Biophys Acta. 1986 Dec 18;868(4):238–242. doi: 10.1016/0167-4781(86)90059-x. [DOI] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D. DNA methylation and late replication probably aid cell memory, and type I DNA reeling could aid chromosome folding and enhancer function. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):285–297. doi: 10.1098/rstb.1990.0012. [DOI] [PubMed] [Google Scholar]
- Selker E. U. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990 Mar;15(3):103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- Szyf M., Bozovic V., Tanigawa G. Growth regulation of mouse DNA methyltransferase gene expression. J Biol Chem. 1991 Jun 5;266(16):10027–10030. [PubMed] [Google Scholar]
- Szyf M., Kaplan F., Mann V., Giloh H., Kedar E., Razin A. Cell cycle-dependent regulation of eukaryotic DNA methylase level. J Biol Chem. 1985 Jul 25;260(15):8653–8656. [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Wilke K., Rauhut E., Noyer-Weidner M., Lauster R., Pawlek B., Behrens B., Trautner T. A. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 1988 Aug;7(8):2601–2609. doi: 10.1002/j.1460-2075.1988.tb03110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G. Organization of restriction-modification systems. Nucleic Acids Res. 1991 May 25;19(10):2539–2566. doi: 10.1093/nar/19.10.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]
- Xiao L., Celano P., Mank A. R., Pegg A. E., Casero R. A., Jr Characterization of a full-length cDNA which codes for the human spermidine/spermine N1-acetyltransferase. Biochem Biophys Res Commun. 1991 Aug 30;179(1):407–415. doi: 10.1016/0006-291x(91)91385-p. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., Nelkin B. D., Celano P., Yen R. W., Falco J. P., Hamilton S. R., Baylin S. B. High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3470–3474. doi: 10.1073/pnas.88.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]