Abstract
The Protein Structure Initiative’s Structural Biology Knowledgebase (SBKB, URL: http://sbkb.org) is an open web resource designed to turn the products of the structural genomics and structural biology efforts into knowledge that can be used by the biological community to understand living systems and disease. Here we will present examples on how to use the SBKB to enable biological research. For example, a protein sequence or Protein Data Bank (PDB) structure ID search will provide a list of related protein structures in the PDB, associated biological descriptions (annotations), homology models, structural genomics protein target status, experimental protocols, and the ability to order available DNA clones from the PSI:Biology-Materials Repository. A text search will find publication and technology reports resulting from the PSI’s high-throughput research efforts. Web tools that aid in research, including a system that accepts protein structure requests from the community, will also be described. Created in collaboration with the Nature Publishing Group, the Structural Biology Knowledgebase monthly update also provides a research library, editorials about new research advances, news, and an events calendar to present a broader view of structural genomics and structural biology.
Keywords: Protein, Protein production, Structural biology, Structural databases, Structural genomics, Theoretical models
Introduction
When the Protein Structure Initiative (PSI) began in 2000, project results were communicated independently through publications [1], the Protein Data Bank (PDB) [2, 3] structure depositions, individual PSI websites, and presentations at meetings. The PSI-1 and -2 centers had focused their efforts on rapidly determining the structures of proteins on a genomic scale with the emphasis on covering sequence-structure space. To accomplish this, the centers developed a wide variety of tools to carefully select targets and many new technologies to determine and annotate the structures. However, there was no centralized access point for this information. It became clear that in order for the biological community to get the maximum benefit from the products of the PSI effort, a single centralized website was needed. In February 2008, the Structural Genomics Knowledgebase [4] was created which combined the PSI products with data from publicly available biological resources in order to show comprehensive information, including experimental data prior to publication, to jumpstart biological research. In September 2008, the Structural Genomics Knowledgebase entered into a collaboration with the Nature Publishing Group (NPG) to become a “Gateway” site, delivering editorial content about the latest structure and technology reports, a research library, events calendar and science news in addition to the searchable protein database.
In keeping with the recent start of the third phase of the Protein Structure Initiative, PSI:Biology, the website changed its name in August 2010 to the Structural Biology Knowledgebase (SBKB, URL http://sbkb.org), and developed more tools to aid in protein research design. It also improved features to foster collaborations between the biological community and the PSI. Editorial content, displayed on the SBKB as the “Structural Biology Update”, is updated on the third Thursday of each month, with database updates occurring on a weekly basis. In this article, we describe all of the features available on the SBKB that can be used to enable biological research, and present some examples of its use.
Navigating the SBKB
The SBKB homepage provides entry points to the following content and functionalities:
Left navigation menu
These menu items give facile access to topics relevant to the biological and biomedical research, such as resources for target, structure, methods, models, and publication information. It also contains menu items for PSI and SBKB information such as FAQs, classroom tutorials, PSI Policies and Reports, links to PSI administrative and Center sites, PSI funding opportunities, and links to NPG online resources.
Structural biology update
This section of the SBKB provides a collection of recent research and technical highlights from the PSI and broader structural biology community, news, upcoming events, and a research library of PSI and other structural biology articles provided by the NPG.
Featured molecule
This highlight [5] consists of simplified explanations and illustrations of interesting PSI protein structures with molecular graphics that allow the user to interact and learn about the molecule’s biological role. A new molecule is selected each month.
Query capabilities
A central search box queries the SBKB database for all sequence, structure, functional or technological data related to a sequence, PDB ID code or text string. Text searches will also return matching highlights published as part of the Structural Biology Update.
Nature e-alerts and RSS feeds
Users can subscribe to a monthly electronic Table of Contents alert service (e-alert) on Nature.com with links to the SBKB’s latest content. Alternatively, two RSS (Really Simple Syndication) feeds, which can be managed by a user’s web browser, keeps readers apprised of (a) the month’s Structural Biology Update content and (b) the latest PSI structures released by the PDB.
Community-Nominated Targets portal
The PSI:Biology Network invites the biological community to nominate proteins of biological relevance for structural determination. This proposal system begins the process of matching your project with one of 13 high-throughput and membrane protein structure determination centers to carry out the study. Access is available from either the SBKB homepage or at http://cnt.sbkb.org/CNT.
Sequence Comparison and Analysis tool
The Sequence Comparison and Analysis (SCA) tool consists of the same functionality as the Community-Nominated Target proposal system described previously, but provides an evaluation report to the author only rather than forwarding it to a selection committee. It also supports batch submission to evaluate 100s of sequences. This tool can also be found at http://cnt.sbkb.org/CNT.
Functional Sleuth
Functional Sleuth enables further research for proteins in the Protein Data Bank archive whose functions are unknown or minimally characterized. These “structures of unknown function” (SUFs) are currently organized by source organism, and users can choose to display the SUFs from any phylogenetic level from domain to species. Making a selection on a “tree-of-life” image will launch an interactive tree browser which will further filter your list of SUFs, and right clicking on the name will display the gallery of protein structures. You can download a comma-separated-variable (.csv) file of PDB IDs from your particular gallery, and a full list of all SUF PDB IDs is available at http://sbkb.org/KB/unkstrucs.txt. This feature is updated weekly in conjunction with the weekly PDB release.
Latest PSI statistics
The PSI Network tracks its measure of success and progress though a series of agreed metrics. These metrics, including statistics such as the number of protein structures solved and calculated modeling leverage, were defined by the Goals and Metrics Committee for the PSI-2 Network.
BioSync
BioSync provides technical details about structural biology beamlines at synchrotron radiation facilities. Originally maintained by the RCSB PDB, it is now a part of the SBKB. Progress on future facilities is tracked and information on decommissioned sites is maintained for historical purposes. Links are also provided to related external resources. Summary statistics, based on PDB depositions, are produced and updated weekly. At the beamline level, galleries of structures, tables of citations and general information are also available. Separate statistics are provided for structures solved by structural genomics efforts. This site can be found on the methods hub page from the left navigation menu.
Tutorial and educational resources
The SBKB also has tutorials and classroom exercises to explain how to use the SBKB. These tutorials not only introduce new users to the SBKB, but also give lessons on how to interpret the data being presented. See http://sbkb.org/about/getting_started.html or contact us at comments@sbkb.org for more specific requests.
Content in the Structural Biology Knowledgebase
The SBKB is a web portal [6] that collects different types of structural and methodological information about proteins from the PSI centers and publicly available resources to provide facile access for biomedical researchers and students. The SBKB has also established additional portals (Table 1) to capture data produced by the PSI including protein selection and status (TargetDB [7]), their trial history and protocols (PepcDB [8]), homology models (Protein Model Portal [9]), developed technologies (PSI Technology Portal), and publications (PSI Publications Portal). To goal of these portals is to organize the PSI results and methodological data into a learning and research design optimization resource, especially since much of data is released prior to publication. This information is accessible by searching the SBKB by protein or DNA sequence, PDB structure ID, or text, and more specific queries can be made using the individual portals’ websites.
Table 1.
List of web addresses to access the features and underlying portals of the Structural Biology Knowledgebase
| SBKB and portal sites | Data | Web address |
|---|---|---|
| Structural Biology Knowledgebase | Query and reporting; editorials and news | http://sbkb.org |
| TargetDB | Protein sequence selection and progress | http://targetdb.sbkb.org |
| PepcDB | Target trial information and protocols | http://pepcdb.sbkb.org |
| Protein Model Portal | Theoretical models | http://www.proteinmodelportal.org |
| PSI Technology Portal | Technology reports | http://technology.lbl.gov/portal/home |
| PSI Publications Portal | PSI publications | http://olenka.med.virginia.edu/psi/ |
| BioSync | X-ray crystallography methods | http://biosync.sbkb.org |
| Community-Nominated Target proposal system | Community requests for protein structures | http://cnt.sbkb.org/CNT |
Searching the SBKB
We describe the resources used in SBKB query and reporting mechanism in the context of commonly used examples.
Finding sequence-level or structure-level information about a protein of interest
Conducting a sequence or PDB ID search of the SBKB will yield the following:
Links to information from publicly available biological resources
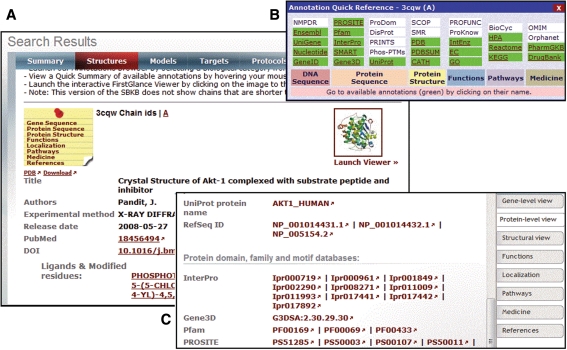
The SBKB web portal draws links, identifiers, and annotation values from over 100 PSI and other public resources for protein sequences found in TargetDB and the PDB archive. From the PSI network, information derived from structural and functional annotation tools developed by the four PSI Large-Scale centers such as TOPSAN and others [10–13] is presented. In addition, the external resources include genomic databases [14–18] and model organism databases [19–27], protein primary sequence resources such as Pfam [28], InterPro [29], and others [30–41], structural databases such as the PDB and others [3, 42–52] and structure comparison resources [53–56], functional annotation resources [57–65] evolutionary relationships [66], interactions and pathways [64, 65, 67–78], protein expression profiles [79–98], genetic variations [99–102], disease and pharmacological relationships [103–106], and PubMed [107] for reference information. Access to this collected data is available from a central list appearing next to each target sequence or 3D structure in the SBKB search results. Available for each structure or target results, an example is shown for (PDB + 3cqw) in Fig. 1. By combining all of the total evidence in one view, the user not only reviews what knowledge exists but also can recognize gaps in the literature on that particular topic to enable future hypotheses and studies.
Fig. 1.
Example of a structure result summary for (PDB + 3cqw). a Search results that contain structural matches will present an annotations panel and molecular visualization tool. b Hovering the mouse over the Chain ID will launch an annotations quick table, summarizing if popular resources have an annotation for the protein sequence in that structure’s chain. c All annotations are accessible from a “post-it” with biological subjects on it—clicking on any subject will take you directly to that page of the “annotations notebook”
Visualizing protein structures in an interactive viewer
If a 3D structure exists for a protein of interest, the SBKB currently utilizes the molecular viewer FirstGlance [108] to explore the molecule and binding partners (other proteins, ligands, nucleic acids, etc.). This Java-based viewer also provides explanations of the images and representations for the novice user, with display options (such as “color by B-factors/uncertainty”) for advanced users as well.
Experimental target tracking databases, TargetDB and PepcDB
The SBKB manages two databases that track structural genomics efforts. TargetDB [7] tracks information on over a quarter million protein sequences, or “targets”, that have been selected for structural determination by worldwide structural genomics projects. This information includes sequence and site information, target experimental status, and timestamps for the latest experimental step (cloned, expressed, purified, etc.). The Protein Expression, Purification, and Crystallization database [8], PepcDB, provides more detailed information about PSI targets registered in TargetDB (275,000 as of April 2011) by reporting on progress for each experimental trial. Information in PepcDB includes the sequence and site information, rationale for the target’s selection (biomedical, community-nominated, technology development, PSI:Biology partnership selection, etc.), each target’s experimental histories including individual trial details, the protocols used for protein production and structure determination, and reasons why work was terminated if a 3D structure was not determined. TargetDB and PepcDB are updated weekly with data provided by the PSI centers, and are searchable by TargetID or other popular accession ID (UniProt [30], GenBank [16], PDB [3], see site for details), or filter results by the discussed data attributes. Regular checks are made to ensure that these databases are consistent with relevant information in the PDB and the PSI:Biology-Materials Repository (psimr.asu.edu).
In 2011, a new target history tracking resource that merges TargetDB and PepcDB will be released. Please visit the TargetDB and PepcDB websites for more information on this transition.
Links to the PSI:Biology-Materials Repository
The SBKB also searches its partner PSI resource center, the PSI:Biology-Materials Repository, which stores, maintains and distributes protein expression plasmids and vectors created by the PSI centers. It currently holds over 40,000 PSI plasmids and nearly 100 empty vectors available for request with an additional PSI plasmids added on a monthly basis.
If a sequence search yielded no experimental structures for the protein of interest, there are other sources of information to help design future experiments:
Theoretical models from the Protein Model Portal
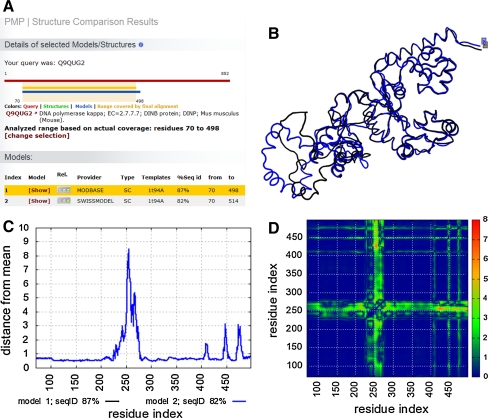
The Protein Model Portal (PMP) was created in 2007 as a single entry point to federate structure information from different resources: theoretical structure models from several modeling resources, i.e. the PSI centers CSMP, JCSG, MCSG, NESG, NMHRCM, NYSGXRC, JCMM, and the large scale comparative modeling resources ModBase [51] and SWISS-MODEL Repository [52], and experimental protein structure information from the PDB. One of the challenges in using model information effectively has been to access all models available for a specific protein in heterogeneous formats at different sites using various incompatible accession code systems. To overcome this problem, protein sequences of the UniProt knowledgebase are used as a unified reference system to organize heterogeneous structural information from the different sites. For the first time, it was now possible to access several modeling resources simultaneously, and be able to compare the results computed by different methods in a comprehensive way. An example is shown in Fig. 2 of two theoretical models of a DNA polymerase.
Fig. 2.
Example of a Protein Model Portal model comparison analysis. Two theoretical models of a DNA polymerase from mouse (UniProt + Q9QUG2) have been selected for further structural variability analysis by the PMP. a The graphical representation indicated the overlapping residue range that can be compared. b The superposition of the two models shows that ModBase (black) and Swiss-model (blue) predict a different structure on the left side of the depiction. c. A graph showing the local (per residue) deviation of individual models/structures from mean of the ensemble of models/structures based on a distance RMSD (dRMSD). d The colors of the spectrum indicate the degree of variability (based on a weighted dRMSD) among the structure models (blue = low, green = medium, red = high)
The current release (Mar 2011) consists of 15.1 million comparative protein models for 3.8 million distinct UniProt sequences. Queries can be entered as protein sequences in one-letter code or UniProt accession IDs. Results are presented in a graphical and intuitive way, indicating regions of the proteins where structural information (experimental or theoretical) is available, complemented with functional and domain annotation. Detailed technical information about each model is also provided, such as the date of creation and “latest verification”, the template structure and the target-template alignment the model was based on, and the expected model accuracy based on the evolutionary distance between the target and the template. Graphs and images that display and assess model quality and reliability are available, and are an essential component to allow users to select the best available structural information for a specific application.
Community requests to the PSI
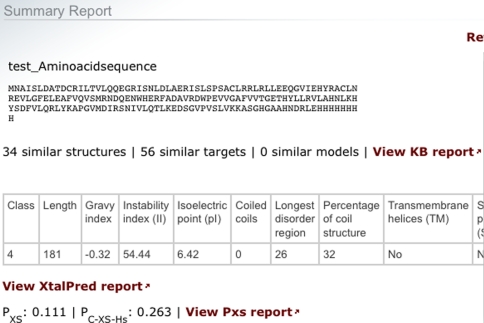
Individual investigators are encouraged to nominate protein sequences for structural determination by the PSI:Biology network’s high-throughput and membrane protein centers (sbkb.org/KB/psi_centers.html). These sequences should match current PSI goals of biological/biomedical relevance and sequence novelty. The Community-Nominated Target (CNT) proposal system performs an analysis of each proposed target sequence and reports information including crystallization propensities from the PXS [109–111] and XtalPred [112] prediction servers. It also provides a list of similar structures, targets, and homology models found in the SBKB. An example report is shown in Fig. 3. All results are presented to the contributing investigator prior to final submission. Once submitted, the CNT proposals are reviewed by a target selection committee using procedures defined by the PSI and NIGMS [113]. Access to the proposal system is available from the right column of the SBKB homepage or at http://cnt.sbkb.org/CNT/.
Fig. 3.
Example of a Sequence Analysis report. The CNT system and SCA-tool will both perform an SBKB query on given sequences to find the number of similar structures, models, and targets. It will also submit the sequence to the PSI-developed XtalPred and Pxs servers to calculate biophysical parameters and crystallization propensities. In this example, the given sequence was calculated to be a class 4 difficulty (hard), so further construct optimization may be required to ensure a successful structure determination
Finding new methods and technologies
The PSI centers have developed and utilized many technologies that facilitated structural determination and analysis. A text search of the SBKB allows a user to find these methods.
The PSI Technology Portal
The PSI Technology Portal provides access to over 200 summaries of key PSI technologies with links to the responsible PSI center or related publication. Reports can be queried by text, by PSI center, or by clicking an image of an experimental pipeline diagram. All tech reports can be “forwarded” to display on a user’s social networking sites, such as Facebook, del.icio.us, and others. A YouTube channel to show videos of PSI technologies in action has also been created (www.youtube.com/user/sbkbtech) and a Nature Network discussion forum (network.nature.com/groups/psikb_tech/) was also created as a means of communicating with the user directly.
The PSI Publications Portal
The PSI has published over 1,500 peer-reviewed articles over the past 10 years. These articles are organized by topic (structural or methodological) or by PSI center, and can be searched by author, title, journal, PDB ID, or other attributes. Information presented includes links to the PubMed abstract, related PDB ID, number of times cited, and can also be downloaded in EndNote format. Text searches of the Publications Portal can also be conducted from the central SBKB search box.
Using the Structural Biology update—research library
As part of the Structural Biology update, SBKB and Nature editors add articles published by leading scientific journals that relate to structural biology and structural genomic. Articles are organized into subjects that range from cloning to automated annotation, it should be noted that the articles listed here will require subscription to the relevant journals.
Tools for the Home Lab: a booklet about methods developed by the PSI
The SBKB presented a four-part “PSI-2 Achievements” series from July 2010 to October 2010 detailing advances in the areas of methods, modeling, structures, and outreach. The methods articles from this series summarized the latest strategies developed for protein isolation and structural determination methods. We have assembled these articles into a handy PDF booklet called “Tools for the Home Lab: New Methods from the Protein Structure Initiative”. This book of ideas, which can be downloaded from http://sbkb.org/pdf/PSI-2methods.pdf, can enable any level of wet-bench scientist interested in protein research.
Architecture
The SBKB uses an architecture that collects IDs and selected annotations into a central portal database that is organized to facilitate queries and the addition of new annotations. Each of the SBKB portals developed their underlying architectures with guidelines for common protocols that would allow for easy data exchange with the central SBKB. With this structure, it is possible to query the information from the SBKB home page or from an individual portal module for more specialized queries.
Remotely accessing the SBKB
The content and functionality of the SBKB may be accessed remotely in a variety of ways. Editorial content may be accessed by linking to SBKB pages that are regularly updated (e.g. http://sbkb.org/update/). The RSS feeds are provided to describe both new editorial content and new structure data. Web services are provide to enable program level access to search features of the SBKB. Remote access is provided via both Common Gateway Interface (CGI) and Simple Object Access Protocol (SOAP) protocols. The SBKB also provides an embeddable widget which can be incorporated into a web page at a remote site. The widget provides a dynamically updated display of new articles, features and structure data on the KB site. The protocol interface details and widget installation instructions are described in http://sbkb.org/about/webservices.html.
Conclusions
With the overarching goal of creating new knowledge about the interrelationships of sequence, structure and function, the Structural Biology Knowledgebase captures and highlights the products of the PSI projects for use by the broader biological communities, creates an information repository and web portal that integrates the products of the PSI with publicly available biological information, and encourages collaborative interactions between the PSI and the biological communities. We welcome feedback from the community; users with questions may contact the SBKB at comments@sbkb.org.
Acknowledgments
The SBKB is a resource center within the Protein Structure Initiative and is supported by grant U01 GM093324-01 from the National Institute of General Medical Sciences.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- 3D
Three-dimensional
- CGI
Common Gateway Interface
- CSMP
PSI Center for Structures of Membrane Proteins
- JCMM
PSI Joint Center for Molecular Modeling
- JCSG
PSI Joint Center for Structural Genomics
- MCSG
PSI Midwest Center for Structural Genomics
- NESG
PSI Northeast Structural Genomics Consortium
- NMHRCM
PSI New Methods in High-Resolution Comparative Modeling
- NIGMS
National Institute of General Medical Sciences
- NPG
Nature Publishing Group
- NYSGXRC
PSI New York SGX Research Center for Structural Genomics
- PepcDB
Protein expression purification and crystallization database
- PDB
Protein Data Bank
- PMP
Protein Model Portal
- PSI
Protein Structure Initiative
- RSS
Really Simple Syndication
- SBKB
Structural Biology Knowledgebase
- SG
Structural Genomics
- SOAP
Simple Object Access Protocol
- WSDL
Web Services Description Language
References
- 1.Smith TL (ed) (2000) Structural Genomics Supplement Issue. Nat Struct Biol 7(11s):927–994
- 2.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10(12):980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 3.Berman HM, et al. The protein data bank. Nucleic Acids Res. 2000;28:p235–p242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berman HM, et al. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37(Database issue):D365–D368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodsell D (2009) PSI featured molecule series. Available from: http://sbkb.org/KB/structures.jsp
- 6.Reddy P (2004) In: Bidgoli H (ed) The internet encyclopedia, vol 2 G-O. Wiley, Hoboken, NJ, pp 298–310
- 7.Chen L, et al. TargetDB: a target registration database for structural genomics projects. Bioinformatics. 2004;20:2860–2862. doi: 10.1093/bioinformatics/bth300. [DOI] [PubMed] [Google Scholar]
- 8.Kouranov A, et al. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold K, et al. The protein model portal. J Struct Funct Genomics. 2009;10(1):1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Open Protein Structure Annotation Network (2009). Available from: http://www.topsan.org/
- 11.Binkowski A (2009) Global protein surface survey. Available from: http://gpss.mcsg.anl.gov/
- 12.Fischer M (2009) NESG function annotation server. Available from: http://luna.bioc.columbia.edu/honiglab/nesg/cgi-bin/browse.pl
- 13.Functional Analysis Server at the NYSGXRC (2009). Available from: http://www.nysgxrc.org/functional/
- 14.Hubbard T, et al. Ensembl 2005. Nucleic Acids Res. 2005;33(Database issue):D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flicek P, et al. Ensembl 2008. Nucleic Acids Res. 2008;36(Database issue):D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson DA, et al. GenBank. Nucleic Acids Res. 2008;36(Database issue):D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perriere G, Duret L, Gouy M. HOBACGEN: database system for comparative genomics in bacteria. Genome Res. 2000;10(3):379–385. doi: 10.1101/gr.10.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhee SY, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31(1):224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guldener U, et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 2005;33(Database issue):D364–D368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chisholm RL, et al. dictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 2006;34(Database issue):D423–D427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudd KE. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28(1):60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crosby MA, et al. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35(Database issue):D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eppig JT, et al. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35(Database issue):D630–D637. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Twigger SN, et al. The Rat Genome Database, update 2007–easing the path from disease to data and back again. Nucleic Acids Res. 2007;35(Database issue):D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bieri T, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35(Database issue):D506–D510. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprague J, et al. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34(Database issue):D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apweiler R, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29(1):37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The UniProt Consortium The Universal Protein Resource (UniProt) Nucleic Acids Res. 2007;35(Database issue):D193–D197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu CH, et al. iProClass: an integrated, comprehensive and annotated protein classification database. Nucleic Acids Res. 2001;29(1):52–54. doi: 10.1093/nar/29.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Attwood TK, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31(1):400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gattiker A, et al. Automated annotation of microbial proteomes in SWISS-PROT. Comput Biol Chem. 2003;27(1):49–58. doi: 10.1016/S1476-9271(02)00094-4. [DOI] [PubMed] [Google Scholar]
- 35.Haft DH, et al. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29(1):41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31(1):371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bru C, et al. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33(Database issue):D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hulo N, et al. The PROSITE database. Nucleic Acids Res. 2006;34(Database issue):D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mihalek I, Res I, Lichtarge O. Evolutionary trace report_maker: a new type of service for comparative analysis of proteins. Bioinformatics. 2006;22(13):1656–1657. doi: 10.1093/bioinformatics/btl157. [DOI] [PubMed] [Google Scholar]
- 40.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeats C, et al. Gene3D: comprehensive structural and functional annotation of genomes. Nucleic Acids Res. 2008;36(Database issue):D414–D418. doi: 10.1093/nar/gkm1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laskowski RA, et al. PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem Sci. 1997;22:488–490. doi: 10.1016/S0968-0004(97)01140-7. [DOI] [PubMed] [Google Scholar]
- 43.Orengo CA, et al. The CATH Database provides insights into protein structure/function relationships. Nucleic Acids Res. 1999;27(1):275–279. doi: 10.1093/nar/27.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuff AL, et al. The CATH classification revisited–architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37(Database issue):D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andreeva A, et al. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32(Database issue):D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murzin AG, et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 47.Huhne R, Koch FT, Suhnel J (2007) A comparative view at comprehensive information resources on three-dimensional structures of biological macro-molecules. Brief Funct Genomic Proteomic 6(3):220–239 [DOI] [PubMed]
- 48.Schultz J, et al. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37(Database issue):D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulrich EL, et al. BioMagResBank. Nucleic Acids Res. 2008;36(Database issue):D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pieper U, et al. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2009;37(Database issue):D347–D354. doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiefer F et al (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37(Database issue):D387–D392 [DOI] [PMC free article] [PubMed]
- 53.Ye Y, Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19(Suppl 2):ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 54.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterhouse AM, et al. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28(1):304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hodis E, et al. Proteopedia—a scientific ‘wiki’ bridging the rift between three-dimensional structure and function of biomacromolecules. Genome Biol. 2008;9(8):R121. doi: 10.1186/gb-2008-9-8-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pal D, Eisenberg D. Inference of protein function from protein structure. Structure. 2005;13(1):121–130. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 60.The Gene Ontology Consortium Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33(Web Server issue):W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barthelmes J, et al. BRENDA, AMENDA and FRENDA: the enzyme information system in 2007. Nucleic Acids Res. 2007;35(Database issue):D511–D514. doi: 10.1093/nar/gkl972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Liu M, Gilson MK. BindingDB: a web-accessible molecular recognition database. Comb Chem High Throughput Screen. 2001;4(8):719–725. doi: 10.2174/1386207013330670. [DOI] [PubMed] [Google Scholar]
- 64.Liu T, et al. BindingDB: a web-accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res. 2007;35(Database issue):D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schaefer CF, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37(Database issue):D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nikolskaya AN, et al. PIRSF family classification system for protein functional and evolutionary analysis. Evol Bioinform Online. 2006;2:197–209. [PMC free article] [PubMed] [Google Scholar]
- 67.Salwinski L, et al. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chatr-aryamontri A, et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35(Database issue):D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chautard E, et al. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics. 2009;25(5):690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goll J, et al. MPIDB: the microbial protein interaction database. Bioinformatics. 2008;24(15):1743–1744. doi: 10.1093/bioinformatics/btn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mewes HW, et al. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 2006;34(Database issue):D169–D172. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang R, et al. The PDBbind database: methodologies and updates. J Med Chem. 2005;48(12):4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 75.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8(5):R95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joshi-Tope G, et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33(Database issue):D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karp PD, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33(19):6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kerrien S, et al. IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 2007;35(Database issue):D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.NextBio (2009). Available from: http://www.nextbio.com/
- 80.Oxford GlycoProteomics 2-DE database (2009). Available from: http://proteomewww.bioch.ox.ac.uk/2d/2d.html
- 81.Human Cornea 2-DE database (2009). Available from: http://www.cornea-proteomics.com/
- 82.DOSAC-COBS 2D-PAGE database (2009). Available from: http://www.dosac.unipa.it/2d/
- 83.Parasite host cell interaction 2D-PAGE database (2009). Available from: http://www.gram.au.dk/2d/2d.html
- 84.Purkyne Military Medical Academy 2D-PAGE database (2009). Available from: http://www.pmma.pmfhk.cz/2d/2d.html
- 85.Reproduction 2D-PAGE (2009). Available from: http://reprod.njmu.edu.cn/cgi-bin/2d/2d.cgi
- 86.Bini L et al (2009) 2D-PAGE database from the Department of Molecular Biology, University of Siena, Italy. Available from: http://www.bio-mol.unisi.it/2d/2d.html
- 87.Celis JE, et al. Human and mouse proteomic databases: novel resources in the protein universe. FEBS Lett. 1998;430(1–2):64–72. doi: 10.1016/S0014-5793(98)00527-4. [DOI] [PubMed] [Google Scholar]
- 88.Evans G, et al. Construction of HSC-2DPAGE: a two-dimensional gel electrophoresis database of heart proteins. Electrophoresis. 1997;18(3–4):471–479. doi: 10.1002/elps.1150180322. [DOI] [PubMed] [Google Scholar]
- 89.Hoogland C, et al. The World-2DPAGE Constellation to promote and publish gel-based proteomics data through the ExPASy server. J Proteomics. 2008;71(2):245–248. doi: 10.1016/j.jprot.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 90.Hoogland C, et al. SWISS-2DPAGE, ten years later. Proteomics. 2004;4(8):2352–2356. doi: 10.1002/pmic.200300830. [DOI] [PubMed] [Google Scholar]
- 91.Imin N, et al. Characterisation of rice anther proteins expressed at the young microspore stage. Proteomics. 2001;1(9):1149–1161. doi: 10.1002/1615-9861(200109)1:9<1149::AID-PROT1149>3.3.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 92.Li XP, et al. A two-dimensional electrophoresis database of rat heart proteins. Electrophoresis. 1999;20(4–5):891–897. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<891::AID-ELPS891>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 93.Parkinson H, et al. ArrayExpress–a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35(Database issue):D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pitarch A, et al. Analysis of the Candida albicans proteome. II. Protein information technology on the Net (update 2002) J Chromatogr B Analyt Technol Biomed Life Sci. 2003;787(1):129–148. doi: 10.1016/S1570-0232(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 95.Praz V, Jagannathan V, Bucher P. CleanEx: a database of heterogeneous gene expression data based on a consistent gene nomenclature. Nucleic Acids Res. 2004;32(Database issue):D542–D547. doi: 10.1093/nar/gkh107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uhlen M, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4(12):1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 97.VanBogelen RA, et al. Escherichia coli proteome analysis using the gene-protein database. Electrophoresis. 1997;18(8):1243–1251. doi: 10.1002/elps.1150180805. [DOI] [PubMed] [Google Scholar]
- 98.Vijayendran C, et al. 2DBase: 2D-PAGE database of Escherichia coli. Biochem Biophys Res Commun. 2007;363(3):822–827. doi: 10.1016/j.bbrc.2007.09.050. [DOI] [PubMed] [Google Scholar]
- 99.Thorisson GA, et al. The International HapMap Project Web site. Genome Res. 2005;15(11):1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Packer BR, et al. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004;32(Database issue):D528–D532. doi: 10.1093/nar/gkh005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Karchin R, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21(12):2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 103.Hamosh A, et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33(Database issue):D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thorn CF, Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. Methods Mol Biol. 2005;311:179–191. doi: 10.1385/1-59259-957-5:179. [DOI] [PubMed] [Google Scholar]
- 105.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liem SL. Orphanet and the Dutch Steering Committee Orphan Drugs. A European and Dutch databank of information on rare diseases. Ned Tijdschr Tandheelkd. 2008;115(11):621–623. [PubMed] [Google Scholar]
- 107.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36(Database issue):D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martz E (2009) FirstGlance in Jmol. Available from: http://firstglance.jmol.org
- 109.Price WN, 2nd, et al. Understanding the physical properties that control protein crystallization by analysis of large-scale experimental data. Nat Biotechnol. 2009;27(1):51–57. doi: 10.1038/nbt.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32(Web Server issue):W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ward JJ, et al. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 112.Slabinski L, et al. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics. 2007;23(24):3403–3405. doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- 113.Framework for Handling PSI-2 Community Nominated Targets (2008). Available from: http://sbkb.org/KB/index1.jsp?pageshow=62