Abstract
Important human phenotypes like height or facial appearance run in families—that has been known for millennia. Systematic studies of the way in which crossing pea plants resulted in changes in important pea plant phenotypes such as flower color or leaf number were defined in the mid-nineteenth century by Mendel [1] and the chemical basis for “inherited inborn errors of metabolism” by Garrod [2] at the turn of the twentieth century. Thus, some of the fundamental and familiar rules that we accept in a contemporary understanding of human genetics were laid down decades ago. However, an understanding of the mechanisms whereby genetic information is transmitted from generation to generation and how this information modulates important physiologic or disease susceptibility traits has been more recent. The fundamental discovery was the double-helix structure of DNA, which immediately led to the inference that DNA replication might replicate itself [3]. The last 50 years has seen the development of increasingly robust techniques for sequencing DNA and for using DNA as a laboratory reagent.
Keywords: Phenotypes, Genotypes, DNA
1 A gene-centered view of human physiology
These advances have led to the identification of genes in which mutations cause rare and well-recognized human genetic diseases, such as the congenital arrhythmia syndromes and many others. The identification of these disease genes and the elucidation of underlying disease mechanisms in these relatively rare conditions have truly revolutionized our understanding of normal and abnormal human physiology. Our understanding of the normal cardiac cycle is now commonly cast in terms of now familiar concepts such as opening and closing of ion channels, propagation through gap junctions, calcium-induced calcium release and actin–myosin cross-bridging, and repolarization and pacemaker activity. These are all now viewed, by basic scientists and clinicians alike, as molecular events, and we are all now familiar with the concept that mutations disrupting individual components of this molecular ballet can alter any one of its components with a resultant highly arrhythmogenic substrate.
2 The genotype–phenotype dilemma
The vast majority of disease-associated mutations identified to date are in the small proportion of DNA that actually encodes protein sequences. When the congenital long QT syndrome, the prototypical familial ion channel disease, was first described, it was very rare and highly lethal in families [4–6]. With increasing recognition of the entity and, in particular, with widespread availability of genetic testing have come the recognition that many individuals, almost certainly the majority of mutation carriers in fact, will never have symptoms, and many of these will not have abnormally long QT intervals [7]. This is the phenomenon of incomplete penetrance [8]; the commonly invoked explanations are the presence of other common or rare genetic variants, environmental factors (that may themselves act through genetic pathways), or interactions between genes or between genes and the environment.
This progression from recognition of a familial rare disease with an obvious and often highly lethal clinical phenotype to recognition that these diseases are commoner, with phenotypes that may vary from nothing (despite the presence of a disease-associated mutation) to severe, has now repeated itself in many other arrhythmia and other diseases such as the Brugada syndrome and arrhythmogenic right ventricular dysplasia (ARVD). We are in the increasingly awkward position of identifying individuals who carry genetic variants that, in some patients (perhaps their own family members), may confer susceptibility to a severe disease and yet in the affected individual have no clinical phenotypes. This is the genotype–phenotype dilemma. This review will discuss tools to identify genetic variation and our current understanding of how this variation modulates physiologic traits, disease susceptibility, and drug responses, with a focus on clinical electrophysiologic phenotypes.
3 Finding mutations that cause rare familial syndromes
The conventional approach to identification of disease genes in familial syndromes of any type is to identify a large kindred with many affected and unaffected individuals. Assignment of phenotype as affected or unaffected allows investigators to then identify polymorphic sites or regions associated with phenotype status. With enough affected individuals (hence the need for large kindreds in initial studies), the genomic region isolated can be quite small. The usual next step is to examine the linked region to determine if it contains any logical candidates (e.g., ion channels for an arrhythmia syndrome); such candidates are then sequenced in affected and unaffected subjects to identify specific mutations that associate with (the formal term is segregate with) the phenotype. If no logical candidates are identified, sequencing all coding regions or the entire region may be required. Early studies of this type [9, 10] were hampered by very crude maps of polymorphic sites across the genome and the absence of a fully annotated genome.
4 Common genetic variation
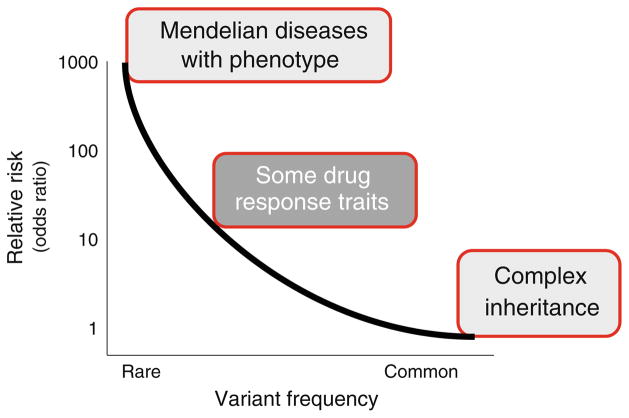
Diseases like long QT syndrome, hypertrophic cardiomyopathy, or cystic fibrosis are caused by rare, function-altering variants, termed mutations, in disease genes. Patients who carry causative mutations and who have manifest clinical phenotypes (like marked QT prolongation or marked ventricular hypertrophy) are at high risk for clinical events (Fig. 1). However, cloning studies have long recognized that human genetic sequences also include common variants, termed polymorphisms. The cloning of the first draft of the human genome was followed by an initial map (the “HapMap”) of common genetic variants that characterized each individual human genome [11, 12]. The commonest such variant is a single nucleotide polymorphism (SNP), a substitution of a single DNA base pair for a different base pair. Other genetic variants include insertions or deletions, which may be as small as two nucleotides and as large as thousands, copy number variations, and regions with highly variable number of tandem repeats.
Fig. 1.
Relationship between the frequency of a genetic variant and relative risk that it confers. Mendelian disorders, such as long QT syndrome or hypertrophic cardiomyopathy, are caused by very rare genetic variants that confer very high risk for clinical events in patients with manifest clinical phenotypes. At the other end of the spectrum are very common variants that may nevertheless be associated with increased susceptibility to common human traits such as atrial fibrillation or sudden cardiac death, as discussed further in the text. This type of variant generally does not confer risk that is high enough to warrant a change in clinical management. Genetic variants that modulate drug responses may be common and also confer high odds for unusual drug responses; this is likely one example of the general principle that some genetic variants may exert large effects in the presence of environmental triggers (such as drugs). One component of the “genotype–phenotype dilemma” is the problem of assigning risk to patients with rare, disease-associated variants but no manifest clinical phenotype
Polymorphisms are by definition common, often defined as occurring in greater than 1% of a given population. We do know that some variations occur across all racial groups (these are termed “cosmopolitan”) and others may be common in one specific racial group but rare or absent in others. Common polymorphisms do not occur independently, that is, the presence of a variant at a specific location in a gene may be highly predictive of a polymorphism at a nearby site. Polymorphisms that display such associations are said to be in “linkage disequilibrium.” We now know that there are large regions of the human genome in which hundreds or thousands of variant sites are in tight linkage disequilibrium. In these regions, genotyping even a single SNP or a handful of SNPs then defines the common sequence variation of the entire region. Moreover, common genetic variants are scattered across the entire genome. Since coding regions account for only a small fraction of the entire genomic sequence, most genetic variants do not affect the sequences of the proteins encoded by specific genes. Rather, non-coding genetic variation is thought to modulate variability in human traits by regulating the timing or amount of protein or specific splice isoforms generated by the coding sequences. Such transcriptional regulation is commonly invoked as a mechanism underlying variability in common human traits.
5 Genome-wide association studies
One of the most exciting uses of HapMap data has been our ability to conduct “genome-wide association” studies (GWAS). We have known for decades that common human diseases like atherosclerosis or diabetes “run in families.” In a GWAS experiment, hundreds of thousands of common SNPs are genotyped in thousands of subjects with and without the target phenotype (early myocardial infarction, atrial fibrillation (AF), diabetes, etc.) [13]. This experiment allows the identification of regions of the genome that are associated with the trait of interest. The GWAS paradigm has been amazingly successful in identifying new genetic pathways that define mechanisms underlying variability in common human physiology as well as disease susceptibility. The approach has been criticized because it has not resulted in identification of a clean series of genes for atherosclerosis, diabetes, or AF. In addition, the nature of the experiment does not allow identification of the specific genetic variants that confer very high risk for the traits under study, but only the region. The major challenge to and opportunity for biologists is to understand the way in which SNPs in a particular region cause susceptibility to these diseases.
The risks associated with GWAS “hits” tend to carry only very modest effects so at this point they do not carry any therapeutic implication (Fig. 1). Evolutionary considerations are thought to dictate why most common genetic variants do not produce large changes in disease susceptibility. In general, a common genetic variation that confers a major survival disadvantage will not propagate over generations and thus will become rare. There are exceptions. For example, variants that modulate risk for diseases in the post-reproductive years (age-related macular degeneration or Alzheimer’s disease) can be common.
6 GWAS for common arrhythmia-related phenotypes
GWAS approaches have been used to analyze variability in both normal ECG intervals (PR, QRS, QT, RR) and arrhythmia susceptibility itself [14]. Initial GWAS studies in this area examined variability in the QT interval across a normal population, and the locus that has the statistically greatest effect is near the NOS1AP gene, which encodes an accessory protein for the nitric oxide synthase type 1 gene [15–17]. The mechanisms whereby NOS1AP variants modulate QT interval are not completely understood, but it has also been reported that variants may predict susceptibility to sudden cardiac death in a broad population [18]. In addition, more recently, NOS1AP variants have been shown to modulate the risk for events in patients with the congenital long QT syndromes [19, 20]. Other genetic variants identified in QT variability include ion channel genes as well as genes with as yet undetermined influences on normal repolarization. Interestingly, one of the latter, GINS3, was also identified in a study examining the role of genetic variation in modulating changes in zebrafish heart action potential duration in response to challenge with the potent QT-prolonging, IKr-blocking agent dofetilide [21]. In addition, as a result of QT GWAS studies, we know that variation in common ion channel genes, such as KCNQ1, KCNH2, and SCN5A, not only determines diseases like congenital long QT syndrome or Brugada syndrome but also modulates common human traits like the duration of the QT interval across a large population [16, 17]. Further, the finding that variation in NOS1AP or GINS3 exerts an important effect on human traits would not have been discovered without the GWAS paradigm.
GWAS approaches to AF have consistently identified a locus at chromosome 4q25 [22, 23], and the nearest gene, PITX2, is known to be important in left/right differentiation during cardiac development [24, 25]. Other GWAS have identified a potassium channel and a transcription factor that may be important in lone AF [26, 27], again through mechanisms that remain to be fully elucidated. 4q25 AF-associated variants predict the likelihood of atrial fibrillation developing after cardiac surgery [28] and the likelihood of successful pharmacologic or ablation therapy for AF [29, 30]. In addition, 4q25 variants also are associated with an increased risk of cryptogenic stroke, suggesting that AF may play a more important role than heretofore appreciated in this disease [31]. The mechanism underlying this susceptibility is now being worked out and might result in better biomarkers or even better drugs. However, an individual patient who carries a variant SNP at the 4q25 locus carries an approximately 50% increase risk for AF. Thus, beyond conventional treatment that might alter AF risk, such as managing hypertension, individuals carrying such risk alleles do not yet earn special interventions or counseling. More recent studies have identified small sets of subjects with combinations of SNPs that may increase risk up to 6-fold [32]. Eventually identification of such marker sets could lead to trials of primary prevention in high-risk subjects.
GWAS has been used to study the risk of ventricular fibrillation early during myocardial infarction. This study uncovered a locus at chromosome 21q21, and the nearest gene is a Coxsackie virus receptor [33]. The mechanisms remain to be elucidated. GWAS analysis of the PR interval consistently identifies variation at a sodium channel gene, SCN10A, heretofore unassociated with cardiac physiology [34–37]. Similarly, analysis of the normal QRS duration pointed to multiple regions, notably the SCN10A–SCN5A locus [38]. Although the mechanisms remain to be worked out, these results again demonstrate the power of the GWAS approach to identify new pathways modulating physiology and disease.
7 Heritability
Another way of looking at the GWAS result is to ask to what extent genetic variants explain known heritability of a trait. This is not often assessed, but when it is, the individual loci identified by GWAS generally account for well under 5% of the heritability of diseases that are recognized to be familial, like diabetes or lone AF; this has been termed the problem of “missing heritability.” It is thought that the remainder of the heritability of such traits lies in interactions among genetic variants (a statistically difficult problem to address), environmental factors, interactions between genetics and environmental factors, or the presence of multiple rare and as yet undescribed genetic sequence variants [39]. Recall that the GWAS experiment only interrogates common SNPs, and we are only now acquiring new sequencing tools that will allow us to understand the extent to which rare genetic variants contribute to human disease.
8 Pharmacogenomic traits and heritability
An interesting exception to this heritability rule is in the field of pharmacogenomics. For example, studies in families have demonstrated that up to 70% of the variability in extent to which clopidogrel inhibits platelet aggregation is heritable. Polymorphisms at one specific locus, involving the cytochrome P450 that bioactivates clopidogrel (CYP2C19), account for approximately 15% of such variation [40]. For a clinician, that number sounds small, but put in the context of other common genetic variants that modulate important human traits, this is quite a high number and in fact is now included in the FDA label for the drug. Similarly, up to 50% of the variability in steady-state warfarin dose can be explained by variation in two genes, CYP2C9 and VKORC1 [41–43]. Thus, common variation may contribute extensively to variability in drug response traits. Such pharmacogenomic traits therefore appear to represent examples of relatively common alleles that nevertheless confer sufficiently high risk that they may be “actionable” (Fig. 1). Pre-emptive genotyping for patients likely to receive a drug with a pharmacogenetic “story” has been proposed as a pathway to clinical implementation [44, 45].
9 Moving genomic information to practice
It is now possible for any individual to have GWAS-genotyping performed and interpreted for several hundred dollars. Critics of such “direct to consumer” genomic services point out, correctly, that the vast majority of such variation, while interesting to the individual, carries no “actionability” at this point; one view is that a risk allele, or set of alleles, should confer a relative risk of ≥10 before any clinical, genotype-based action should be considered [46]. That said, the field is turning increasingly to the use of genomic and other markers to identify small sets of individuals predicted to be at especially high risk for subsequent events. Thus, single variants at chromosome 4q25 may increase the odds ratios for subsequent atrial fibrillation by 50%; further work in the area is identifying combinations of SNPs at this and other loci that may allow us to identify patients at much higher risk for subsequent atrial fibrillation and thus deserving of primary prevention interventions. It is also worth emphasizing that many diseases studied by arrhythmia specialists (including atrial fibrillation [47, 48] and sudden cardiac death [49–51]) include family history as a risk factor; thus, family history should be incorporated systematically into the record of patients seen even for common conditions. At this point, the only common genetic variants that appear to have clinical implications are those associated with variable drug responses.
10 Using next-generation sequencing to identify disease loci
Sequencing the first human genome cost several billion dollars. Advanced technological advances over the last decade have occurred at a breathtaking pace, and it is now possible to purchase the sequence of a whole human genome for less than $5,000. In addition, technologies have been developed to sequence the coding regions of the genome (the “exome”), and this is now feasible for less than $2,000. The challenges in obtaining, analyzing, and understanding such vast amounts of data are formidable, but the costs are dropping so fast that the idea of embedding each individual’s genome in their medical record is raised repeatedly. The power and limitations of the technique were underlined by a 2009 report of 12 human exome sequences [52]. Included in this group were four individuals with a known rare genetic disease, the Freeman Sheldon syndrome (FSS). Sequencing the exomes in each of the four probands identified hundreds of non-synonymous rare SNPs, i.e., SNPs that change the amino acid sequence of the encoded protein, that had never been seen before, and that might, therefore, represent causative mutations. By examining the overlaps across genes in the four probands, it was possible to identify a single gene mutated in all four probands, and this is the disease gene for FSS. Thus, we now understand that we all harbor hundreds of variants that are rare and that change the encoding amino acid. Indeed, looking across whole genomes, rather than exomes, we all harbor tens of thousands of rare variants, and the extent to which these modulate common disease susceptibilities and cause rare diseases is under very active investigation [53, 54].
In early 2010, the first fully annotated genome of a human being was published [55]. The individual, Stephen Quake, is one of the co-founders of Helicos, a company developing next-generation sequencing methodologies. The Quake genome includes 2,795 rare variants predicted to change the encoded amino acid. Interestingly, Stephen Quake has a cousin with ARVD and he himself carries three rare variants, one in myosin-binding protein C (previously reported), one in desmoplakin not previously reported, and one in TMEM43 also not previously reported that could predispose him to ARVD. The complexities of interpreting even a single genome highlight the increasing need for clinicians trained in genomics, partnering with specialized genetic counselors, to advise families and develop strategies to optimally manage risk and benefits of genetic testing and the further evaluations and interventions that may flow from it.
11 Back to the genotype–phenotype dilemma
Modern genomics is progressing at breathtaking speed to defining hundreds or thousands of rare genetic variants that each of us harbor. The potential associations between these variants and disease are enormous, and the fact of the matter is that the vast majority of such associations are likely to be false positives. If all genes are the “genome,” all genes encoding exons are the “exome,” then the collection of genetic variants that have no clinical implications but are discovered incidentally has been termed the “incidentalome” [56]. Developing mechanisms to understand which genetic variants do not require intervention and which might carry clinical implications represents the major challenge to modern genetics and indeed to contemporary medical practice over the next several decades. This problem must be solved. Otherwise, with the increasing availability of genetic information will come an explosion in needless diagnostic testing and therapeutic interventions. The electrophysiology community stands at the precipice of this problem. The phenotypes with which we deal, notably sudden cardiac death in the young, are so poignant that the compulsion to act on a genetic variant, even in a patient without a clinical phenotype, is sometimes overwhelming. The solution lies in more data: more clinical data relating to genotypes to clinical phenotypes and outcomes and more investment in basic and translational sciences to develop rules to understand the functional consequences of individual new rare genetic variants. Solving this paradox is a vital next step to our field’s demonstrated ability to translate between basic sciences and improvement in clinical care.
Acknowledgments
This study was supported in part by grants from the US National Institutes of Health (U01 HL65962, RC2 GM092618, U01 HG04603).
Footnotes
Conflicts of interest Dr. Roden is a consultant to Sanofi-Aventis, one of the sponsors of the meeting at which this work was presented. Dr. Roden retained full control of this manuscript throughout its generation; as this is a review, there are no primary data.
References
- 1.Mendel G. Experiments in plant hybridization. Verhandlungen des naturforschenden Vereines in Brunn, bd IV fur das Jahr 1865, Abhandlungen. 1866:3–47. [Google Scholar]
- 2.Garrod AE. Inborn errors of metabolism. 2. Oxford: Oxford University Press; 1923. [Google Scholar]
- 3.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 4.Jervell A, Lange-Nielsen F. Congenital deaf-mutism, functional heart disease with prolongation of the Q-T interval and sudden death. American Heart Journal. 1957;54:59–68. doi: 10.1016/0002-8703(57)90079-0. [DOI] [PubMed] [Google Scholar]
- 5.Romano C, Gemme G, Pongiglione R. Aritmie cardiache rare in età pediatrica. Clinical Pediatrics. 1963;45:656–683. [PubMed] [Google Scholar]
- 6.Ward OC. A new familial cardiac syndrome in children. J Irish Med Assoc. 1964;54:103–106. [PubMed] [Google Scholar]
- 7.Priori SG, Schwartz PJ, Napolitano C, Bloise R, Ronchetti E, Grillo M, et al. Risk stratification in the long-QT syndrome. The New England Journal of Medicine. 2003;348:1866–1874. doi: 10.1056/NEJMoa022147. [DOI] [PubMed] [Google Scholar]
- 8.Priori SG, Napolitano C, Schwartz PJ. Low penetrance in the long-QT syndrome: Clinical impact. Circulation. 1999;99:529–533. doi: 10.1161/01.cir.99.4.529. [DOI] [PubMed] [Google Scholar]
- 9.Collins FS, Drumm ML, Cole JL, Lockwood WK, Vande Woude GF, Iannuzzi MC. Construction of a general human chromosome jumping library, with application to cystic fibrosis. Science. 1987;235:1046–1049. doi: 10.1126/science.2950591. [DOI] [PubMed] [Google Scholar]
- 10.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, et al. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 11.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manolio TA. Genomewide association studies and assessment of the risk of disease. The New England Journal of Medicine. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 14.Milan DJ, Lubitz SA, Kaab S, Ellinor PT. Genome-wide association studies in cardiac electrophysiology: Recent discoveries and implications for clinical practice. Heart Rhythm. 2010;7:1141–1148. doi: 10.1016/j.hrthm.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nature Genetics. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 16.Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nature Genetics. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PIW, Yin X, Estrada K, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nature Genetics. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao WH, Arking DE, Post W, Rea TD, Sotoodehnia N, Prineas RJ, et al. Genetic variations in nitric oxide synthase 1 adaptor protein are associated with sudden cardiac death in US white community-based populations. Circulation. 2009;119:940–951. doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crotti L, Monti MC, Insolia R, Peljto A, Goosen A, Brink PA, et al. NOS1AP is a genetic modifier of the long-QT syndrome. Circulation. 2009;120:1657–1663. doi: 10.1161/CIRCULATIONAHA.109.879643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomas M, Napolitano C, De GL, Bloise R, Subirana I, Malovini A, et al. Polymorphisms in the NOS1AP gene modulate QT interval duration and risk of arrhythmias in the long QT syndrome. Journal of the American College of Cardiology. 2010;55:2745–2752. doi: 10.1016/j.jacc.2009.12.065. [DOI] [PubMed] [Google Scholar]
- 21.Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, et al. A drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120:553–559. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 23.Kaab S, Darbar D, van Noord C, Dupuis J, Pfeufer A, Newton-Cheh C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. European Heart Journal. 2009;30:813–819. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mommersteeg MTM, Brown NA, Prall OWJ, de Gierde Vries C, Harvey RP, Moorman AFM, et al. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circulation Research. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nature Genetics. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nature Genetics. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Body SC, Collard CD, Shernan SK, Fox AA, Liu KY, Ritchie MD, et al. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009;2:499–506. doi: 10.1161/CIRCGENETICS.109.849075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. Journal of the American College of Cardiology. 2010;55:747–753. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 30.Rowan SB, Estrada JC, Stubblefield T, Kucera G, Carter S, Roden DM, et al. A single nucleotide polymorphism at 4q25 associated with atrial fibrillation modulates symptomatic response to antiarrhythmic drug therapy. Heart Rhythm. 2009;6:PO04-7. [Google Scholar]
- 31.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Annals of Neurology. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 32.Lubitz SA, Sinner MF, Lunetta KL, Makino S, Pfeufer A, Rahman R, et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation. 2010;122:976–984. doi: 10.1161/CIRCULATIONAHA.109.886440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bezzina CR, Pazoki R, Bardai A, Marsman RF, de Jong JS, Blom MT, et al. Genome-wide association study identifies a susceptibility locus at 21q21 for ventricular fibrillation in acute myocardial infarction. Nature Genetics. 2010;42:688–691. doi: 10.1038/ng.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nature Genetics. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 35.Chambers JC, Zhao J, Terracciano CMN, Bezzina CR, Zhang W, Kaba R, et al. Genetic variation in SCN10A influences cardiac conduction. Nature Genetics. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 36.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, et al. Genome-wide association study of PR interval. Nature Genetics. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, et al. Identification of genomic predictors of atrioventricular conduction: Using electronic medical records as a tool for genome science. Circulation. 2010;122:2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotoodehnia N, Isaacs A, de Bakker PI, Dorr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nature Genetics. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. Journal of the American Medical Association. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veenstra DL, You JH, Rieder MJ, Farin FM, Wilkerson HW, Blough DK, et al. Association of vitamin K epoxide reductase complex 1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenetics and Genomics. 2005;15:687–691. doi: 10.1097/01.fpc.0000174789.77614.68. [DOI] [PubMed] [Google Scholar]
- 42.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genetics. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roden DM, Brown NJ. Preprescription genotyping: not yet ready for prime time, but getting there. Circulation. 2001;103:1608–1610. doi: 10.1161/01.cir.103.12.1608. [DOI] [PubMed] [Google Scholar]
- 45.Collins F. Opportunities and challenges for the NIH—an interview with Francis Collins. Interview by Robert Steinbrook. The New England Journal of Medicine. 2009;361:1321–1323. doi: 10.1056/NEJMp0905046. [DOI] [PubMed] [Google Scholar]
- 46.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. American Journal of Epidemiology. 2004;159:882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 47.Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen WK, et al. Familial atrial fibrillation is a genetically heterogeneous disorder. Journal of the American College of Cardiology. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 48.Ellinor PT, Yoerger DM, Ruskin JN, Macrae CA. Familial aggregation in lone atrial fibrillation. Human Genetics. 2005;118:179–184. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 49.Friedlander Y, Siscovick DS, Weinmann S, Austin MA, Psaty BM, Lemaitre RN, et al. Family history as a risk factor for primary cardiac arrest. Circulation. 1998;97:155–160. doi: 10.1161/01.cir.97.2.155. [DOI] [PubMed] [Google Scholar]
- 50.Jouven X, Desnos M, Guerot C, Ducimetiere P. Predicting sudden death in the population: The Paris Prospective Study I. Circulation. 1999;99:1978–1983. doi: 10.1161/01.cir.99.15.1978. [DOI] [PubMed] [Google Scholar]
- 51.Dekker LRC, Bezzina CR, Henriques JPS, Tanck MW, Koch KT, Alings MW, et al. Familial sudden death is an important risk factor for primary ventricular fibrillation: A case–control study in acute myocardial infarction patients. Circulation. 2006;114:1140–1145. doi: 10.1161/CIRCULATIONAHA.105.606145. [DOI] [PubMed] [Google Scholar]
- 52.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio DD, Chen DC, Nazareth L, et al. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. The New England Journal of Medicine. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohane IS, Masys DR, Altman RB. The incidentalome: A threat to genomic medicine. Journal of the American Medical Association. 2006;296:212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]