Abstract
Transforming growth factor-β-activated kinase-1 (TAK1) is a mitogen activated protein kinase kinase kinase that is involved in diverse biological roles across species. Functioning downstream of TGF-β and BMP signaling, TAK1 mediates the activation of the c-Jun N-terminal kinase signaling pathway, serves as the target of pro-inflammatory cytokines, such as TNF-α, mediates NF-κβ activation, and plays a role in Wnt/Fz signaling in mesenchymal stem cells. Expression of TAK1 in the cochlea has not been defined. Data mining of previously published murine cochlear gene expression databases indicated that TAK1, along with TAK1 interacting proteins 1 (TAB1), and 2 (TAB2), is expressed in the developing and adult cochlea. The expression of TAK1 in the developing cochlea was confirmed using RT-PCR and immunohistochemistry. Immunolabeling of TAK1 in embryonic, neonatal, and mature cochleas via DAB chromogenic and fluorescent immunohistochemistry indicated that TAK1 is broadly expressed in both the developing otocyst and periotic mesenchyme at E12.5 but becomes more restricted to specific types of supporting cells as the organ of Corti matures. By P1, TAK1 immunolabeling is found in cells of the stria vascularis, hair cells, supporting cells, and Kölliker’s organ. By P16, TAK1 labeling is limited to cochlear supporting cells. In the adult cochlea, TAK1 immunostaining is only present in the cytoplasm of Deiters’ cells, pillar cells, inner phalangeal cells, and inner border cells, with no expression in any other cochlear cell types. While the role of TAK1 in the inner ear is unclear, TAK1 expression may be used as a novel marker for specific sub-populations of supporting cells.
Keywords: cochlea, Deiters’ cell, phalangeal cell, pillar cell, development, gene expression
Introduction
Cochlear hair cells are mechanosensory cells that transduce acoustic stimuli into neural impulses. Death or dysfunction of cochlear hair cells results in sensorineural hearing loss. Stem cell or gene therapies have been applied to the treatment of hearing loss by attempting to regenerate hair cells and restore hearing (reviewed in Parker 2011; Parker and Cotanche 2004; Edge and Chen 2008; Raphael et al. 2007). Ideally, hair cell regeneration would follow the endogenous developmental pattern of cell signaling that occurs during hair cell differentiation.
Cochlear supporting cells are of interest for new approaches to hair cell regeneration because they act as hair cell progenitors in lower vertebrates and may have some capacity to differentiate into hair cells in mammals. Each hair cell in the cochlea is surrounded by nonsensory supporting cells that provide trophic (Santos-Sacchi and Dallos 1983) and structural support for the hair cells (Raphael and Altschuler 2003), trophic support for spiral ganglion neurons (Montcouquiol et al. 1998; Stankovic et al. 2004), and are essential in maintaining proper ionic concentrations in the organ of Corti (Wangemann 2006; Zdebik et al. 2009) (Fig. 1). During development, hair and supporting cells develop from a common progenitor (Driver and Kelley 2009), and the appearance of a hair cell signals surrounding cells to develop into supporting cells through contact inhibition via the Notch signaling pathway (Kelley 2006). In animals that exhibit spontaneous hair cell regeneration after damage, the death of a hair cell triggers the adjacent supporting cell either to directly transdifferentiate into a regenerated hair cell, or to undergo mitosis to produce a new supporting cell and a regenerated hair cell (Stone and Cotanche 2007). In mammals, cochlear supporting cells can be induced to develop into hair cells in certain experimental conditions such as inhibition of the Notch pathway in newborn mice (Doetzlhofer et al. 2009; Jeon et al. 2008), in vitro culture (Sage et al. 2006), or forced expression of the pro-hair cell gene Atoh1 (Kawamoto et al. 2003; Zheng and Gao 2000). However, the capacity for cochlear supporting cell-to-hair cell transdifferentiation in mammals decreases postnatally (Davies et al. 2007; Kwan et al. 2009). While significant advances have been made in the understanding of signaling pathways in cochlear and hair cell development, understanding of these pathways remains incomplete (Driver and Kelley 2009).
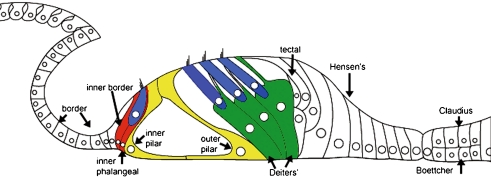
FIG. 1.
Schematic of the adult organ of Corti. The sensory epithelium of the organ of Corti consists of the mechanosensory hair cells (blue), which transduce mechanical motion of the basilar membrane into neural impulses, and their supporting cells, which provide nutritional and structural support for the hair cells. Both supporting cells and hair cells originate from a common progenitor during development, and supporting cells may act as hair cell progenitors. The inner hair cell is surrounded by an inner border cell (red) on its medial side and an inner phalangeal cell in its lateral side (red) which also extends a phalangeal process superiorly to separate one inner hair cell from the next along the length of the organ of Corti. The inner and outer pillar cells (yellow) are the first cell type to differentiate in the developing organ of Corti and provide structural support to the organ of Corti. The basal portion of each outer hair cell rests within an individual Deiters’ cell (green) which also extends a phalangeal process apically to separate one Deiters’ cell from the next along the length of the organ of Corti. During development, each of the cells types presented here exhibits TAK1 immunolabeling, however, in the adult cochlea, only the colored supporting cells label with TAK1. Image adapted from (Rio et al. 2002).
The essential role played by supporting cells in hair cell regeneration underscores a need for a more complete understanding of their biology. The identification of molecular markers that are specific to a given supporting cell sub-population may lead to a better understanding of their development and function. There are few molecular markers that identify supporting cell sub-populations, and many of the available markers are only expressed in the embryo. For example, the cell cycle inhibitor p27kip1 is expressed in many cochlear supporting cell types (Chen and Segil 1999). Prospero homeobox protein 1 (Prox1), is expressed in discrete sub-populations of supporting cells during embryonic and perinatal development; however, its expression is down-regulated in the mature cochlea (Bermingham-McDonogh et al. 2006). Finally, markers such as glutamate-aspartate transporter (GLAST) (Glowatzki et al. 2006) and the SRY-box containing gene (Sox2) (Dabdoub et al. 2008; Hume et al. 2007; Kiernan et al. 2005; Oesterle et al. 2008; Puligilla et al. 2010) are expressed in discrete populations of supporting cells in the organ of Corti, but are also expressed in other cochlear tissue as well (in this case, in the spiral ganglion).
In this paper, we report the expression of a molecular marker that specifically labels pillar cells and the supporting cells which contact the inner and outer hair cells in the adult cochlea. Tumor growth factor (TGF)-β-activated kinase-1 (TAK1) is a mitogen activated protein kinase kinase kinase (MAP3K) that is involved in a diverse range of biological functions across species (Yamaguchi et al. 1995). TAK1 is ubiquitously expressed during early embryogenesis (E3–E6) after which its expression becomes restricted to dorsal root ganglia, heart, mesenchymal cells, and epithelia (Jadrich et al. 2003). Expression of TAK1 in the cochlea has not been defined. Our results indicate that TAK1 is broadly expressed throughout the cochlear sensory epithelium, spiral limbus, spiral ganglion and stria vascularis at birth. By P16, TAK1 immunolabeling becomes restricted to cochlear supporting cells. In adult cochleas, TAK1 immunolabeling is limited to Deiters’ cells, inner phalangeal cells, and inner border cells.
Methods
Gene chip data mining Cochlear gene expression data obtained from the Gene Expression Omnibus (NCBI 2010), which is a publicly accessible MIAME-compliant gene expression database, was searched for expression of TAK1 and related cochlear markers. Gene expression in the developing otocyst was obtained from E9–E15 embryos at 0.5-day intervals as described by Sajan et al. (2007b; data accessible at Sajan et al. 2007a). Adult gene expression data were mined from an Affymetrix 430 2.0 expression database that analyzed total RNA extracted from the dissected soft tissue (including spiral ganglion) of eight normal CBA/CaJ male 9-week-old mice (data accessible at Adams 2008a, 2008b). Since there are slight differences in the version of Affymetrix gene array used between the embryonic (v. 430A 2.0) and adult tissue (v. 430 2.0), only identical probe sets were used for comparison. In both cases, the called presence/absence of gene expression was based on the published results of the respective arrays.
Animals Wild-type mice from CBA/CaJ, C57BL/6J, and B6D2F1 (C57Bl/6 DBA) backgrounds were used in these experiments. Female mice in mating cages were checked daily for the presence of a plug, which indicated embryonic day 1.
Western blot Ten organs of Corti were isolated from P0–P3 pups as previously described (Parker et al. 2010), and total protein was isolated using the following protocol. Organs of Corti were digested in 100 μl RIPA buffer, 5 μl of 0.2 M phenylmethanesulfonyl fluoride, and 1 μl of Protease Inhibitor Cocktail (all from Sigma, St Louis, MO) on ice for 30 min. Samples were then centrifuged at 8,000 rpm for 2 min at 4°C. Protein determination on the recovered supernatant was conducted using 5μL of supernatant in a protein determination assay (BioRad; Hercules, CA). Next, 1:10 NuPage Sample Reducing Agent and 1:4 NuPage LDS sample buffer (both from Invitrogen) were added to the recovered supernatant which was then boiled at 100°C for 5 min, then frozen at −20°C, and stored for later analysis. Fifteen-micrograms per lane of this total protein was loaded into a NuPage 10% bis-tris gel (Invitrogen), run for 45 min at 120 V and transferred to nitrocellulose membrane using 0.5 A for 1 h (Mini Trans-Blot Electrophoretic Transfer Cell, BioRad). Next, membranes were blocked in 10% milk in TBS-T (50 mM Tris, 150 mM NaCl, 0.05% Tween 20, HCl to pH 7.6) for 1 h at 4°C, incubated in polyclonal anti-Tak1 primary antibody (made in rabbit, Sigma) diluted 1:200 in TBS-T at 4°C overnight (ON) under agitation, washed three times for 10 min in TBS-T, incubated in HRP-conjugated mouse anti-rabbit secondary antibody (Millipore; Billerica, MA) diluted 1:2,000 in TBS-T supplemented with 1% milk for 2 h at room temperature (RT), washed again as described, incubated in 0.1 ml/cm2 Amersham ECL Western Block Detection Kit (GE Healthcare; Pittsburgh, PA) for 5 min at RT, and then analyzed using a ChemiDoc XRS illumination system equipped with a CCD camera (BioRad). To test the specificity of the TAK1 antibody, control membranes obtained from Western blotting were incubated in either 1:200 anti-Tak1 incubated 30 min with 1:1 molar concentration of a blocking peptide specific to TAK1 (CKKQLEVIRSQQQKRQGTS); or 1:200 anti-Tak1 incubated 30 min with 1:1 molar composition of a random peptide (HAVEHGFMQTLLKVTLE) to act as a negative control. An equivalent volume of dimethyl sulfoxide used to dilute the blocking peptides was added to the TAK1 primary antibody prior to incubation.
Fluorescent immunohistochemistry For E12.5 mice, whole embryos were dissected, fixed in 10% formalin plus 0.1% glutaraldehyde at 4°C ON. Heads were then dissected and processed as described below. For E16 to adult tissue, the animals were killed using CO2, decapitated, and their cochleas were dissected, round oval windows were exposed and flushed with 10% formalin plus 0.1% glutaraldehyde at 4°C ON. Next, the tissue was washed in phosphate-buffered saline (PBS), and incubated on ice in 10%, sucrose solution for 0.5 h or until the tissue sank, then in15% sucrose solution for 0.5 h or until the tissue sank, and then incubated in 20% sucrose at 4°C ON. The tissue was then incubated in 1:1 Cryo-OCT Compound (VWR; Batavia, IL) and 20% sucrose at 4°C ON. Next, the tissue was washed in 100% OCT for 10 min and then frozen in dry ice/ethanol bath, cryosectioned in 16 mM sections, and subsequently stored at −20°C. For single TAK1 labeling, sections were then processed for TAK1 immunolabeling using the fluorescent Tyramide Signal Amplification-plus System (Perkin Elmer; Waltham, MA). Sections were blocked for 30 min in PBS containing 0.5% TSA Blocking reagent (Perkin Elmer), 0.3% Triton X-100 (Sigma), and four drops per milliliter of Avidin solution (Vector Laboratories, Burlingame, CA). Next, the sections were incubated in a solution containing four drops of Biotin per milliliter PBS, 1:1,000 dilution of anti-TAK1 polyclonal antibody made in rabbit, and 1:200 dilution of either anti-Sox2, anti-Jag1, anti-myosin 7a, or anti-calbindin (described below) at 4°C ON in a humidified chamber. The sections were washed for three times for a total of 10 min in PBS containing 0.3% Triton X-100, and then incubated in this same solution supplemented with 0.1% glutaraldehyde for 5 min to reduce background staining. Next, sections were incubated in 1:200 Biotin-SP-AffiniPure goat anti-rabbit IgG (H+L) antibody (Jackson ImmunoResearch; West Grove, PA) diluted in TSA Blocking reagent for 1 h at RT in a humidified chamber. The sections were washed as described and incubated with Avidin-Biotin Complex reagent for 1 h (Jackson ImmunoResearch) at RT in a humidified chamber. The sections were washed and then incubated with either Cy5-conjugated TSA-plus reagent for 3 min at RT in a humidified chamber. Sections were washed again and then mounted using Prolong-Gold Anti-fade reagent plus DAPI (Invitrogen). Initial experiments included negative control sections that were processed in parallel and either included 1:1 molar concentrations of TAK1 blocking peptide, lacked primary antibodies, secondary antibodies, or TSA reagent. For co-labeling experiments, the tissue was incubated in Antigen Unmasking Solution as directed (Vector Laboratories), washed, and blocked in PBS supplemented with 15% normal donkey serum (NDS) plus % Triton X-100 for 1 h. Next, tissue was incubated in either 1:200 anti-Sox2 antibody (made in goat; Santa Cruz Biotechnology; Santa Cruz, CA), 1:200 anti-Jag1 antibody (made in goat; Santa Cruz Biotechnology), 1:100 anti-Myosin 7a antibody (made in mouse; Developmental Studies Hybridoma Bank; University of Iowa), or 1:200 anti-calbindin monoclonal antibody (made in mouse, Sigma) diluted in PBS supplemented with 10% NDS plus 0.1% Triton X-100 ON at 4°C. Next the tissue was washed, and then incubated in either anti-goat or anti-mouse AlexaFlour-568 diluted 1:250 in PBS supplemented with 10% NDS plus 0.1% Triton X-100 for 2 h at RT. The tissue was washed and blocked in Avidin Blocking Solution (Vector) for 30 min at RT, washed again, and then incubated in Biotin Blocking Solution (Vector) for 30 min. Next, the tissue was blocked in PBS supplemented with 15% normal goat serum (NGS) plus % Triton X-100 for 1 h, washed, and then incubated in anti-TAK1 diluted 1:200 in PBS supplemented with 10% NGS plus 0.1% Triton X-100 ON at 4°C. Tissue was washed, and incubated in Biotin-SP-AffiniPure goat anti-rabbit IgG (H+L) antibody (Jackson ImmunoResearch) diluted 1:200 in PBS supplemented with 10% NGS plus 0.1% Triton X-100 for 2 h at RT. Sections were washed, then incubated in streptavidin-647 (Invitrogen) diluted 1:750 in PBS for 15 min at RT, washed again, and then mounted using Prolong-Gold Anti-fade reagent plus DAPI. Images were recorded using an Axioskop2 mot plus Zeiss upright fluorescent microscope (Carl Zeiss; Maple Grove, MN) and controlled by Axiovision40 v4.5.0.0 software.
DAB immunohistochemistry Adult mice (older than 30 days) were anesthetized with ketamine (60 mg/kg)/xylazine (5 mg/kg), intracardially perfused with10% formalin plus 0.1% glutaraldehyde, and left in the fixative ON. Animals were then decapitated and their brains removed using blunt dissection. The cochleas were then dehydrated in graded alcohols, cleared with Xylenes Substitute Histological Clearing Reagent (Sigma), and incubated in melted paraffin ON. Blocks were solidified, sectioned in 6–10 μM sections using a rotary microtome, and stored at RT. For immunostaining, sections were deparaffinized in Xylenes, and then hydrated in a descending ethanol series. Sections were then blocked for 15 min in 5% normal horse serum (Sigma) diluted in PBS and then incubated in a 1:3,000 dilution of the anti-TAK1 antibody (Sigma) ON in a humidified chamber. Next, sections were washed three times for 10 min in PBS, and then incubated at RT in a 1:300 dilution of biotinylated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch) for 1 h in a humidified chamber. Sections were then washed in PBS as described above and incubated in Avidin-Biotin Complex reagent (Jackson ImmunoResearch) for 1 h at RT in a humidified chamber. Sections were rinsed as described and then incubated in diaminobenzidine/ hydrogen peroxide in PBS supplemented with 4% 3,3′,4,4′-biphenyltetramine,3,3′,4,4′-tetraaminobiphenyl (Sigma) for 2 to 10 min. Slides were then dehydrated in gradient alcohol washes, cleared with Xylenes, and mounted for analysis.
Reverse transcriptase-polymerase chain reaction The cochlear duct (E12.5) or organ of Corti (E 12.5, E 17.5, E 18.5, P2, and P13) were dissected in HBSS (Invitrogen) and stored in RNLater (Ambion) at −80°C until further use. Total RNA was collected using the RNAeasy Maxi Kit (Qiagen; Valencia, CA) and 1 μg of RNA was used to generate cDNA with ImProm-II reverse transcriptase (Promega; Madison, WI) and random hexamer primers (New England Biolabs; Ipswich, MA) according to Promega reverse transcription protocol. 1 μl of cDNA was employed to perform PCR using an iCycler (BioRad) with the following cycling parameters: 94°C, 30 s; 62°C, 60 s for 37 cycles. Primers against TAK1 (p1: GGGGCCACCGTAAAACCGCT; p2: GCCTTGTCGTTTCTGCTGTTGGC), glyceraldehydes-3-phosphate dehydrogenase (GAPDH) (p1: AACGGGAAGCCCATCACCATCTT; p2: CAGCCTTGGCAGCACCAGTGG) Jagged1 (p1: CAGAATGACGCTTCCTGTCG; p2: TGCAGCTGTCAATCACTTCG) and Sox2 (p1: TCCCCTTCTCCAGTTCGCAGTCCA; p2: CACCCGGGCCTCAACGCTCACG), were designed according to published sequences or newly designed with primer Blast and purchased from Integrated DNA technologies. PCR product was visualized on a 1% agarose gel and imaged with ChemiDoc XRS (BioRad). More product was loaded in from the P13 samples in order to visualize the presence of fainter bands.
Results
TAK1 expression in the cochlea
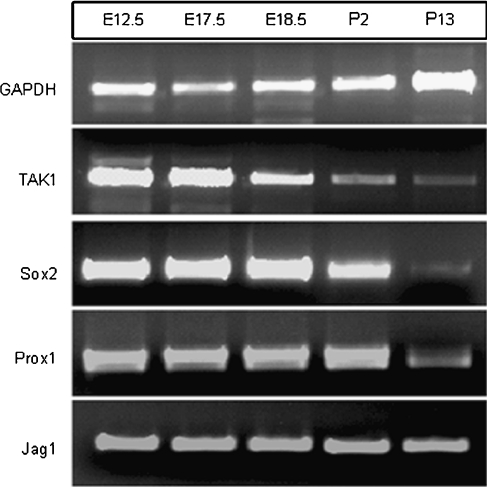
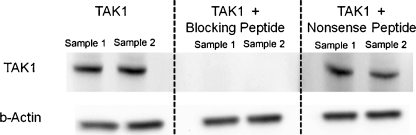
Data mining conducted on two publically accessible murine cochlear Affymetrix gene expression databases indicated that TAK1 (Gene Symbol: Map3k7) was expressed in developing otocysts as early as E9 (Sajan et al. 2007b) and continued to be expressed into adulthood (Adams 2008a) (Table 1). Interestingly, TAK1 associated binding protein-1 (TAB1; Gene Symbol: Map3k7ip1) and TAK1-associated binding protein-2 (TAB2; Gene Symbol Map3k7ip2), which modify TAK1 signaling in several tissues (Sakurai et al. 2002; Shim et al. 2005), were also expressed in both the developing and the adult murine cochlea at these stages. This suggested that known modulators of the TAK1 signaling pathway were present in the cochlea as well. The expression patterns of known cochlear markers were also examined as controls. As expected, Notch 1 and Sox2 were expressed at all stages and p27kip1 and myosin 7a were upregulated in the developed cochlea. To confirm the gene chip findings, TAK1 expression in the cochlea was measured using reverse transcriptase-polymerase chain reaction (RT-PCR) and Western analysis. RT-PCR results indicated that similar to Sox2 and Prox1 expression, TAK1 was expressed at higher levels at E12.5 and that its expression decreased as the cochlea matures (Fig. 2). Western analysis confirmed that TAK1 protein was expressed in the cochleas of P0 mice and that TAK1 immunolabeling could be blocked using a synthetic peptide directed against the antibody which confirmed the specificity of the TAK1 antibody (Fig. 3).
TABLE 1.
Summary of TAK1 gene expression in the cochlea during development
| Gene title | Gene symbol | Affymetrix probe set ID | Developmental age | |||
|---|---|---|---|---|---|---|
| E9 | 12.5 | E15 | Adult | |||
| TAK1 | MAP3K7 | 1419988_at | P | P | P | P |
| TAK1 associated binding protein-1 | TAB1 | 1426898_at | 1/2 | P | P | P |
| TAK1 associated binding protein-2 | TAB2 | 1423462_at | P | P | P | P |
| Notch gene homolog 1 | Notch1 | 1418634_at | P | P | P | P |
| SRY-box containing gene 2 | SOX2 | 1416967_at | P | P | P | P |
| Cyclin-dependent kinase inhibitor 1B (p27kip1) | CDKN1B | 1419497_at | A | P | P | P |
| Myosin 7a | MYO 7A | 1421385_a_at | A | A | A | P |
Two publically accessible Affymetrix microarray data obtained from the developing and adult cochlea were mined for expression of TAK1. Gene expression data obtained from CBA/J murine otocyst dissections at E9–E15 were searched for the expression of TAK1 and associated markers (Sajan et al. 2007a). P = called present in two replicates of a given age, 1/2 = called present in one replicate but absent in the second, A = called absent in both replicates. Gene expression in the adult was obtained from cochleas isolated from eight male wild-type CBA/CaJ 9-week-old mice (Adams 2008b). P = called present in all eight samples
FIG. 2.
TAK1 expression in the cochlea decreases during development. Total RNA was extracted from the developing cochlear ducts of prenatal and postnatal mice and assayed for TAK1 expression using RT-PCR. Similar to Sox2 and Prox1, TAK1 expression in the cochlea is relatively higher in the embryonic stages (E12.5–E17.5) and decreases as the cochlea matures. By P13, TAK1 was expressed in relatively lower levels in the cochlea.
FIG. 3.
TAK1 protein expression in the organ of Corti. Western blot analysis of total protein isolated from ten neonatal organs of Corti indicated that TAK1 protein was expressed in this tissue (left column). In order to test the antibody specificity, the TAK1 antibody was incubated with 1:1 molar concentration of TAK1 blocking peptide (middle column), or nonsense peptide (right column). Incubation of the TAK1 primary antibody TAK1-specific blocking eliminated the signal, whereas 1:1 incubation with nonsense peptide produced no change in TAK1 antibody labeling. Samples 1 and 2 were obtained from repeat experiments.
TAK1 immunolabeling in the cochlea
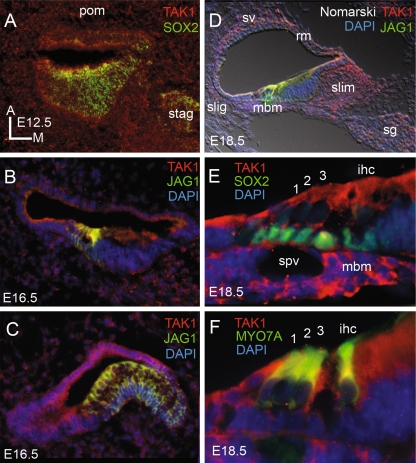
In order to identify which cell types expressed TAK1, cochleas were obtained from embryonic (E12.5–E18) neonatal (P0–3), adolescent (P16), and adult (P30+) mice and were immunolabeled for TAK1. The results of TAK1 immunolabeling in the murine cochlea are summarized in Table 2. Between embryonic days 12.5 and 16.5, low levels of TAK1 immunolabeling was present in the otocyst, statoacoustic ganglion, and periotic mesenchyme (Fig. 4). Double labeling with Sox2, which labels the developing otocyst and statoacoustic ganglion (Kiernan et al. 2005; Mak et al. 2009; Puligilla et al. 2010), confirmed TAK1 expression in this tissue. At E16.5, TAK1 immunolabeling was also present in all cell types of the developing vestibular system and periotic mesenchyme. Double labeling with Jagged1 (Jag1), which labels the developing prosensory domain of the developing otocyst, and Sox2 which additionally labels the developing statoacoustic ganglion indicated that TAK1 was expressed in each of these cell types.
TABLE 2.
Summary of developmental changes in TAK1 immunolabeling in the murine cochlea
| Developmental age | TAK1-positive cell types |
|---|---|
| Embryonic day 12.5 | Periotic mesenchyme |
| Otocyst (ubiquitous) | |
| Embryonic day 16.5 | Periotic mesenchyme |
| Cochlear duct (ubiquitous) | |
| Vestibular duct (ubiquitous) | |
| Embryonic day 18.5 | Spiral ganglion |
| Spiral limbus | |
| Spiral ligament | |
| Stria vascularis Reissner’s membrane | |
| Mesenchyme of the basilar membrane | |
| Vestibular hair and supporting cells | |
| Postnatal Day1 | Stria vascularis |
| Boettcher cells Henson's cells | |
| Tectal cells | |
| Pillar cells | |
| Hair cells | |
| Kölliker’s organ | |
| Mesenchymal cells of the basilar membrane | |
| Reissner’s membrane | |
| Vestibular hair and supporting cells (weak) | |
| Postnatal day 16 | Claudius cells |
| Henson's cells | |
| Deiters’ cells | |
| Inner phalangeal cells | |
| Border cells | |
| Inner sulcus cells (weak) | |
| Adult | Base: (strong) Deiters’ cells |
| Base: (weak) inner pillar inner phalangeal, inner border cells | |
| Mid-turn: Deiters’ cells, inner and outer pillar cells, inner phalangeal cell, inner boarder cells | |
| Apex: (weak) Deiters’ cells, outer pillar cell | |
| Apex: (strong) inner pillar cell, inner phalangeal cell, inner border cell |
This table lists the cell types that exhibited TAK1 immunolabeling at various developmental stages of the cochlea
FIG. 4.
Ubiquitous TAK1 immunolabeling in the developing cochlea. A At E12.5, TAK1 immunolabeling (red) was observed in all cells types in the developing otocyst and statoacoustic ganglions (stag), both of which co-label (yellow) for Sox2 (green), as well as the surrounding periotic mesenchyme (pom). At E16.5, TAK1 immunolabeling is still present in these tissues as well as the developing sensory epithelium of the cochlea (B) and vestibular system (C), which co-label (yellow) for Jag1 (green). D By E18.5, TAK1 labeling (red) becomes more restricted to the cochlea and is present in the spiral ganglion (sg), spiral limbus (slim), mesenchymal cells on the basal surface of the basilar membrane (mbm), spiral ligament (slig), stria vascularis (sv), and Reissner’s membrane (rm). Higher magnification of the organ of Corti shows TAK1 immunolabeling (red) in mbm and spiral vesicle (spv), and in both supporting cells (E), which co-label with sox2 (green), and hair cells (F), which co-label (yellow) with Myo7a (green). TAK1 immunolabeling seen in the cytoplasm of both inner and outer hair cells and in the developing streocilia of the outer hair cells at this stage. ihc inner hair cell; 1, 2, 3, rows of outer hair cells. DAPI, blue.
At E18, there was a decrease in TAK1 immunolabeling in the surrounding tissue and bone, but labeling was retained in the developing stria vascularis, spiral limbus, spiral ganglion, and sensory epithelia of the cochlea (Fig. 4). Bright TAK1 immunolabeling was also seen in the mesenchymal cells located below the basilar membrane and surrounding the developing spiral vesicle. Double labeling with myosin 7a and Sox2 at this age indicated that TAK1-labeled hair cells and supporting cells respectively. TAK1 immunolabeling was also present in both hair and supporting cells of the vestibular system at this age (data not shown).
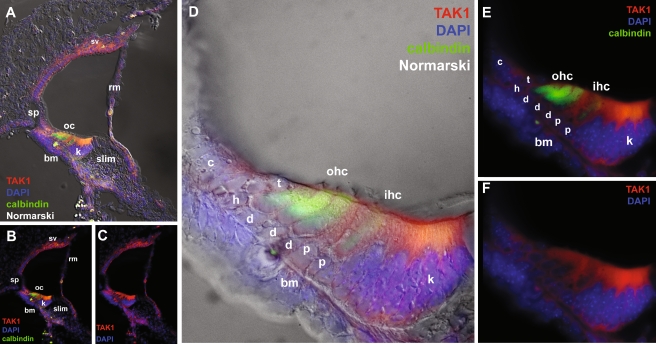
At postnatal day 1 (P1), TAK1 immunolabeling was reduced in the spiral ganglion, spiral limbus, and vestibular sensory epithelia, however labeling was observed in many cell types that border the scala media (Fig. 5). TAK1 labeling was seen in the stria vascularis, Claudius cells, Boettcher cells, mesenchymal cells of the basilar membrane and basement membrane of the spiral limbus , and Reissner’s membrane. In the developing organ of Corti region at this stage, all cell types were stained for TAK1 including Hensen’s cells, tectal cells, outer and inner hair cells, Deiters’ cells, and pillar cells. However, the strongest TAK1 immunolabeling was observed in Kölliker’s organ (also known as the greater epithelial ridge) consisting of multivillous columnar epithelial cells which fill the space that will develop into the inner sulcus (Hinojosa 1977; Kikuchi and Hilding 1965).
FIG. 5.
TAK1 immunostaining in the P1 cochlea. A–C At P1, the intensity of TAK1 immunostaining increases in the organ of Corti and is observed in many cell types bordering the scala media including the stria vascularis (sv), and Reissner’s membrane (rm). D–F Higher magnification of the organ of Corti. In the sensory epithelia, strong TAK1 immunostaining was seen in Kolliker’s organ (k) and progressively weaker immunostaining was seen in the inner hair cells (ihc) and outer hair cells (ohc) that also label with calbindin, pillar cells (p), Deiters’ cells (d), Hensen’s cells (h), and tectal cells (t). No labeling is seen in the spiral prominence cells (sp) and Claudius cells (c) at this stage. Cell nuclei are counterstained with DAPI (blue). bm basilar membrane. The gamma was adjusted in these images in order to reduce nonspecific background autofluorescence.
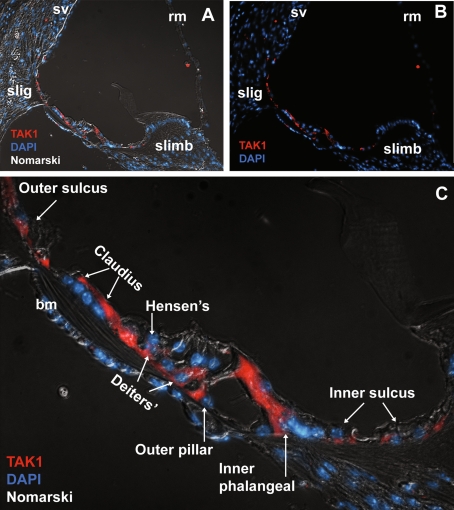
At P 16, which is the approximate time of functional maturity of the organ of Corti in mice (Alford and Ruben 1963; Pujol and Hilding 1973; Tritsch et al. 2007), TAK1 immunolabeling was limited to cochlear supporting cells (Fig. 6). Weak TAK1 labeling was observed in the inner sulcus cells, the inner border cells which contact the medial surfaces of each hair cell (Zhang 1999), inner phalangeal cells which are located beneath the inner hair cells (Hartman et al. 2009) and separate the inner hair cells from the pillar cells (Zhang 1999), inner and outer pillar cells, Deiters’ cells, Hensen’s cells, tectal cells (data not shown) and Claudius cells. Notably, TAK1 immunolabeling was down-regulated in the stria vascularis, Reissner’s membrane, and hair cells.
FIG. 6.
Down-regulation of TAK1 in the P16 cochlea. A, B By P16, TAK1 immunostaining was down-regulated in the stria vascularis, Reissner’s membrane, spiral limbus and both inner and outer hair cells. TAK1 labeling was present in the fibrocytes in the spiral ligament (sl) and supporting cells of the organ of Corti. C Higher magnification reveals that TAK1 immunostaining was limited to the supporting cells of the organ of Corti as well as the Claudius cells. TAK1 immunolabeling was also seen in the cells of the inner and outer sulcus. Cell nuclei are counterstained with DAPI (blue). The gamma was adjusted in these images in order to reduce nonspecific background autofluorescence.
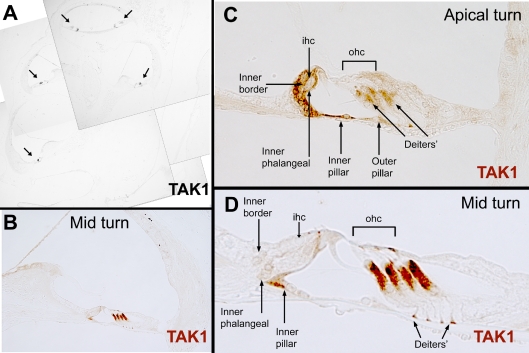
In the adult cochlea (P30+), definitive TAK1 immunolabeling was limited to specific sub-populations of supporting cells (Figs. 7 and 8). TAK1 immunolabeling varied slightly along the basal-to-apical axis of the organ. In the basal turns of the organ of Corti, the Deiters’ cells exhibited strong TAK1 labeling and weaker labeling was observed in the inner pillar cells, inner phalangeal cells, and inner border cells surrounding the inner hair cell. The reverse pattern of labeling intensity appeared in the apex, where inner border, inner phalangeal, and inner pillar cells exhibited strong TAK1 labeling, and Deiters’ cells exhibited weaker staining. In addition to those cells labeled in the base, apical outer pillar cells exhibited a low level of TAK1 immunolabeling. TAK1 was expressed in a gradient fashion in these cells along the length of the cochlea. The mid-turns of the cochlea exhibited strong and relatively uniform labeling of these cells.
FIG. 7.
Limited expression of TAK1 in the adult cochlea. A In the adult cochlea, TAK1 (DAB, black, arrows) is immunostained in discrete cell types within the organ of Corti. B Higher magnification shows that TAK1 (DAB, brown) is selectively immunostained in the pillar cells and supporting cells adjacent to the hair cells. The intensity of TAK1 immunolabeling in these cell changes along the length of the organ of Corti. C In the apical turn, TAK1 immunostaining is stronger in the inner border and inner phalangeal cells and weaker in the Deiters’ cells. Additionally, there is weak labeling of the outer pillar cells in the apical turn. D In the low and mid-turns of the cochlea, TAK1 immunostaining is strong in the Deiters’ cells, and weak in the inner pillar, inner phalangeal, and inner border cells. D Higher magnification of (B).
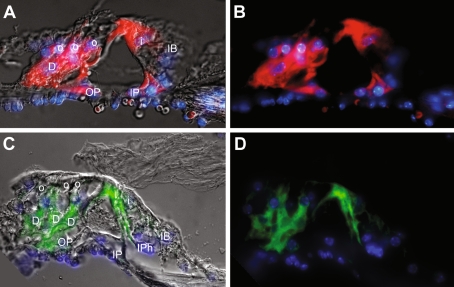
FIG. 8.
TAK1 labels Deiters’ cells, pillar cells, inner phalangeal cells, and inner border cells in the adult cochlea. A, B Apical sections of an adult cochlea show that TAK1 (Cy3, red) labels supporting cells that contact the inner (i) and outer hair cells (o). TAK1 immunostaining is seen in the Deiters’ cells (D), outer pillar cells (OP), inner pillar cells (IP), and inner border cell (IB). Red blood cells beneath the basilar membrane exhibit non-specific autofluorescence. The image in (A) is the same image as in (B) with a DIC overlay. C, D TAK1 immunostaining (AlexaFluor 488, green) is observed in inner phalangeal cells (IPh) adjacent to an inner hair cell, inner pillar cells, inner border cells, and Deiters’ cells. The image in (C) is the same image as in (D) with a DIC overlay. Cell nuclei are counterstained with DAPI (blue). The gamma was adjusted in these images in order to reduce nonspecific background autofluorescence.
Discussion
These results indicate that TAK1 is expressed in the cochlea and that its expression pattern changes as the cochlea matures. TAK1 is broadly expressed in both the developing otocyst and periotic mesenchyme in the murine embryo. However, TAK1 expression decreases in the periotic mesenchyme and remains in the statoacoustic ganglion, and sensory epithelium of the developing inner ear by E18.5. Discrete populations of supporting cells in the neonatal, adolescent, and adult cochlea exhibit TAK1 immunostaining (Table 1). By adulthood, TAK1 labeling is limited to the pillar cells and the supporting cells in direct contact with inner and outer hair cells. Furthermore, the intensity of TAK1 labeling exhibits a base-to-apex gradient, with a relatively stronger immunolabeling in Deiters’ cells in the basal turns compared with the apical turn. In comparison, the inner phalangeal and inner border cells exhibit a relatively weaker TAK1 labeling in the basal turn and an increased immunolabeling in the apical turn. In the apical turns, the apical portion of the inner hair cells appears to weakly immunolabel for TAK1; however, this signal is attributed to the adjacent inner border and inner phalangeal cells. In addition, the cuticular plate and cilia of both inner and outer hair cells may also weakly label for TAK1; however, this staining is pattern is ambiguous. Finally, TAK1 labels both hair cells and supporting cells in the embryonic vestibular system. However, vestibular labeling decreases into adulthood. TAK1 labeling completely absent in vestibular hair cells at this age and is so faint in as to render its expression as ambiguous.
The specific expression pattern of TAK1 in adulthood suggests that it may be a valuable molecular marker for the cochlear supporting cells that contact hair cells. Given the heterogeneous morphology of the supporting cell population, there are surprisingly few molecular markers that are specifically expressed in a given supporting cell type in the adult animal. For example, the commonly used supporting cell markers p27kip1 (Chen and Segil 1999), Notchicd (Lanford et al. 1999), Musashi (Murata et al. 2004), Sox2 (Oesterle et al. 2008), and Cx26 (Lautermann et al. 1998) are ubiquitously expressed in cochlear supporting cells. Additionally, there are markers such as GLAST (Glowatzki et al. 2006), TrkC (Glikeria et al. 1999), p75NGFR (Glikeria et al. 1999), and GFAP (Rio et al. 2002) that label specific sub-populations of supporting cells in the organ of Corti, but also label other cell types in the cochlea such as the spiral limbus, spiral ligament, or cells within the spiral ganglion. Finally, there are markers that are expressed in sub-populations of cochlear supporting cells during embryonic development, but are down-regulated in the adult animal. For example, Prox1 is differentially expressed in Deiters’ and pillar cells during late embryonic development (E17) (Bermingham-McDonogh et al. 2006). However, Prox1 is down-regulated in the mouse by the time the cochlea reaches functional maturity at P14. Similarly, members of the Notch signaling pathway Notch1, Jagged 1, Hes 1, and Hes 5 are expressed in specific sub-populations of supporting cells during development, but their expression diminishes in the adult organ of Corti (Batts et al. 2009). The work presented here documents a novel molecular marker that selectively labels sub-populations of cochlear supporting cells in the adult mammal.
The specificity of TAK1 expression may prove useful for studying hearing loss and treatment methods using gene targeting. The TAK1 promoter could be used to drive the expression of pro-hair cell or other genes-of-interest in specific supporting cell populations. For example, it has been shown that virally mediated delivery of the pro-hair cell gene Atoh1 into the damaged guinea pig cochlea results in the ectopic expression of hair cells (Izumikawa et al. 2005; Kawamoto et al. 2003). In order to reduce the numbers of ectopic hair cells produced by Atoh1 infection in the adult animal, the Atoh1 gene could be placed under control of the TAK1 promoter. In such a system, only those cells that express TAK1 (i.e. directly beneath the hair cells) would express Atoh1 and potentially transdifferentiate into hair cells even though the virus would infect all cochlear supporting cells. Preliminary studies to test the strength of the TAK1 promoter would have to be undertaken to ascertain the effectiveness of such a system. If proven effective, the TAK1 promoter could be used to control the expression of other genes-of-interest in these specific supporting cell populations as well.
Additionally, this finding may lead to a greater understanding of supporting cell biology by utilizing conditional gene targeting experiments in knock-out mice. Previous experiments have shown that targeted deletions of TAK1 are lethal at E10.5–12 (Jadrich et al. 2006; Sato et al. 2005; Shim et al. 2009, 2005), primarily due to deficient vascular development. These TAK1 knock-out animals may provide information relating to the role of TAK1 in the development of the early otocyst. Additionally, conditional TAK1 knock-out animals have been engineered by crossing transgenic mice containing a floxed TAK1 allele with a transgenic mouse containing the Cre-recombinase gene under the control of promoters for Mx1 (mesenchymal stem cell marker) (Tang et al. 2008), K5 (keratinocyte marker) and collagen 2 (cartilage marker) (Shim et al. 2009). These experiments have described the functional roles of TAK1 in later developmental stages by conditional deletion of TAK1 in late embryonic and adult mice. Similar to these experiments, the floxed TAK1 mice could be crossed with mice expressing the CRE-recombinase gene under control of supporting cell markers such as Prox1 (Fritzsch et al. 2010) or GLAST (Mori et al. 2006) in order to examine the role of TAK1 in cochlear supporting cells during development and adulthood.
The function of TAK1 in these cells remains to be determined. However, given that TAK1 is activated by several ligands that are present in the cochlea, such as BMP, tumor necrosis factor (TNF)-α, Wnt, and TGF-β, there are several signaling pathways in which TAK1 could be involved. Additionally, the particular signaling pathway and role of TAK1 in a given cell type may change as the organ matures.
One possibility is that TAK1 could play a role in cell cycle regulation in the cochlea through TGF-β signaling. TGF-β is expressed in the developing sensory epithelia and mesenchymal cells of the E10–18.5 otocyst (Schmid et al. 1991). Furthermore, TGF-β signaling in avian cochlear supporting cell proliferation has been reported, albeit with opposing functions. Warchol (1999) reported that TGF-β1 inhibited supporting cell proliferation in cultured avian utricles. However, more recently it has been reported that activin A, which is a ligand for the TGF-β receptors Acvr2a and Acvr2b, acts through downstream Smad2 signaling to potentiate supporting cell proliferation in the avian cochlea (McCullar, et al. 2010). The apparent discrepancy between the proliferative and inhibitory effects of TGF-β may be attributed to the numerous ligands, receptors, and signaling cascades included in the TGF-β superfamily. The anti-proliferative effects of TGF-β signaling in epithelial cells been shown to act through a p27kip1-mediated pathway that leads to cell cycle arrest at G1 (Toyoshima and Hunter 1994). Since TAK1 is activated by TGF-β (Yamaguchi et al. 1995), further experiments could be conducted to determine whether TAK1 mediates the p27kip1 signaling in the developing or adult cochlea. In contrast, the proliferative effects of TGF-β in the chick cochlea may be attributed to signaling through the activin A/Acvr2a/Smad2 pathway described by McCullar, et al. (2010). Interestingly, this proliferative TGF-β pathway only appears to be present in supporting cells of organs that exhibit hair cell regeneration, such as mouse vestibular epithelia and chick inner ear tissue, and is absent in supporting cells of the adult mouse cochlea that doesn’t exhibit robust proliferation. Since TAK1 has been shown to inhibit the nuclear accumulation of activated Smad2 (Hoffmann et al. 2005), would be interesting to determine whether TAK1 plays an inhibitory role in TGF-β proliferative signaling in the adult mammalian cochlea.
Another possible role of TAK1 is that it may be involved in the inflammatory response within the cochlea after ototoxic insult. Pro-inflammatory cytokines, such as IL1 and TNF-α induce both pro-apoptotic and anti-apoptotic signaling in target cells depending upon the activation of TAK1 (Tang et al. 2008). In the cochlea, TNF-α is expressed in response to noise overstimulation (Fujioka et al. 2006), cisplatin treatment (So et al. 2008), or mechanical trauma (Zou et al. 2005). The specific expression of TAK1 in supporting cells suggests that it may be involved in the survival of these cells after ototoxin induced apoptosis of hair cells. However, more work is required in order to identify the functional role of TAK1 in the cochlea.
Acknowledgements
The authors would like to thank Amy Osborne for her artwork and the National Institute of Deafness and other Communicative Disorders (R03DC010065-Parker; RO1DC007174-Edge; P30DC05209-MEEI Core Support for Hearing Research) for supporting this work.
References
- Adams, J (2008a) CBA/CaJ mouse cochlea gene expression profile. Retrieved from the National Center for Biotechnology Information. Available at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13421. Accessed 22 November 2010
- Adams, J (2008b) Genes expressed in the mouse cochlea. Retrieved from the Massachusetts Eye & Ear Infirmary. Available at: http://research.meei.harvard.edu/Otopathology/tbimages/mouse.html. Accessed 22 November 2010
- Alford BR, Ruben RJ. Physiological, behavioral and anatomical correlates of the development of hearing in the mouse. Ann Otol Rhinol Laryngol. 1963;72:237–247. doi: 10.1177/000348946307200119. [DOI] [PubMed] [Google Scholar]
- Batts SA, Shoemaker CR, Raphael Y. Notch signaling and Hes labeling in the normal and drug-damaged organ of Corti. Hear Res. 2009;249(1–2):15–22. doi: 10.1016/j.heares.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM, Hayashi T. Expression of Prox1 during mouse cochlear development. J Comp Neurol. 2006;496(2):172–186. doi: 10.1002/cne.20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Segil N. p27(Kip1) links cell proliferation to morphogenesis in the developing organ of Corti. Development. 1999;126(8):1581–1590. doi: 10.1242/dev.126.8.1581. [DOI] [PubMed] [Google Scholar]
- Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci USA. 2008;105(47):18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D, Magnus C, Corwin JT. Developmental changes in cell extracellular matrix interactions limit proliferation in the mammalian inner ear. Eur J Neurosci. 2007;25(4):985–998. doi: 10.1111/j.1460-9568.2007.05355.x. [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A, Basch ML, Ohyama T, Gessler M, Groves AK, Segil N. Hey2 regulation by FGF provides a Notch-independent mechanism for maintaining pillar cell fate in the organ of Corti. Dev Cell. 2009;16(1):58–69. doi: 10.1016/j.devcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driver EC, Kelley MW. Specification of cell fate in the mammalian cochlea. Birth Defects Res C Embryo Today. 2009;87(3):212–221. doi: 10.1002/bdrc.20154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge AS, Chen ZY. Hair cell regeneration. Curr Opin Neurobiol. 2008;18(4):377–382. doi: 10.1016/j.conb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Dillard M, Lavado A, Harvey NL, Jahan I. Canal cristae growth and fiber extension to the outer hair cells of the mouse ear require Prox1 activity. PLoS ONE. 2010;5(2):e9377. doi: 10.1371/journal.pone.0009377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Kanzaki S, Okano HJ, Masuda M, Ogawa K, Okano H. Proinflammatory cytokines expression in noise-induced damaged cochlea. J Neurosci Res. 2006;83(4):575–583. doi: 10.1002/jnr.20764. [DOI] [PubMed] [Google Scholar]
- Glikeria G, Barbara W, Ulrike Z, Mark P, Karin R, Iris K, et al. Differential expression of trkB.T1 and trkB.T2, truncated trkC, and p75NGFR in the cochlea prior to hearing function. J Comp Neurol. 1999;414(1):33–49. doi: 10.1002/(SICI)1096-9861(19991108)414:1<33::AID-CNE3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Glowatzki E, Cheng N, Hiel H, Yi E, Tanaka K, Ellis-Davies GCR, et al. The glutamate-aspartate transporter GLAST mediates glutamate uptake at inner hair cell afferent synapses in the mammalian cochlea. J Neurosci. 2006;26(29):7659–7664. doi: 10.1523/JNEUROSCI.1545-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman BH, Basak O, Nelson BR, Taylor V, Bermingham-McDonogh O, Reh TA. Hes5 expression in the postnatal and adult mouse inner ear and the drug-damaged cochlea. J Assoc Res Otolaryngol. 2009;10(3):321–340. doi: 10.1007/s10162-009-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinojosa R. A note on development of Corti's organ. Acta Otolaryngol. 1977;84(3–4):238–251. doi: 10.3109/00016487709123963. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Preobrazhenska O, Wodarczyk C, Medler Y, Winkel A, Shahab S, et al. Transforming growth factor-I-activated kinase-1 (TAK1), a MAP3K, interacts with smad proteins and interferes with osteogenesis in murine mesenchymal progenitors. J Biol Chem. 2005;280(29):27271–27283. doi: 10.1074/jbc.M503368200. [DOI] [PubMed] [Google Scholar]
- Hume CR, Bratt DL, Oesterle EC. Expression of LHX3 and SOX2 during mouse inner ear development. Gene Expr Patterns. 2007;7(7):798–807. doi: 10.1016/j.modgep.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005;11(3):271–276. doi: 10.1038/nm1193. [DOI] [PubMed] [Google Scholar]
- Jadrich JL, O'Connor MB, Coucouvanis E. Expression of TAK1, a mediator of TGF-[beta] and BMP signaling, during mouse embryonic development. Gene Expr Patterns. 2003;3(2):131–134. doi: 10.1016/S1567-133X(03)00012-7. [DOI] [PubMed] [Google Scholar]
- Jadrich JL, O'Connor MB, Coucouvanis E. The TGF beta activated kinase TAK1 regulates vascular development in vivo. Development. 2006;133(8):1529–1541. doi: 10.1242/dev.02333. [DOI] [PubMed] [Google Scholar]
- Jeon S-J, Fujioka M, Edge A (2008) Inhibition of notch signaling increases differentiation of stem cells to hair cells through upregulation of math1. Paper presented at the Association for Research in Otolaryngology Midwinter Meeting. Available at: http://www.aro.org/archives/2008/2008_461_52c7a7f5.html
- Kawamoto K, Ishimoto S-I, Minoda R, Brough DE, Raphael Y. Math1 gene transfer generates new cochlear hair cells in mature guinea pigs in vivo. J Neurosci. 2003;23(11):4395–4400. doi: 10.1523/JNEUROSCI.23-11-04395.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MW. Hair cell development: commitment through differentiation. Brain Res. 2006;1091(1):172–185. doi: 10.1016/j.brainres.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KKH, Tang ASP, Bell DM, Tease C, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Hilding D. The development of the organ of Corti in the mouse. Acta Otolaryngol. 1965;60(3):207–222. doi: 10.3109/00016486509127003. [DOI] [PubMed] [Google Scholar]
- Kwan T, White PM, Segil N. Development and regeneration of the inner ear. Ann NY Acad Sci. 2009;1170:28–33. doi: 10.1111/j.1749-6632.2009.04484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, et al. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21(3):289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- Lautermann J, Cate WJ, Altenhoff P, Grummer R, Traub O, Frank H, et al. Expression of the gap-junction connexins 26 and 30 in the rat cochlea. Cell Tissue Res. 1998;294(3):415–420. doi: 10.1007/s004410051192. [DOI] [PubMed] [Google Scholar]
- Mak AC, Szeto IY, Fritzsch B, Cheah KS. Differential and overlapping expression pattern of SOX2 and SOX9 in inner ear development. Gene Expr Patterns. 2009;9(6):444–453. doi: 10.1016/j.gep.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullar JS, Ty S, Campbell S, Oesterle EC. Activin potentiates proliferation in mature avian auditory sensory epithelium. J Neurosci. 2010;30(2):478–490. doi: 10.1523/JNEUROSCI.5154-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Valat J, Travo C, Sans A. A role for BDNF in early postnatal rat vestibular epithelia maturation: implication of supporting cells. Eur J Neurosci. 1998;10(2):598–606. doi: 10.1046/j.1460-9568.1998.00070.x. [DOI] [PubMed] [Google Scholar]
- Mori T, Tanaka K, Buffo A, Wurst W, Kuhn R, Gotz M. Inducible gene deletion in astroglia and radial glia—a valuable tool for functional and lineage analysis. Glia. 2006;54(1):21–34. doi: 10.1002/glia.20350. [DOI] [PubMed] [Google Scholar]
- Murata J, Murayama A, Horii A, Doi K, Harada T, Okano H, et al. Expression of Musashi1, a neural RNA-binding protein, in the cochlea of young adult mice. Neurosci Lett. 2004;354(3):201–204. doi: 10.1016/j.neulet.2003.10.036. [DOI] [PubMed] [Google Scholar]
- NCBI (2010). Gene expression omnibus. Publication. Retrieved from the National Center for Biotechnology Information. Available at: http://www.ncbi.nlm.nih.gov/geo/. Accessed 9 November2010
- Oesterle EC, Campbell S, Taylor RR, Forge A, Hume CR. Sox2 and JAGGED1 expression in normal and drug-damaged adult mouse inner ear. J Assoc Res Otolaryngol. 2008;9(1):65–89. doi: 10.1007/s10162-007-0106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MA (2011) Biotechnology in the treatment of hearing loss: foundations and Future of hair cell regeneration. Journal of Speech, Lanuage and Hearing Research (in press) [DOI] [PMC free article] [PubMed]
- Parker MA, Cotanche DA. The potential use of stem cells for cochlear repair. Audiol Neurootol. 2004;9(2):72–80. doi: 10.1159/000075998. [DOI] [PubMed] [Google Scholar]
- Parker M, Brugeaud A, Edge AS. J Vis Exp. (2010) Feb 4; Primary culture and plasmid electroporation of the murine organ of Corti. (36). pii: 1685. doi:10.3791/1685 [DOI] [PMC free article] [PubMed]
- Pujol R, Hilding D. Anatomy and physiology of the onset of auditory function. Acta Otolaryngol. 1973;76(1):1–10. doi: 10.3109/00016487309121476. [DOI] [PubMed] [Google Scholar]
- Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci. 2010;30(2):714–722. doi: 10.1523/JNEUROSCI.3852-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raphael Y, Altschuler RA. Structure and innervation of the cochlea. Brain Res Bull. 2003;60(5–6):397–422. doi: 10.1016/S0361-9230(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Raphael Y, Kim YH, Osumi Y, Izumikawa M. Non-sensory cells in the deafened organ of Corti: approaches for repair. Int J Dev Biol. 2007;51(6–7):649–654. doi: 10.1387/ijdb.072370yr. [DOI] [PubMed] [Google Scholar]
- Rio C, Dikkes P, Liberman MC, Corfas G. Glial fibrillary acidic protein expression and promoter activity in the inner ear of developing and adult mice. J Comp Neurol. 2002;442(2):156–162. doi: 10.1002/cne.10085. [DOI] [PubMed] [Google Scholar]
- Sage C, Huang M, Vollrath MA, Brown MC, Hinds PW, Corey DP, et al. Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc Natl Acad Sci. 2006;103(19):7345–7350. doi: 10.1073/pnas.0510631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajan SA, Warchol ME, Lovett M (2007a) Microarray expression profiling of discrete microdissected mouse inner ear tissues from E9 to E15 at half-day intervals. Retrieved from National Center for Biotechnology Information. Available at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7536. Accessed 23 November 2010
- Sajan SA, Warchol ME, Lovett M. Toward a systems biology of mouse inner ear organogenesis: gene expression pathways, patterns and network analysis. Genetics. 2007;177(1):631–653. doi: 10.1534/genetics.107.078584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Nishi A, Sato N, Mizukami J, Miyoshi H, Sugita T. TAK1-TAB1 fusion protein: a novel constitutively active mitogen-activated protein kinase kinase kinase that stimulates AP-1 and NF-[kappa]B signaling pathways. Biochem Biophys Res Commun. 2002;297(5):1277–1281. doi: 10.1016/S0006-291X(02)02379-3. [DOI] [PubMed] [Google Scholar]
- Santos-Sacchi J, Dallos P. Intercellular communication in the supporting cells of the organ of Corti. Hear Res. 1983;9(3):317–326. doi: 10.1016/0378-5955(83)90034-5. [DOI] [PubMed] [Google Scholar]
- Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6(11):1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- Schmid P, Cox D, Bilbe G, Maier R, McMaster GK. Differential expression of TGF beta 1, beta 2 and beta 3 genes during mouse embryogenesis. Development. 1991;111(1):117–130. doi: 10.1242/dev.111.1.117. [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, et al. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19(22):2668–2681. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JH, Greenblatt MB, Xie M, Schneider MD, Zou W, Zhai B, et al. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28(14):2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So H, Kim H, Kim Y, Kim E, Pae HO, Chung HT, et al. Evidence that cisplatin-induced auditory damage is attenuated by downregulation of pro-inflammatory cytokines via Nrf2/HO-1. J Assoc Res Otolaryngol. 2008;9(3):290–306. doi: 10.1007/s10162-008-0126-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovic K, Rio C, Xia A, Sugawara M, Adams JC, Liberman MC, et al. Survival of adult spiral ganglion neurons requires erbB receptor signaling in the inner ear. J Neurosci. 2004;24(40):8651–8661. doi: 10.1523/JNEUROSCI.0733-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JS, Cotanche DA. Hair cell regeneration in the avian auditory epithelium. Int J Dev Biol. 2007;51(6–7):633–647. doi: 10.1387/ijdb.072408js. [DOI] [PubMed] [Google Scholar]
- Tang M, Wei X, Guo Y, Breslin P, Zhang S, Wei W, et al. TAK1 is required for the survival of hematopoietic cells and hepatocytes in mice. J Exp Med. 2008;205(7):1611–1619. doi: 10.1084/jem.20080297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78(1):67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- Tritsch NX, Yi E, Gale JE, Glowatzki E, Bergles DE. The origin of spontaneous activity in the developing auditory system. Nature. 2007;450(7166):50–55. doi: 10.1038/nature06233. [DOI] [PubMed] [Google Scholar]
- Wangemann P. Supporting sensory transduction: cochlear fluid homeostasis and the endocochlear potential. J Physiol. 2006;576(Pt 1):11–21. doi: 10.1113/jphysiol.2006.112888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warchol ME. Immune cytokines and dexamethasone influence sensory regeneration in the avian vestibular periphery. J Neurocytol. 1999;28(10–11):889–900. doi: 10.1023/A:1007026306730. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, et al. Identification of a member of the MAPKKK family as a potential mediator of TGF-b signal transduction. Science. 1995;270(5244):2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- Zdebik AA, Wangemann P, Jentsch TJ. Potassium ion movement in the inner ear: insights from genetic disease and mouse models. Physiology. 2009;24(5):307–316. doi: 10.1152/physiol.00018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-X. An atlas of histology. 1. Berlin: Springer; 1999. [Google Scholar]
- Zheng JL, Gao WQ. Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat Neurosci. 2000;3(6):580–586. doi: 10.1038/75753. [DOI] [PubMed] [Google Scholar]
- Zou J, Pyykkö I, Sutinen P, Toppila E. Vibration induced hearing loss in guinea pig cochlea: expression of TNF-[alpha] and VEGF. Hear Res. 2005;202(1–2):13–20. doi: 10.1016/j.heares.2004.10.008. [DOI] [PubMed] [Google Scholar]