Abstract
A new, fully automated approach for indexing documents is presented based on associating textwords in a training set of bibliographic citations with the indexing of journals. This journal-level indexing is in the form of a consistent, timely set of journal descriptors (JDs) indexing the individual journals themselves. This indexing is maintained in journal records in a serials authority database. The advantage of this novel approach is that the training set does not depend on previous manual indexing of hundreds of thousands of documents (i.e., any such indexing already in the training set is not used), but rather the relatively small intellectual effort of indexing at the journal level, usually a matter of a few thousand unique journals for which retrospective indexing to maintain consistency and currency may be feasible. If successful, JD indexing would provide topical categorization of documents outside the training set, i.e., journal articles, monographs, WEB documents, reports from the grey literature, etc., and therefore be applied in searching. Because JDs are quite general, corresponding to subject domains, their most probable use would be for improving or refining search results.
Introduction
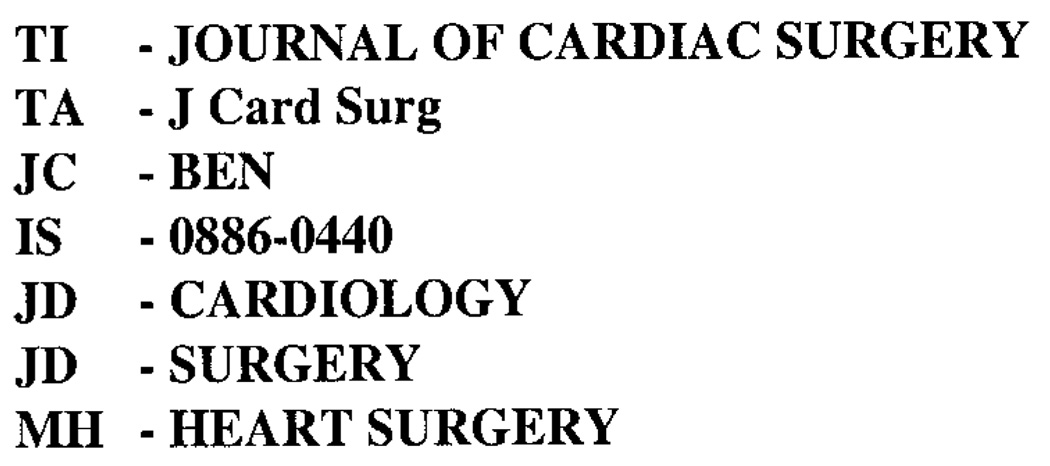
This paper describes a preliminary investigation of a fully automated approach for general categorization of documents. This novel approach is based on associating textwords in bibliographic citations (words in titles and abstracts extracted from documents) with journal-level indexing from a serials authority database. A segment of the MEDLINE citation database created at the National Library of Medicine (NLM) may form a training set to furnish the textwords and the journals (represented by unique journal codes in the citations). The journal-level indexing would be the journal descriptors (JDs) in journal records, corresponding to the training set journals, in NLM’s serials authority database known as SERLINE. For example, the SERLINE record for the Journal of Cardiac Surgery in Figure 1 shows the JDs CARDIOLOGY and SURGERY along with the fields TI (Title), TA (Title Abbreviation), JC (Journal Code), IS (ISSN), and MH (MeSH Heading, for terms selected by indexers from NLM’s Medical Subject Headings thesaurus). The associations between the textwords in MEDLINE citations and JDs in SERLINE records would be the basis for automatically indexing, at a general level, not only the documents (journal articles) represented by citations in the training set, but also documents outside the training set (other journal articles, monographs, WEB documents, etc.).
FIG. 1.
SERLINE record for Journal of Cardiac Surgery.
For example, words in titles and abstracts of MEDLINE citations for articles published in the American Journal of Cardiology can be said to be associated with CARDIOLOGY, which is the JD for this journal in SERLINE. If the association is very strong, i.e., if certain words in a citation appear more often in citations for articles in Cardiology journals than in journals in other disciplines, based on the JDs which index journals in general, we can then use the descriptor CARDIOLOGY as an indexing term in the citation (i.e., to index the article). If the words in this citation are also strongly associated with journals having the JD PHARMACOLOGY, we can also consider this descriptor to be an indexing term in the citation. Once these associations are computed for the training set, they may then be used for indexing any text that has many words in common with those in the training set. For example, an uncataloged report might automatically receive the descriptors CARDIOLOGY and PHARMACOLOGY if it has many words matching the words in the training set that are most strongly associated with these journal descriptors. The JDs are envisioned as corresponding to universally accepted subject areas and therefore translatable among languages.
Motivation, Objective, and Possible Applications
In order to understand why we considered this approach of automatically generating general document descriptors, we briefly review some of the problems with current ways information can be organized for retrieval in real-world systems.
Thesaurus-based human indexing is expensive and labor-intensive. The NLM’s indexing scheme is complex, requiring special training. A beginner’s indexing is normally reviewed by senior indexers for one year. A review of 37 indexing systems (Milstead, 1990) reports that thesauri typically contained 15,000 concepts, and are still growing to keep up with new information. Indexers cannot master or retain the entire thesaurus. Indexer training is both costly and a never-ending task. Systems using machine-aided indexing relying on text analysis techniques reveal two fundamental problems, namely, that machine understanding of text still eludes us and that these efforts are limited by the difficulty of developing large enough dictionaries to capture the richness of language. Research systems have been developed based on automated clustering of documents by topic resulting in graphical semantic maps (Lin, 1997). However, such displays remain to be integrated into real retrieval environments where their functionality and usefulness can be demonstrated, if not tested.
Computer indexing by natural language is also problematic. Some words may be ambiguous, resulting in way too many hits. Other words may be too specialized for retrieving concepts, resulting in too few hits. Nowhere is this more evident than in searching the WEB. Using search engines effectively requires a major time and intellectual investment (Williams, 1996): “If you’re really serious about your searching, you’ll use all the different engines and their various search tricks.” These difficulties have resulted in a renewed appreciation of the librarian and library classification. Mitch Kapor, cofounder of Lotus and the Electronic Frontier Foundation, has called for an “overarching classification scheme to avoid knowledge chaos” and Boston Globe Magazine columnist John Yemma says, “ask the librarian” (Marcus, 1996). A letter to the editor proclaims the virtues of libraries (Hoyt, 1996): “I too have had fun on the Internet, but I still feel the best search engine is the local library. There I have random access to thousands of texts neatly categorized and filed for my convenience. … for those seriously searching information, I suggest they try our libraries first.” Several companies have invested in the development of classification schemes, i.e., the “Net Search” systems featured in Netscape, despite generating paltry revenues and losing money (Maloney, 1996).
All of the above considerations (humans cannot index everything, using text alone is problematic, general categorizations are regarded as useful) have caused us to wonder, what if, instead of applying intellectual efforts to human indexing of individual documents, we instead focus on the much less daunting task of indexing journals, and seeing if this indexing can be used for describing the documents in the journals. Our goal is to automatically index new documents using a consistent, timely set of descriptors, which we feel can realistically be maintained if we focus on journal-level indexing.
If successful, the approach would result in general topical indexing of documents which may have several potential uses for information retrieval, such as providing a general search parameter for intersection with words in search strategies, especially in the case of ambiguous words; partitioning large databases into topical areas at regular intervals (daily, weekly, monthly) for current awareness within the topics; and disambiguation to avoid undesirable results in other automated approaches, such as natural language processing.
It may be possible to describe entire databases, based on a consensus of topical indexing of documents in them, or possibly treating all the text in the database as a single document. We may then process a query as if it were a document, and suggest appropriate databases to search, based on the ranking of the query descriptors that are generated. For example, TOXICOLOGY may be generated as the top-ranked query descriptor. TOXICOLOGY may also be a descriptor for several databases. The database where TOXICOLOGY is ranked highest (not necessarily the top-ranked descriptor for the database) would seem to be the best database to use initially for the query. There may be applications in nonretrieval areas, such as knowledge discovery. For example, computing the JDs associated with a drug would reveal disciplines, possibly quite disparate ones, in which the drug is being used or investigated.
In summary, our goal is to automatically index new documents using a consistent, timely set of descriptors.
Methodology
Previous approaches have been reported based on associations between words in text and manually assigned indexing terms using very large training sets of hundreds of thousands of citations representing individual documents (Biebricher, Fuhr, Lustig, Schwantner, & Knorz, 1988; Cooper & Miller, 1998; Lewis & Gale, 1994; Plaunt & Norgard, 1998)). Approaches based on associations between words in a dictionary and relatively few general subject codes (Liddy, Paik, & Woelfel, 1993; Liddy & Paik, 1992) have also been reported, specifically, SFCs (Subject Field Codes) comprised of 124 major fields (e.g., Anatomy, Cricket, Knots) and 250 subfields that have been manually assigned by lexicographers to more than 35,000 words (actually more than 50,000 word senses) in Longman’s Dictionary of Contemporary English (LDOCE). Words from a collection of machine-readable documents (a corpus of Wall Street Journal articles was used) are then tagged with the appropriate SFCs according to the associations, and statistical algorithms are applied to cluster documents into meaningful groupings not directly encoded in SFCs (for example, grouping together documents about AIDS). Other work using SFCs is cited by this report, including the first such effort, using stories from the New York Times News Service (Walker & Amsler, 1986).
The JD indexing approach we propose has several advantages compared to previous approaches with respect to our goal of automatic indexing using a consistent, timely set of descriptors. Compared to document indexing, developing an initial training set using JD indexing would involve significantly less intellectual effort since it would be based on indexing far fewer items (journals rather than documents). Also, with fewer items to index, it might be feasible to reindex according to changes in the JD scheme, including new JDs and changes in indexing policy. By contrast, the volume in document indexing normally prohibits assigning new descriptors retrospectively or reindexing to reflect changes in indexing policy, and therefore the indexing would become inconsistent over time. Concerning the SFC tagging approach, updating the SFC scheme would mean the retagging of tens of thousands of word senses requiring the specialized knowledge of lexicographers, which would be a greater intellectual effort than updating the indexing of a few thousand journals to conform to an updated JD scheme. Perhaps the most important difference between the SFC and JD approaches is that the former is designed to use SFCs “as an intermediate level representation of a text’s contents” (Liddy, Paik, & Woelfel, 1993), whereas JDs are a final representation. That is, the SFC-based system produces document clusters that, by inspection, correspond to topics such as “airlines” and “medical treatment” but these would not be system-generated descriptors.
Since maintaining journal-level descriptors and assigning them to journals are normal functions at NLM, one may ask, why not simply use them directly as document indexing terms? One reason is that NLM has assigned JDs, which number about 135, for only selected journals, i.e., for the subject section of the publication List of Journals Indexed in Index Medicus (LJI). Furthermore, JDs only partially describe documents published in a journal. For example, a document in the American Journal of Cardiology may also deserve the descriptor PHARMACOLOGY. Some JDs are too general to be useful, e.g., MEDICINE, the JD for the New England Journal of Medicine. Some journals have multiple JDs, not all of which would describe a particular document.
Furthermore, as a practical matter, no system that searches the MEDLINE database, including NLM’s own Web-based IGM (Internet Grateful Med) and PubMed, uses JDs as a search parameter. This may be due to the fact that JDs are not part of the NLM-produced MEDLINE citation (i.e., not mapped to MEDLINE records from SERLINE as are the journal title abbreviation, unique journal code, and ISSN for the journal) as well as absence of JDs for many journals as noted earlier. At this time, in order to search journals in a particular domain, one must locate the journals and then use the union of journal title abbreviations or journal codes as the search parameter. Professional search intermediaries (librarians) have been doing this for years using the JD headers in the subject section of LJI or the JD field in SERLINE to locate some of the journals according to domain. In IGM, alphabetic journal title menus, displaying journal titles ten at a time, are available from which no more than 15 may be selected as the journal search parameter. Users may request journal title menus based on a single keyword matched as a substring of titles. The problem with this, in addition to having to think of all the words that must be used for a complete result, is illustrated by the entry MENTAL for selecting titles with this individual word, like Community Mental Health Journal, but also listing the following titles in which this entry is embedded in a word: Developmental Biology, Environmental Research, Fundamental and Applied Toxicology, Journal of Experimental Biology, etc. PubMed has a journal browser for selecting journal titles or ISSNs, but only one journal at a time will work as a search parameter.
The JD indexing of documents based on associations between textwords and JDs in a training set can be viewed as a way of further extending this latent capability of using JDs if only they were mapped to MEDLINE records from SERLINE. The extension would be to supplement those JDs already in the citation by virtue of the would-be mapping from SERLINE and to generate JDs for indexing any biomedical document, not just those published in journals having JDs in SERLINE.
The approach taken in this research is to use as a training set a dataset of MEDLINE citations, and to compute the association of individual textwords, from document titles and abstracts, with journal-level indexing (i.e., the JDs), an association which we call the word JD profile. We then use the word JD profiles for a document to compute a ranked list of JDs for the document, or document JD profile, as discussed further on. Our training set, which comes from another ongoing research project, is a sample taken from MEDLINE indexing input during 1993, comprised of 3,995 citations from 1,466 different journals. Every citation in the training set must have been associated with at least one JD. The journals have total JD counts (taken from SERLINE) as follows: 1,016 journals have one JD, 370 have two JDs, 69 have three JDs, and 11 have four JDs. There are 31,983 unique words in the training set, extracted from titles and abstracts with the exception of one- and two-character words.
Figure 2 shows the result of computing the word JD profile for the textword MITRAL which occurs 26 times in 11 citations in the training set. We initially compute the rankings based on the number of occurrences of MITRAL for each JD, divided by the total number of occurrences of this word in the training set. For example, MITRAL occurs 11 times in journals described by the JD CARDIOLOGY; we divide this by 26, which is the total number of occurrences of this word, giving us the ranking of 0.423077 for this JD. We do this for each JD, ranking all the JDs for journals in which MITRAL occurs, as displayed under OCCURRENCES OF WORD PER JD / TOTAL OCCURRENCES, BY COUNT. Alternatively, we also compute the rankings based on the number of citations containing MITRAL for each JD, divided by the total number of citations containing this word in the training set. For example, MITRAL occurs in 6 citations in journals described by the JD CARDIOLOGY; we divide this by 11, which is the total number of citations in which this word occurs, giving us the ranking of 0.545455 for this JD. Again, we do this for each JD, ranking all the JDs for journals in which MITRAL occurs, as displayed under CITATION COUNT FOR WORD PER JD / TOTAL CITATION COUNT, BY COUNT. We intend to study the relative merits of the two computations (based on word counts versus citation counts). To provide more detail as to the origins of the computations, under the header JOURNAL TITLES WITH THEIR JD’S are displayed on separate lines the title abbreviations for each of the journals in which MITRAL appears. The first number in each line is the number of occurrences of the word in the journal. The second number is the number of citations in which the word occurs for the journal. For example, MITRAL appears seven times in two citations in the journal Indian Heart J, which has the JD CARDIOLOGY.
FIG. 2.
Word JD profile for textword MITRAL.
Computing the profiles for the set of textwords in a citation to a document develops a JD profile for that document. Suppose, in addition to MITRAL, a citation also contains the textword VALVE. The word JD profile for VALVE (based on substituting the set of variants VALVE/VALVES) is displayed in Figure 3. The top ranking of CARDIOLOGY in the JD profile for VALVE as well as the JD profile for MITRAL reinforces this JD as a descriptor likely to be appropriate for this document with both textwords in the citation.
FIG. 3.
Word JD profile for textword VALVE (including variant VALVES).
To compute a ranked list of JDs for a document (the document JD profile) we average the rankings for each JD in the word JD profiles in the training set of textwords that occur in the citation to the document (to be profiled, a word must be in the training set). A metaphor for this procedure would be to consider the 135 JDs as candidates in an election. Each word “votes” by submitting a “ballot” of the candidates in preferred order, assigning a ranking for each candidate. The “winner” is the candidate with the highest average ranking. Of course, the textwords are more like committed delegates rather than free voters, as the rankings are predetermined by their associations with each candidate. This computation will be illustrated later on.
We can get an indication of the possible usefulness of document JD profiling by an example categorizing a document that is outside the training set, represented by a MEDLINE- like citation followed by the document JD profile (Fig. 4). The fields taken from the MEDLINE citation are the UI (Unique Indicator), TI (Title), MH (MeSH Headings, which include stars as central concept indicators and subheadings), TA (Journal Title Abbreviation), JC (Journal Code, a unique code for the journal), and AB (abstract). In addition, our system maps to the citation the JD field from the SERLINE record for the journal. The FIELDS value of TI, AB indicates that the word JD profiles for computing the document JD profile are for the set of textwords in both the title and the abstract. The document JD profile is computed in two ways. The first list of ranked JDs (JD’S AND RANK BASED ON WORD/VARIANTS OCCURRENCES, BY RANK) uses word JD profiles based on textword occurrences in journals having particular JDs; the second list (JD’S AND RANK BASED ON CITATION COUNTS FOR WORD/VARIANTS, BY RANK) uses word JD profiles based on citation counts for textwords in journals having particular JDs.
FIG. 4.
Sample document JD profile based on title and abstract.
In this sample citation, since CARDIOLOGY is the JD for the journal, the top-ranked CARDIOLOGY in the document JD profile can serve as a test for this methodology. It would seem that the program should at least return highly-ranked JDs matching the JD of the journal in which the cited document is published. However, in addition, the results give us the highly-ranked descriptor PHARMACOLOGY. We can verify that this is a good descriptor as well by noting the consistency with the MeSH indexing (MH field) performed by humans. But remember, we are trying to categorize documents without the benefit of this indexing.
To illustrate the computation, we can compute the document JD profile for this document based, for the sake of brevity, only on the title (Fig. 5), and then describe how the ranking for the top four JDs is arrived at. Excluding words on our stopword list (discussed in the next section), words used from the title are BLOCKADE, FORMATION, CONDUCTANCE (including the variant CONDUCTANCES), CORONARY (including the variant NONCORONARY), and VESSELS (including the variant VESSEL). The word JD profiles for these words in the training set with respect to the top four JDs in the document JD profile (CARDIOLOGY, PHYSIOLOGY, NEUROSCIENCES, PHARMACOLOGY) are shown in Figure 6. The rankings for CARDIOLOGY, NEUROSCIENCES, PHYSIOLOGY, and PHARMACOLOGY in the document JD profile in Figure 5 were computed by averaging the rankings for the respective JDs based on the word JD profiles in Figure 6, for example for CARDIOLOGY based on word occurrences:
The numbers in the numerator in the preceding equation are taken from the ranking of CARDIOLOGY under OCCURRENCES OF WORD (or WORD VARIANTS) PER JD/TOTAL OCCURRENCES, BY COUNT in the five word JD profiles in Figure 6. The result is the average of these numbers, which matches the ranking for CARDIOLOGY in the document JD profile in Figure 5 under JD’S AND RANK BASED ON WORD/VARIANTS OCCURRENCES, BY RANK.
FIG. 5.
Sample document JD profile based on title.
FIG. 6.
JD rankings for CARDIOLOGY, NEUROSCIENCES, PHYSIOLOGY, and PHARMACOLOGY in word JD profiles used for computing sample document JD profile based on title (Fig. 5).
Our programs can also compute an MH JD profile for a MeSH indexing term in citations in the training set. For example, the MH JD profile for Coronary Vessels/*DRUG EFFECTS is shown in Figure 7, where the value of SEARCH corresponds to this indexing term, and UI-LIST has as its value a list of the three Unique Identifiers for citations indexed under this MeSH term in the training set. We compute the rankings in an analogous fashion to the word JD profile based on citation count described earlier and illustrated by Figure 2, except here we are profiling an indexing term instead of a textword. For example, this indexing term appears in two citations in journals described by the JD PHARMACOLOGY; we divide this by three, which is the total number of citations containing this term, giving us the ranking of 0.666667 for this JD. We do this for each JD, giving us the ranking of all the JDs for this term.
FIG. 7.
MH JD profile for MeSH indexing term Coronary Vessels/*DRUG EFFECTS.
A document JD profile can be computed using the MH JD profiles for the set of MeSH indexing terms for the document in an analogous fashion to the document JD profile based on citation count described earlier and illustrated by Figure 4 to Figure 6, except here we would be profiling the document based on MH JD profiles of MeSH terms in the citation for the document instead of word JD profiles of textwords in the citation. It would be interesting to compare document profiles based on human indexing against those based on textwords in titles and abstracts. For example, we can compare the document JD profile based on word JD profiles (Fig. 4) with the document JD profile for the same document based on MH JD profiles (Fig. 8). As seen by the modified MH field in Figure 8 compared to Figure 4, the document JD profile in Figure 8 is based on MH JD profiles for the set of MHs after the removal of stars (central concept indicators), subheadings, and high-frequency MHs known as checktags (Animal, Comparative Study, Dogs, and Support, Non-U.S. Gov’t). CARDIOLOGY and PHARMACOLOGY are the top-ranked descriptors in both document JD profiles, but they seem to stand out more in the profile based on MH JD profiles (Fig. 8) than the one based on word JD profiles (Fig. 4).
FIG. 8.
Sample document JD profile based on MeSH indexing terms, ignoring stars (central concept indicators), subheadings, and checktags (high-frequency terms).
Methodology for attempting to solve the normalization problem is described in the next section.
Problem Areas
We necessarily applied a stopword list in developing the system. Intuitively, it seemed that words like THE, AND, etc., would not be useful. We also applied a word frequency constraint, ignoring words with total frequency less than 13 in the entire training set. It may be worthwhile to attempt a brute force method simply using all words as they are. In the meantime, we very liberally added to our stopword list when a word returned a JD profile with an even distribution across JDs. It should be noted that different high-ranking JDs in a word JD profile are not necessarily a bad result, since the descriptors ultimately assigned to the document are, in effect, a consensus of the JDs for the words. If we continue to explore non-brute force, we would need to develop statistical criteria for useful words, study the effect of word frequency in the training set as well as in the document being profiled, and explore using established lexicons as the basis for word variants (determined on an ad hoc basis in the current system). Since our computations can generate the actual sentences in which words occur in training set documents, we can potentially use the part of speech of a word as a criterion for word selection.
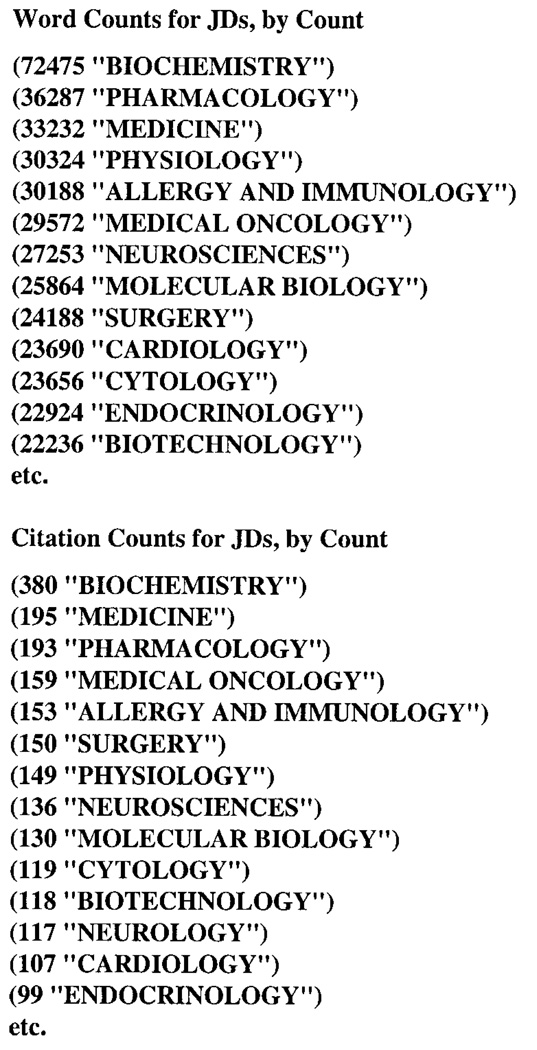
Another problem to be resolved is caused by infrequent or overly frequent JDs in the training set, which in turn is caused, respectively, by too few or too many journals in certain domains. An example of infrequent JDs is illustrated by the poor showing of the word JD profile for ANATOMY (Fig. 9). As can be seen by the journal title abbreviations with their JDs, the high ranking for CARDIOLOGY based on occurrences is due to the word ANATOMY occurring seven times in one issue of J Am Coll Cardiol, plus once in an issue of Am J Cardiol, and the high ranking for SURGERY based on citations is due to ANATOMY occurring in three surgery journals. The JD ANATOMY makes a poor showing because the word appears only twice in one issue of J Morphol. If there were more citations in journals with the JD ANATOMY, this JD would probably be ranked higher, assuming that the word ANATOMY is used often in Anatomy journals. The problem of overly frequent JDs is illustrated by the word JD profile for THE (Fig. 10), showing BIOCHEMISTRY as the outstanding top-ranked JD compared to the rest. It seems doubtful that authors in biochemistry journals use THE more than twice as often as authors in other fields. Contributory to this result is the top journal in this profile, J Biol Chem, which can have 100 documents in a single issue, and is a weekly publication.
FIG. 9.
Word JD profile for textword ANATOMY.
FIG. 10.
Word JD profile for textword THE.
More general evidence for this problem of overly frequent domains in the training set is demonstrated in Figure 11, displaying listings, in descending order by count, of total word counts and citation counts associated with JDs, showing BIOCHEMISTRY having about twice as many words as the next JD. One can assume that the word JD profile for any word equally in the domain of BIOCHEMISTRY and some other domain represented by a JD would have BIOCHEMISTRY ranked higher than the JD for the other domain, simply on the grounds that BIOCHEMISTRY has far more words and citations than the other JD.
FIG. 11.
Word and citation counts associated with JDs in descending order by count.
In an attempt to counteract the effects of the uneven distribution of domains in the training set, we attempted to normalize rankings in word JD profiles based on citation count. We reasoned that the normalization factor for a specified JD, which would be multiplied by the non-normalized ranking in JD profiles, can be expressed as the inverse of the citation count for the specified JD, as follows:
That is, the JD with the highest citation count should have the lowest normalization factor, and the JD with the lowest citation count should have the highest normalization factor. We employed as a constant the average citation count for all JDs, which can be calculated as follows:
We found it useful to incorporate this constant, noting that the normalization factor becomes greater than 1 for a JD when the citation count for the JD is less than this average:
For our final formula, we substituted the ratio expressing the average citation count for all JDs, as follows:
For BIOCHEMISTRY, the factor would be computed as follows, using the above formula:
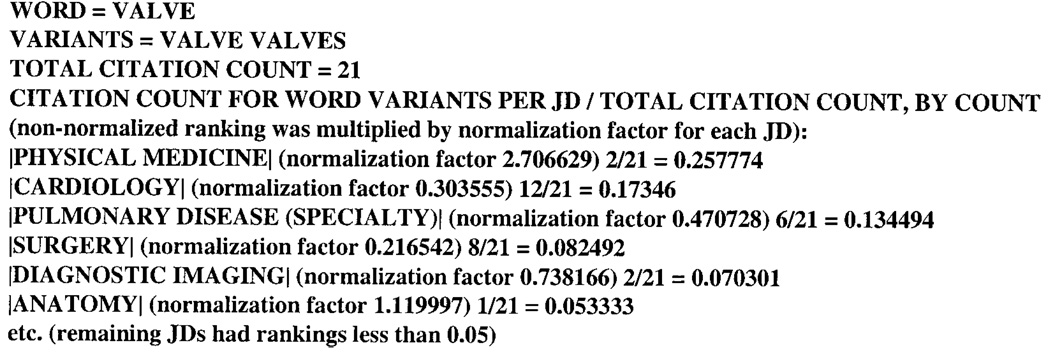
To illustrate the effect of this normalization, we can compare the normalized word JD profile for VALVE/ VALVES based on citation count (Fig. 12) with the non-normalized word JD profile (Fig. 3): The promotion of PULMONARY DISEASE (SPECIALTY) over SURGERY, due to normalization, seems acceptable. The undesirable promotion of PHYSICAL MEDICINE to the top-ranked JD may be due to the low citation count of 12 for PHYSICAL MEDICINE. One may therefore question the validity of the normalized rank for JDs with low (<32) citation counts. Ignoring JDs for this reason would eliminate 70 of the 123 JDs for consideration for any ranking. Perhaps this problem will be helped by using a larger test set where presumably practically all of the JDs would be adequately represented. The normalization process warrants further research, especially in exploring a brute force approach (not using stopwords, variants, etc.).
FIG. 12.
Word JD profile for textword VALVE (including variant VALVES) after applying a normalization algorithm.
We also need to study the effect of current JDs in SERLINE. The same journal may have several JDs, some of which are not descriptive of all documents in the journal, for example, the JDs MEDICAL ONCOLOGY and NEOPLASMS, EXPERIMENTAL for J Cancer Res Clin Oncol. Our programs automatically associate a textword in this journal with both JDs, for example, a document titled “phase II study of 5-fluoruracil, leucovorin, and azidothymidine in patients with metastatic colorectal cancer” (clearly NEOPLASMS, EXPERIMENTAL is inappropriate). Perhaps we need the concept of primary and secondary JDs, where a primary JD would be representative of each and every document in the journal, and a secondary JD would be highly representative of many but not all documents. For this example, both current JDs might be secondary descriptors, and the additional JD of NEOPLASMS might be the primary JD. Another enhancement might be to include as JDs the MHs assigned to serials by catalogers, thus greatly expanding the pool of candidate descriptors from the 135 JDs to virtually all MeSH descriptors (although by the nature of cataloging, most are too specific). For example, the SERLINE record for the J Cardiac Surg in Figure 1 shows the MH HEART SURGERY, which may be useful in providing greater specificity than the official JDs CARDIOLOGY and SURGERY.
For some applications, it would be useful to specify the best JDs from the rankings. We suspect that a specific percentage cutoff applied across the board will not work. We would like to develop algorithms that separate a ranked list into chunks. For example, we would like to automate the grouping of ranked JDs based on word/variants occurrences for the document JD profile in Figure 4 which, by inspection, fall into three groupings, with the best JDs in Group 1, intermediate quality JDs in Group 2, and the rest in Group 3 (Fig. 13).
FIG. 13.
Desired grouping of rankings from sample document JD profile with best JDs in Group 1, intermediate quality JDs in Group 2, and the rest in Group 3.
Future Work
Our plans for future investigation include the following:
Use a larger training set such as a complete month’s input to MEDLINE
Develop and test normalization algorithms to counteract uneven word and citation counts associated with JDs, aimed toward possibly using a brute force approach
Develop algorithms to group rankings in a result, with the set of best JDs at the top
Develop criteria for word selection, e.g., word frequency, grouping word variants, part of speech, for non-brute force approach
Compare and possibly combine textword-based profiles with profiles based on MeSH indexing terms for the same documents
Incorporate into the JD indexing approach the use of JDs associated with journal titles in the end-references as possible descriptors for the document
Develop and test the use of JDs in information retrieval (IR) applications
Develop and test other types of application, for example, knowledge discovery (Swanson, 1990)
Develop training sets in non-biomedical domains associated with journal-based bibliographic databases containing at least abstracts and journal-level descriptions.
To elaborate the point about IR applications, these might include: search terms as alternative to detailed human indexing (National Library of Medicine, 1996), text representation using natural language processing (Aronson, Rindflesch, & Browne, 1994; Hersh & Hickam, 1995; Rindflesch & Aronson, 1994; Srinivasan, 1996; Yang, 1994), referral to information sources in multisource systems (Rodgers, 1995), referral to similar documents (Wilbur & Coffee, 1994), retrieving sections of long texts such as monographs (Hearst & Plaunt, 1993), and accessing texts not routinely indexed such as grey literature (Alberani, De Castro Pietrangeli, & Mazza, 1990) that may nowadays be distributed via CD-ROM or on the Web (Levine, 1997). Because JD indexing is at such a general level, its most likely use, if it performs successfully, may be as a system embedded within some IR application in order to improve or refine the results of the greater application.
Acknowledgments
The author expresses her appreciation to Tom Rindflesch, Alan Aronson, and Larry Hunter at the National Library of Medicine, Larry Wright at the National Cancer Institute, Chris Plaunt at NASA Ames Research Center, and Miguel Ruiz at the University of Iowa for their helpful comments.
References
- Alberani V, De Castro Pietrangeli P, Mazza AMR. The use of grey literature in health sciences: A preliminary survey. Bulletin of the Medical Library Association. 1990;78:358–363. [PMC free article] [PubMed] [Google Scholar]
- Aronson AR, Rindflesch TC, Browne AC. Exploiting a large thesaurus for information retrieval. RIAO 94 Conference Proceedings; Paris. C.I.D.-C.A.S.I.S.; 1994. pp. 197–216. [Google Scholar]
- Biebricher P, Fuhr N, Lustig G, Schwantner M, Knorz G. The automatic indexing system AIR/PHYS—from research to application. In: Chiaramella Y, editor. ACM SIGIR 11th International Conference on Research & Development in Information Retrieval; New York. Association for Computing Machinery; 1988. pp. 333–342. [Google Scholar]
- Cooper GF, Miller RA. An experiment comparing lexical and statistical methods for extracting MeSH terms from clinical free text. Journal of the American Medical Informatics Association. 1998;5:62–75. doi: 10.1136/jamia.1998.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearst MA, Plaunt C. Subtopic structuring for full-length document access. In: Korfhage R, Rasmussen E, Willett P, editors. SIGIR ’93, Proceedings of the Sixteenth Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; New York. Association for Computing Machinery; 1993. pp. 59–68. [Google Scholar]
- Hersh WR, Hickam D. An evaluation of interactive Boolean and natural language searching with an online medical textbook. Journal of the American Society for Information Science. 1995;46:478–489. [Google Scholar]
- Hoyt M. Letters to the editor—Libraries still are the best. The Washington Post. 1996 December 28;:A22. [Google Scholar]
- Levine E. Developing the world’s digital collection on peaceful uses of atomic energy. In: Schwartz C, Rorvig M, editors. Proceedings of the 60th ASIS Annual Meeting; Silver Spring, MD. American Society for Information Science; 1997. pp. 183–185. (published by Information Today, Medford, NJ) [Google Scholar]
- Lewis DD, Gale WA. A sequential algorithm for training text classifiers. In: Croft WB, van Rijsbergen CJ, editors. SIGIR ”94, Proceedings of the Seventeenth Annual International ACM-SIGIR Conference on Research and Development in Information Retrieval; London. Springer-Verlag; 1994. pp. 3–12. [Google Scholar]
- Liddy ED, Paik W, Woelfel JK. Use of subject field codes from a machine-readable dictionary for automatic classification of documents. In: Fidel R, Kwasnik BH, Smith PJ, editors. Advances in Classification Research, Proceedings of the 3rd ASIS SIG/CR Classification Research Workshop; Silver Spring, MD. American Society for Information Science; 1993. pp. 83–100. (published by Learned Information, Medford, NJ) [Google Scholar]
- Liddy ED, Paik W. Statistically-guided word sense disambiguation. Intelligent probabilistic approaches to natural language, Papers from the 1992 Fall Symposium, Technical Report FS-92-04; Menlo Park, CA. AAAI Press; 1992. pp. 98–107. [Google Scholar]
- Lin X. Map displays for information retrieval. Journal of the American Society of Information Science. 1997;48:40–54. [Google Scholar]
- Maloney J. Yahoo! Still searching for profits on the Internet. Fortune. 1996;134:174–182. [Google Scholar]
- Marcus SJ. First line: Ask the librarian. Technology Review. 1996;99:5. [Google Scholar]
- Milstead JM. Methodologies for subject analysis in bibliographic databases. Brookfield, CT: The JELEM Company; 1990. (Distributed by Department of Energy, Office of Scientific and Technical Information, Oak Ridge, TN, Publication No. ETDE/OA-58.) [Google Scholar]
- National Library of Medicine. National Library of Medicine Programs and Services, Fiscal Year 1996, 40. 1996. Next generation indexing project. (Available from NTIS, Springfield, VA.) [Google Scholar]
- Plaunt C, Norgard BA. An association-based method for automatic indexing with a controlled vocabulary. Journal of the American Society for Information Science. 1998;49:888–902. [Google Scholar]
- Rindflesch TC, Aronson AR. Ambiguity resolution while mapping free text to the UMLS Metathesaurus. In: Ozbolt JG, editor. Proceedings of the Eighteenth Annual Symposium on Computer Applications in Medical Care; Bethesda, MD. American Medical Informatics Association; 1994. pp. 240–244. (published by Hanley & Belfus, Philadelphia) [PMC free article] [PubMed] [Google Scholar]
- Rodgers RPC. Automated retrieval from multiple disparate information sources: The World Wide Web and the NLM’s Sourcerer Project. Journal of the American Society for Information Science. 1995;46:755–764. [Google Scholar]
- Srinivasan P. Optimal document-indexing vocabulary for MEDLINE. Information Processing & Management. 1996;32:503–514. [Google Scholar]
- Swanson DR. Medical literature as a potential source of new knowledge. Bulletin of the Medical Library Association. 1990;78:29–37. [PMC free article] [PubMed] [Google Scholar]
- Walker DE, Amsler RA. The use of machine-readable dictionaries in sublanguage analysis. In: Grishman R, Kittredge R, editors. Analyzing language in restricted domains: Sublanguage description and processing. Hillsdale, NJ: Lawrence Erlbaum; 1986. pp. 69–83. [Google Scholar]
- Wilbur WJ, Coffee L. The effectiveness of document neighboring in search enhancement. Information Processing & Management. 1994;30:253–266. [Google Scholar]
- Williams M. Networkings—Robot programs for help mastering searches on the Web. The Washington Post. 1996 July 22;:F20. [Google Scholar]
- Yang Y. Expert Network: Effective and efficient learning from human decisions in text categorization and retrieval. In: Croft WB, van Rijsbergen CJ, editors. SIGIR ’94, Proceedings of the Seventeenth Annual International ACM-SIGIR Conference on Research and Development in Information Retrieval; London. Springer-Verlag; 1994. pp. 13–22. [Google Scholar]