Abstract
7-Deaza-2′-deoxyadenosine (1, c7Ad) and 3-deaza-2′-deoxyadenosine (2, c3Ad) have been incorporated into d(AAAAAA) tracts replacing dA at various positions within oligonucleotides. For this purpose suitably protected phosphonates have been prepared and oligonucleotides were synthsized on solid-phase. The oligomers were hybridized with their cognate strands. The duplexes were phosphorylated at OH-5′ by polynucleotide kinase and self-ligated to multimers employing T4 DNA ligase. Oligomerized DNA-fragments were analyzed by polyacrylamide gel electrophoresis and the bending was determined from anomalies of electrophoretic mobility. Replacement of dA by c3Ad decreased the bending more than replacement by c7Ad. Reduction of bending was much stronger when the modified nucleosides replaced one or several dA residues at the 3′-site of an d(AAAAAA)-tract whereas replacement at the 5′-site showed no significant influence [1, 2].
Full text
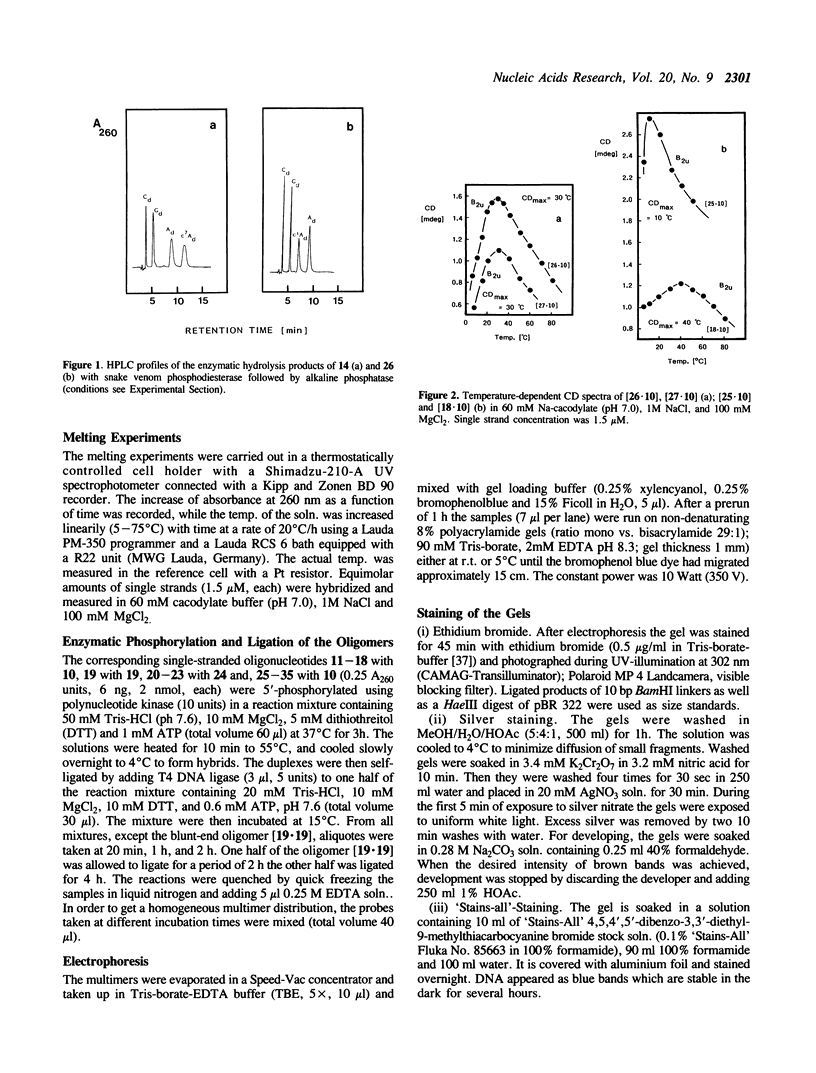
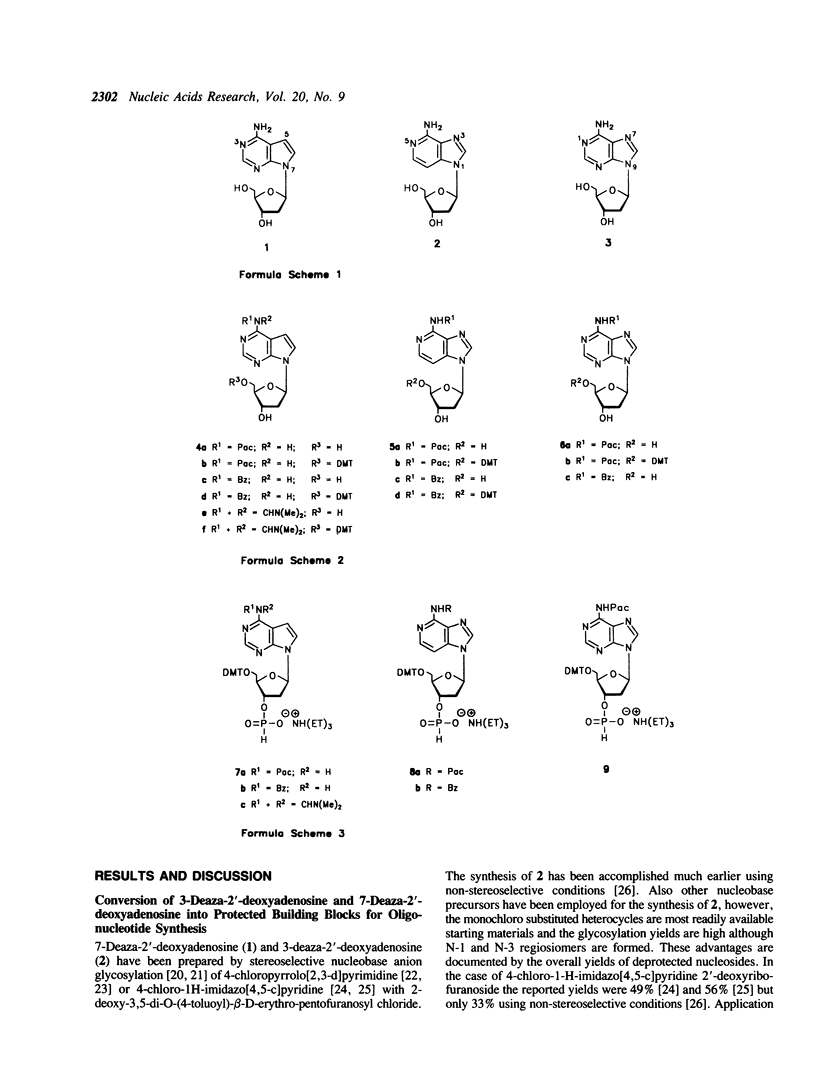
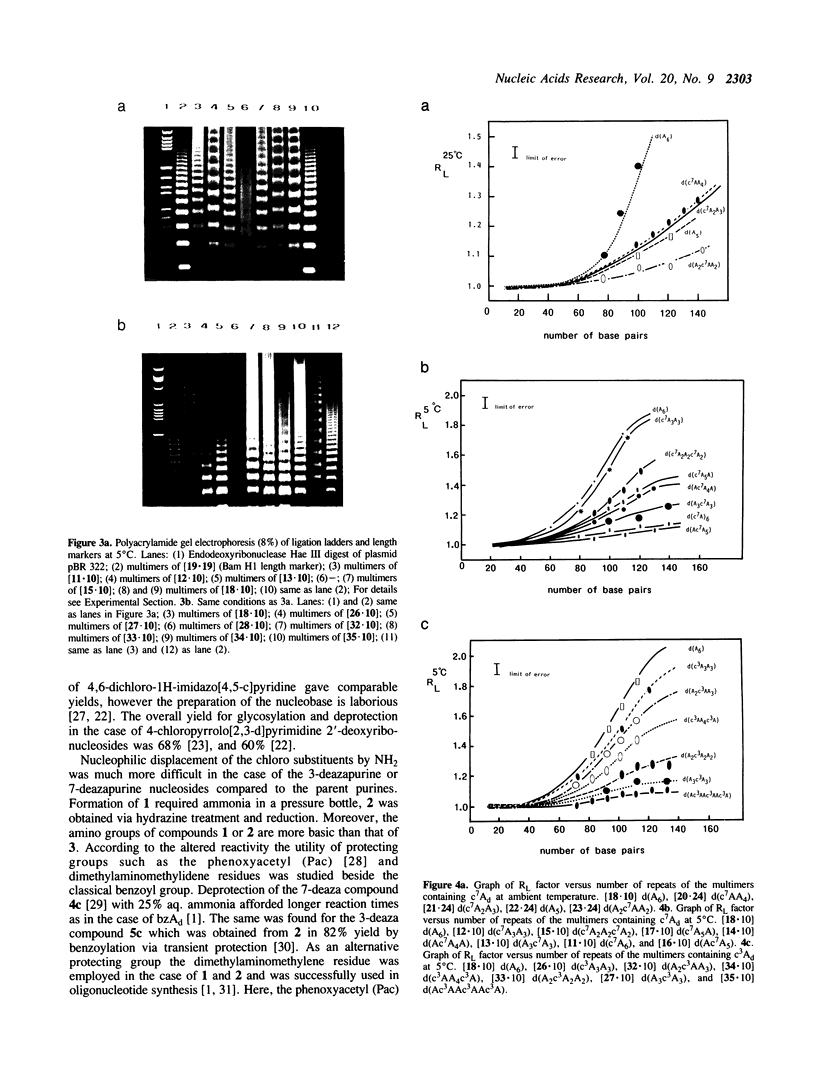
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beidler J. L., Hilliard P. R., Rill R. L. Ultrasensitive staining of nucleic acids with silver. Anal Biochem. 1982 Nov 1;126(2):374–380. doi: 10.1016/0003-2697(82)90530-9. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P. Anomalous structure and properties of poly (dA).poly(dT). Computer simulation of the polynucleotide structure with the spine of hydration in the minor groove. Nucleic Acids Res. 1987 Jan 12;15(1):293–311. doi: 10.1093/nar/15.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuprina V. P. Regularities in formation of the spine of hydration in the DNA minor groove and its influence on the DNA structure. FEBS Lett. 1985 Jul 1;186(1):98–102. doi: 10.1016/0014-5793(85)81347-8. [DOI] [PubMed] [Google Scholar]
- Cosstick R., Li X., Tuli D. K., Williams D. M., Connolly B. A., Newman P. C. Molecular recognition in the minor groove of the DNA helix. Studies on the synthesis of oligonucleotides and polynucleotides containing 3-deaza-2'-deoxyadenosine. Interaction of the oligonucleotides with the restriction endonuclease EcoRV. Nucleic Acids Res. 1990 Aug 25;18(16):4771–4778. [PMC free article] [PubMed] [Google Scholar]
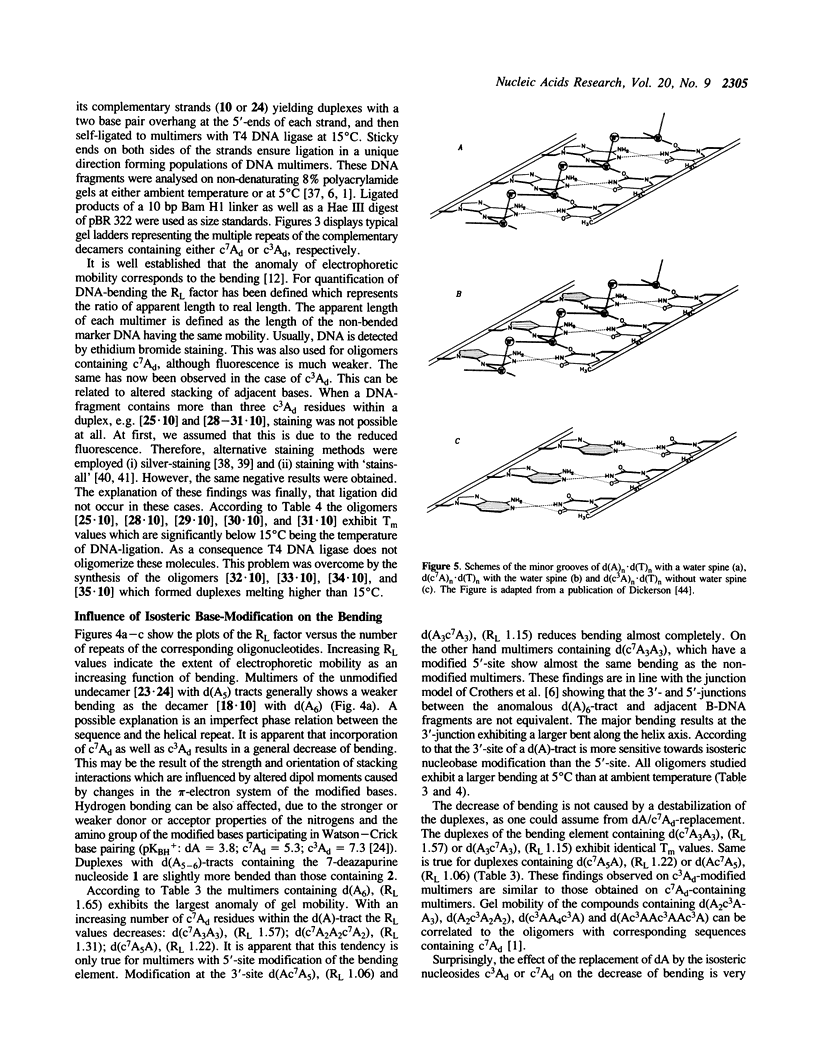
- Dickerson R. E., Drew H. R., Conner B. N., Wing R. M., Fratini A. V., Kopka M. L. The anatomy of A-, B-, and Z-DNA. Science. 1982 Apr 30;216(4545):475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Froehler B. C., Ng P. G., Matteucci M. D. Synthesis of DNA via deoxynucleoside H-phosphonate intermediates. Nucleic Acids Res. 1986 Jul 11;14(13):5399–5407. doi: 10.1093/nar/14.13.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry. 1985 Dec 3;24(25):7033–7037. doi: 10.1021/bi00346a001. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Chemical determinants of DNA bending at adenine-thymine tracts. Biochemistry. 1987 Jun 16;26(12):3745–3748. doi: 10.1021/bi00386a070. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Fratini A. V., Drew H. R., Dickerson R. E. Ordered water structure around a B-DNA dodecamer. A quantitative study. J Mol Biol. 1983 Jan 5;163(1):129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Ono A., Ueda T. Minor-groove-modified oligonucleotides: synthesis of decadeoxynucleotides containing hypoxanthine, N2-methylguanine and 3-deazaadenine, and their interactions with restriction endonucleases Bgl II, Sau, 3AI, and Mbo I (Nucleosides and Nucleotides Part 75). Nucleic Acids Res. 1987 Apr 10;15(7):3059–3072. doi: 10.1093/nar/15.7.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulhof J. C., Molko D., Teoule R. The final deprotection step in oligonucleotide synthesis is reduced to a mild and rapid ammonia treatment by using labile base-protecting groups. Nucleic Acids Res. 1987 Jan 26;15(2):397–416. doi: 10.1093/nar/15.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz H., Reinert K. E. DNA-bending on ligand binding; change of DNA persistence length. J Biomol Struct Dyn. 1991 Oct;9(2):315–329. doi: 10.1080/07391102.1991.10507915. [DOI] [PubMed] [Google Scholar]
- Seela F., Berg H., Rosemeyer H. Bending of oligonucleotides containing an isosteric nucleobase: 7-deaza-2'-deoxyadenosine replacing dA within d(A)6 tracts. Biochemistry. 1989 Jul 25;28(15):6193–6198. doi: 10.1021/bi00441a010. [DOI] [PubMed] [Google Scholar]
- Seela F., Kehne A. Palindromic octa- and dodecanucleotides containing 2'-deoxytubercidin: synthesis, hairpin formation, and recognition by the endodeoxyribonuclease EcoRI. Biochemistry. 1987 Apr 21;26(8):2232–2238. doi: 10.1021/bi00382a024. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Why bend DNA? Cell. 1990 Jan 26;60(2):177–180. doi: 10.1016/0092-8674(90)90729-x. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Sequence-dependent deformational anisotropy of chromatin DNA. Nucleic Acids Res. 1980 Sep 11;8(17):4041–4053. doi: 10.1093/nar/8.17.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kitzing E., Diekmann S. Molecular mechanics calculations of dA12.dT12 and of the curved molecule d(GCTCGAAAAA)4.d(TTTTTCGAGC)4. Eur Biophys J. 1987;15(1):13–26. doi: 10.1007/BF00255031. [DOI] [PubMed] [Google Scholar]