Abstract
Transposable elements (TEs) are increasingly being recognized as powerful facilitators of evolution. We propose the TE-Thrust hypothesis to encompass TE-facilitated processes by which genomes self-engineer coding, regulatory, karyotypic or other genetic changes. Although TEs are occasionally harmful to some individuals, genomic dynamism caused by TEs can be very beneficial to lineages. This can result in differential survival and differential fecundity of lineages. Lineages with an abundant and suitable repertoire of TEs have enhanced evolutionary potential and, if all else is equal, tend to be fecund, resulting in species-rich adaptive radiations, and/or they tend to undergo major evolutionary transitions. Many other mechanisms of genomic change are also important in evolution, and whether the evolutionary potential of TE-Thrust is realized is heavily dependent on environmental and ecological factors. The large contribution of TEs to evolutionary innovation is particularly well documented in the primate lineage. In this paper, we review numerous cases of beneficial TE-caused modifications to the genomes of higher primates, which strongly support our TE-Thrust hypothesis.
Introduction
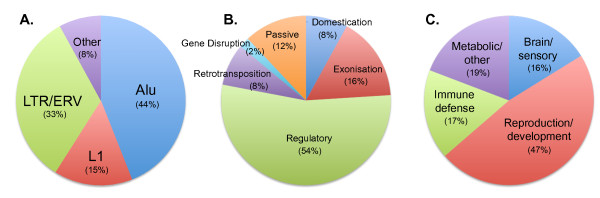
Building on the groundbreaking work of McClintock [1] and numerous others [2-14], we further advanced the proposition of transposable elements (TEs) as powerful facilitators of evolution [15] and now formalise this into 'The TE-Thrust hypothesis'. In this paper, we present much specific evidence in support of this hypothesis, which we suggest may have great explanatory power. We focus mainly on the well-studied higher primate (monkey, ape and human) lineages. We emphasize the part played by the retro-TEs, especially the primate-specific non-autonomous Alu short interspersed element (SINE), together with its requisite autonomous partner long interspersed element (LINE)-1 or L1 (Figure 1A). In addition, both ancient and recent endogenizations of exogenous retroviruses (endogenous retroviruses (ERVs)/solo long terminal repeats (sLTRs) have been very important in primate evolution (Figure 1A). The Alu element has been particularly instrumental in the evolution of primates by TE-Thrust. This suggests that, at least in some mammalian lineages, specific SINE-LINE pairs have a large influence on the trajectory and extent of evolution on the different clades within that lineage.
Figure 1.
Summary of the effect of TEs on primate evolution. (A) Transposable elements (TEs) implicated in the generation of primate-specific traits. (B) Types of events mediated by TEs underlying primate-specific traits. Passive events entail TE-mediated duplications, inversions or deletions. (C) Aspects of primate phenotype affected by TEs. Based on the published data shown in Tables 3 to 6.
The TE-Thrust Hypothesis
The ubiquitous, very diverse, and mostly extremely ancient TEs are powerful facilitators of genome evolution, and therefore of phenotypic diversity. TE-Thrust acts to build, sculpt and reformat genomes, either actively by TE transposition and integration (active TE-Thrust), or passively, because after integration, TEs become dispersed homologous sequences that facilitate ectopic DNA recombination (passive TE-Thrust). TEs can cause very significant and/or complex coding, splicing, regulatory and karyotypic changes to genomes, resulting in phenotypes that can adapt well to biotic or environmental challenges, and can often invade new ecological niches. TEs are usually strongly controlled in the soma, where they can be damaging [16,17], but they are allowed some limited mobility in the germline and early embryo [18-20], where, although they can occasionally be harmful, they can also cause beneficial changes that can become fixed in a population, benefiting the existing lineage, and sometimes generating new lineages.
There is generally no Darwinian selection for individual TEs or TE families, although there may be exceptions, such as the primate-specific Alu SINEs in gene-rich areas [21,22]. Instead, according to the TE-Thrust hypothesis, there is differential survival of those lineages that contain or can acquire suitable germline repertoires of TEs, as these lineages can more readily adapt to environmental or ecological changes, and can potentially undergo, mostly intermittently, fecund radiations. We hypothesize that lineages lacking a suitable repertoire of TEs are, if all else is equal, are liable to stasis, possibly becoming 'living fossils' or even becoming extinct.
TE activity is usually intermittent [23-27], with periodic bursts of transposition due to interplay between various cellular controls, various stresses, de novo syntheses, de novo modifications, new infiltrations of DNA-TEs (by horizontal transfer), or new endogenizations of retroviruses. However, the vast majority of viable TEs usually undergo slow mutational decay and become non-viable (incapable of activity), although some superfamilies have remained active for more than 100 Myr. Episodic TE activity and inactivity, together with differential survival of lineages, suggests an explanation for punctuated equilibrium, evolutionary stasis, fecund lineages and adaptive radiations, all found in the fossil record, and for extant 'fossil species' [15,28].
TE-Thrust is expected to be optimal in lineages in which TEs are active and/or those that possess a high content of homogeneous TEs, both of which can promote genomic dynamism [15]. We hypothesize four main modes of TE-Thrust (Table 1), but as these are extremes of continuums, many intermediate modes are possible.
Table 1.
Hypothesized major modes of transposable element (TE)-thrust
| Mode | TE activity | TE homogeneity | TE population size | Evolutionary outcome | Type of TE thrust |
|---|---|---|---|---|---|
| 1 | Viable and intermittently active | Heterogeneous | Large | Stasis with punctuation events | Active |
| Small | Stasis with punctuation events | Active | |||
| 2 | Viable and intermittently active | Homogeneous | Large | Gradualism with punctuation events | Active and passive |
| Small | Stasis with punctuation events | Active | |||
| 3 | Non-viable/Inactive | Heterogeneous | Large | Stasisa,b | Minimalc |
| Small | Stasisa,b | Minimalc | |||
| 4 | Non-viable/Inactive | Homogeneous | Large | Gradualisma | Passivec |
| Small | Stasisa,b | Minimalc |
aUnless new infiltrations or reactivation of TEs occur.
bFossil taxa are a possible outcome of prolonged stasis.
cInactive/non-viable TEs can be exapted in a delayed fashion, which could cause some resumption of active TE-Thrust.
• Mode 1: periodically active heterogeneous populations of TEs result in stasis with the potential for intermittent punctuation events.
• Mode 2: periodically active homogenous populations of TEs result in: 1) gradualism as a result of ectopic recombination, if the TE population is large, with the potential for periodic punctuation events, or 2) stasis with the potential for periodic punctuation events if the TE population is small.
• Mode 3: non-viable heterogeneous populations of TEs, in the absence of new infiltrations, result in prolonged stasis, which can sometimes result in extinctions and/or 'living fossils'.
• Mode 4: non-viable homogenous populations of TEs, in the absence of new infiltrations, can result in: 1) gradualism as a result of ectopic recombination, if the TE population is large or 2) stasis if the TE population is small.
These modes of TE-Thrust are in agreement with the findings of palaeontologists [29] and some evolutionary biologists [30] that punctuated equilibrium is the most common mode of evolution, but that gradualism and stasis also occur. Many extant 'living fossils' are also known.
We acknowledge that TE-Thrust acts by enhancing evolutionary potential, and whether that potential is actually realized is heavily influenced by environmental, ecological and other factors. Moreover, there are many other 'engines' of evolution besides TE-Thrust, such as point mutation, simple sequence repeats, endosymbiosis, epigenetic modification and whole-genome duplication [31-35], among others. These often complement TE-Thrust; for example, point mutations can endow duplicated or retrotransposed genes with new functions [36,37]. There may also be other, as yet unknown, or hypothesized but unconfirmed, 'engines' of evolution.
Higher primate genomes are very suited to TE-Thrust as they possess large homogeneous populations of TEs
Human and other extant higher primate genomes are well endowed with a relatively small repertoire of TEs (Table 2). These TEs, which have been extensively implicated in engineering primate-specific traits (Table 3; Table 4; Table 5; Table 6), are largely relics of an evolutionary history marked by periodic bursts of TE activity [25,38,39]. TE activity is presently much reduced, but extant simian lineage genomes remain well suited for passive TE-Thrust, with just two elements, Alu and L1, accounting for over 60% of the total TE DNA sequence [21,40,41]. In humans, there are 10 times as many mostly homogeneous class I retro-TEs as there are very heterogeneous class II DNA-TEs [21]. Only L1, Alu, SVA (SINE-R, variable number of tandem repeats (VNTR), Alu) and possibly some ERVs, remain active in humans [42].
Table 2.
Summary of the major transposable elements (TEs) found in humans
| Family | Percentage of genome | Number in genome | Average length, bp | Maximum length, kb | Viable | Potentially autonomous | |
|---|---|---|---|---|---|---|---|
| Type I: retro-TEs | LTRa/ERVb | 8.3 | 443,000 | 510 | 10 | No | Yes (via reverse transcriptase) |
| LINE1c | 16.9 | 516,000 | 900 | 6 | Some | Yes (via reverse transcriptase) | |
| LINE2 | 3.2 | 315,000 | 280 | 5 | No | Yes (via reverse transcriptase) | |
| Alu SINEd | 10.6 | 1,090,000 | 270 | 0.3 | Yes | No | |
| MIRe SINE | 2.2 | 393,000 | 150 | 0.26 | No | No | |
| SVAf SINE-like composite | 0.2 | 3,000 | 1,400 | 3 | Yes | No | |
| Type II: DNA-TEs | Many | 2.8 | 294,000 | 260 | 3 | No | Some (via transposase) |
aLTR = long terminal repeat
bERV = endogenous retrovirus
cLINE = long interspersed nuclear element
dSINE = short interspersed nuclear element
eMIR = mammalian-wide interspersed repeat
fSVA = SINE-VNTR-Alu
Table 3.
Specific examples of transposable elements (TEs) implicated in primate-specific traits: brain and sensory
| TE generated trait | Gene affected | Gene function | TE responsible | Distributiona | Type of event | Effect | Tissue expression | Type of TE-Thrust | Reference |
|---|---|---|---|---|---|---|---|---|---|
| snaRs | Cell growth and translational regulation | Alu | Afr. great ape/ human | Domestication | Novel genes | Brain, testis | Active | Parrott and Mathews, 2009 [105] | |
| BCYRN1 | Translational regulation of dendritic proteins | Alu | Simian | Domestication | Novel gene | Brain | Active | Watson and Sutcliffe, 1987 [106] | |
| FLJ33706 | Unknown | Alu | Human | Domestication | Novel gene | Brain | Active | Li et al., 2010 [107] | |
| Neuronal stability? | SETMAR | DNA repair and replication | Hsmar1 | Simian | Exonization | Novel fusion gene | Brain, various | Active | Cordaux et al., 2006 [108] |
| Survivin | Anti-apoptotic/brain development | Alu | Ape | Exonization | Novel isoform | Brain, spleen | Active | Mola et al., 2007 [109] | |
| ADARB1 | RNA editing/neurotransmitter receptor diversity | Alu | >Human | Exonization | Novel isoform | Brain, various | Active | Lai et al., 1997 [110] | |
| CHRNA1 | Synaptic transmission | MIRb | Great ape | Exonization | Novel isoform | Neuromuscular | Active | Krull et al., 2007 [47] | |
| ASMT | Melatonin synthesis | LINE-1c | >Human | Exonization | Novel isoform | Pineal gland | Active | Rodriguez et al., 1994 [111] | |
| CHRNA3 | Synaptic transmission | Alu | Great ape | Regulatory | Major promoter | Nervous system | Active | Fornasari et al., 1997 [112] | |
| CHRNA6 | Synaptic transmission | Alu | >Human | Regulatory | Negative regulation | Brain | Active | Ebihara et al., 2002 [113] | |
| NAIP | Anti-apoptosis (motor neuron) | Alu | >Human | Regulatory | Alternative promoters | CNS, various | Active | Romanish et al., 2009 [114] | |
| CNTNAP4 | Cell recognition/adhesion | ERVd | >Human | Regulatory | Alternative promoter | Brain, testis | Active | van de Lagemaat et al., 2003 [73] | |
| CCRK | Cell cycle-related kinase | Alu | Simian | Regulatory | CpG island | Brain | Active | Farcas et al., 2009 [86] | |
| Enhanced cognitive capacity/memory? | GLUD2 | Neurotransmitter recycling | Unknown | Ape | Retrotransposition | Novel gene | Brain | Active | Burki and Kaessmann, 2004 [37] |
| Altered auditory perception? | CHRNA9 | Cochlea hair development/ modulation of auditory stimuli | Alu | Human | Deletion | Exon loss | Cochlea, sensory ganglia | Passive | Sen et al., 2006 [62] |
| Trichromatic colour vision | OPN1LW | Cone photoreceptor | Alu | Old World primate | Duplication | Novel gene | Retina | Passive | Dulai et al., 1999 [36] |
a > = Maximum known distribution.
bMIR = mammalian-wide interspersed repeat
cLINE = long interspersed nuclear element
dERV = endogenous retrovirus
Table 4.
Specific examples of transposable elements (TEs) implicated in primate-specific traits: reproduction and development
| TE generated trait | Gene affected | Gene function | TE responsible | Distributiona | Type of event | Effect | Tissue expression | Type of TE-Thrust | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Placental morphogenesis | Syncytin-1 | Trophoblast cell fusion | ERVb | Ape | Domestication | Novel gene | Placenta | Active | Mi et al., 2000 [92] |
| Placental morphogenesis | Syncytin-2 | Trophoblast cell fusion | ERV | Simian | Domestication | Novel gene | Placenta | Active | Blaise et al., 2003 [93] |
| HERVV1 | Unknown | ERV | Simian | Domestication | Novel gene | Placenta | Active | Kjeldbjerg et al., 2008 [115] | |
| HERVV2 | Unknown | ERV | Simian | Domestication | Novel gene | Placenta | Active | Kjeldbjerg et al., 2008 [115] | |
| ERV3 | Development and differentiation? | ERV | Old World primate | Domestication | Novel gene | Placenta, various | Active | Larsson et al., 1994 [116] | |
| DNMT1 | DNA methylation | Alu | >Afr. great ape | Exonization | Novel isoform | Fetal, various | Active | Hsu et al., 1999 [117] | |
| LEPR | Leptin receptor | SVA | Human | Exonization | Novel isoform | Fetal liver | Active | Damert et al., 2004 [118] | |
| IL22RA2 | Regulation of inflammatory responses/interleukin-22 decoy receptor | LTRc | Great ape | Exonization | Novel isoform | Placenta | Active | Piriyapongsa et al., 2007 [119] | |
| PPHLN1 | Epithelial differentiation/nervous-system development | ERV/Alu/LINE-1d | Ape | Exonization | Novel isoforms | Fetal, various | Active | Huh et al., 2006 [120] | |
| CGB1/2 | Chorionic gonadotropin | Alu (snaR-G1/2) | Afr. great ape | Regulatory | Major promoter | Testis | Active | Parrott and Mathews, 2009 [105] | |
| GSDMB | Epithelial development | Alu | Ape | Regulatory | Major promoter | Stomach | Active | Komiyama et al., 2010 [121] | |
| HYAL4 | Hyaluronidase | LINE-1/Alu | >Human | Regulatory | Major promoter | Placenta | Active | van de Lagemaat et al., 2003 [73] | |
| Placental oestrogen synthesis | HSD17B1 | Oestrogen synthesis | ERV | >Human | Regulatory | Major promoter | Ovary, placenta | Active | Cohen et al., 2009 [122] |
| Placental development | INSL4 | Regulation of cell growth and metabolism | ERV | Old World primate | Regulatory | Major promoter | Placenta | Active | Bieche et al., 2003 [123] |
| DSCR4 | Unknown reproductive function | ERV | Ape | Regulatory | Major promoter | Placenta, testis | Active | Dunn et al., 2006 [124] | |
| DSCR8 | Unknown reproductive function | ERV | >Ape | Regulatory | Major promoter | Placenta, testis | Active | Dunn et al., 2006 [124] | |
| CGA | Common subunit of chorionic gonadotropin, luteinizing, follicle-stimulating and thyroid-stimulating hormones | Alu | >Simian | Regulatory | Negative regulation | Placenta, pituitary gland | Active | Scofield et al., 2000 [125] | |
| Globin switching | HBE1 | Embryonic oxygen transport | Alu | >Human | Regulatory | Negative regulation | Fetal | Active | Wu et al., 1990 [126] |
| GH | Growth hormone | Alu | >Human | Regulatory | Negative regulation | Pituitary gland | Active | Trujillo et al., 2006 [127] | |
| WT1 | Urogenital development | Alu | >Human | Regulatory | Negative regulation | Urogenital | Active | Hewitt et al., 1995 [128] | |
| Efficient placental gas exchange | HBG1 | Fetal oxygen transport | LINE-1 | Old World primate | Regulatory | Tissue-specific enhancer | Fetal | Active | Johnson et al., 2006 [91] |
| Placental leptin secretion | LEP | Metabolic regulatory hormone | LTR | >Human | Regulatory | Tissue-specific enhancer | Placenta | Active | Bi et al., 1997 [129] |
| MET | Hepatocyte growth-factor receptor | LINE-1 | > Afr. great ape | Regulatory | Alternative promoter | Liver, Pancreas, Lung | Active | Nigumann et al., 2002 [71] | |
| BCAS3 | Embryogenesis/erythropoiesis | LINE-1 | > Afr. great ape | Regulatory | Alternative promoter | Fetal, various | Active | Wheelan et al., 2005 [130] | |
| CHRM3 | Synaptic transmission | LINE-1 | Human | Regulatory | Alternative promoter | Placenta | Active | Huh et al., 2009 [131] | |
| CLCN5 | Chloride transporter | LINE-1 | >Human | Regulatory | Alternative promoter | Placenta | Active | Matlik et al., 2006 [132] | |
| SLCO1A2 | Organic anion transporter | LINE-1 | >Human | Regulatory | Alternative promoter | Placenta | Active | Matlik et al., 2006 [132] | |
| CHRM3 | Synaptic transmission | LTR | Human | Regulatory | Alternative promoter | Testis | Active | Huh et al., 2009 [131] | |
| IL2RB | Growth-factor receptor | LTR | >Human | Regulatory | Alternative promoter | Placenta | Active | Cohen et al., 2009 [122] | |
| Placental development | ENTPD1 | Thromboregulation | LTR | >Human | Regulatory | Alternative promoter | Placenta | Active | van de Lagemaat et al., 2003 [73] |
| MKKS | Molecular chaperone | LTR/LINE-2 | >Human | Regulatory | Alternative promoter | Testis, fetal | Active | van de Lagemaat et al., 2003 [73] | |
| NAIP | Anti-apoptosis | ERV | >Human | Regulatory | Alternative promoter | Testis | Active | Romanish et al., 2007 [133] | |
| EDNRB | Placental development/circulation | ERV | >Human | Regulatory | Alternative promoter | Placenta | Active | Medstrand et al., 2001 [134] | |
| Placental development | PTN | Growth factor | ERV | Ape | Regulatory | Alternative promoter | Trophoblast | Active | Schulte et al. 1996 [135] |
| MID1 | Cell proliferation and growth | ERV | Old World primate | Regulatory | Alternative promoter | Placenta, fetal kidney | Active | Landry et al., 2002 [136] | |
| NOS3 | Endothelial nitric oxide synthesis | ERV | >Human | Regulatory | Alternative promoter | Placenta | Active | Huh et al., 2008 [137] | |
| GSDMB | Epithelial development | ERV | Ape | Regulatory | Alternative promoter | Various | Active | Sin et al., 2006 [138] | |
| Placental oestrogen synthesis | CYP19 | Oestrogen synthesis | ERV | Simian | Regulatory | Alternative promoter | Placenta | Active | van de Lagemaat et al., 2003 [73] |
| AMACs | Fatty-acid synthesis | SVA | Afr. great ape | Retrotransposition | Novel genes | Placenta, testis | Active | Xing et al., 2006 [139] | |
| POTEs | Pro-apoptosis/spermatogenesis | LINE-1 | Ape | Retrotransposition | Novel fusion genes | Testis, ovary, prostate, placenta | Active | Lee et al., 2006 [140] | |
| PIPSL | Intracellular protein trafficking | LINE-1 | >Great ape | Retrotransposition | Novel fusion gene | Testis | Active | Babushok et al., 2007 [141] | |
| CDYs | Chromatin modification | Unknown | Simian | Retrotransposition | Novel genes | Testis | Active | Lahn and Page, 1999 [142] | |
| ADAM20/21 | Membrane metalloprotease | Unknown | >Human | Retrotransposition | Novel genes | Testis | Active | Betran and Long, 2002 [143] | |
| Placental growth hormone secretion | GH | Placental growth hormone | Alu | Simian | Duplication | Novel genes | Placenta | Passive | De Mendoza et al., 2004 [88] |
| Chr19 miRNAs | Unknown | Alu | Simian | Duplication | Novel genes | Placenta | Passive | Zhang et al., 2008 [144] | |
| Enhanced immune tolerance at fetal-maternal interface | LGALS13/14/16 | Carbohydrate recognition/immune regulation | LINE-1 | Simian | Duplication | Novel genes | Placenta | Passive | Than et al., 2009 [145] |
| Efficient placental gas exchange | HBG2 | Fetal oxygen transport | LINE-1 | Simian | Duplication | Novel gene | Fetal | Passive | Fitch et al., 1991 [90] |
a > = Maximum known distribution.
bERV = endogenous retrovirus
cLTR = long terminal repeat
dLINE = long interspersed nuclear element
Table 5.
Specific examples of transposable elements (TEs) implicated in primate-specific traits: immune defence
| TE generated trait | Gene affected | Gene function | TE responsible | Distributiona | Type of event | Effect | Tissue expression | Type of TE-Thrust | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Soluble CD55 | CD55 | Complement regulation | Alu | >Human | Exonization | Novel isoform | Various | Active | Caras et al., 1987 [146] |
| Intracellular TNFR | P75TNFR | Tumour necrosis factor receptor | Alu | Old World primate | Exonization | Novel isoform | Various | Active | Singer et al., 2004 [147] |
| Altered infectious-disease resistance? | IRGM | Intracellular pathogen resistance | ERVb | Afr. Great Ape | Regulatory | Major promoter | Various | Active | Bekpen et al., 2009 [148] |
| Altered infectious-disease resistance? | IL29 | Antiviral cytokine | Alu/LTRc | >Human | Regulatory | Positive regulation | Dendritic cells, epithelial cells | Active | Thomson et al., 2009 [149] |
| FCER1G | IgE/IgG Fc receptor/T cell antigen receptor | Alu | Ape | Regulatory | Positive/negative regulation | T cells, basophils | Active | Brini et al., 1993 [150] | |
| CD8A | T cell interaction with class I MHC | Alu | Ape | Regulatory | Tissue-specific enhancer | T cells | Active | Hambor et al., 1993 [151] | |
| Red cell ABH antigen expression | FUT1 | Fucosyltransferase | Alu | Ape | Regulatory | Alternative promoter | Erythrocytes | Active | Apoil et al., 2000 [96] |
| TMPRSS3 | Membrane serine protease | Alu/LTR | >Human | Regulatory | Alternative promoter | Peripheral blood leukocytes | Active | van de Lagemaat et al., 2003 [73] | |
| Colon Le antigen expression | B3GALT5 | Galactosyltransferase | ERV | Old World primate | Regulatory | Alternative promoter | Colon, small intestine, breast | Active | Dunn et al., 2003 [152] |
| Prolactin potentiation of the adaptive immune response | PRL | Regulation of lactation and reproduction | ERV | Old World primate | Regulatory | Alternative promoter | Lymphocytes, endometrium | Active | Gerlo et al., 2006 [153] |
| ST6GAL1 | Sialyltransferase | ERV | >Human | Regulatory | Alternative promoter | B lymphocytes | Active | van de Lagemaat et al., 2003 [73] | |
| Vitamin D regulation of cathelicidin antimicrobial peptide gene | CAMP | Antimicrobial peptide | Alu | Simian | Regulatory | Vitamin D responsiveness | Myeloid cells, various | Active | Gombart et al., 2009 [98] |
| MPO | Myeloperoxidase/microbicidal enzyme | Alu | >Human | Regulatory | Thyroid hormone/retinoic acid responsiveness | Myeloid cells | Active | Piedrafita et al., 1996 [154] | |
| Altered infectious-disease resistance? | IFNG | Antiviral/immunoregulatory factor | Alu | Old World primate | Retrotransposition | Novel positive regulatory element | Natural killer cells, T cells | Active | Ackerman et al., 2002 [155] |
| Absence of N-glycolylneuraminic acid/altered infectious-disease resistance? | CMAH | N-glycolylneuraminic acid synthesis | Alu | Human | Gene disruption | Gene loss | Various | Active | Hayakawa et al., 2001 [104] |
| IRGM | Intracellular pathogen resistance | Alu | Old and New World monkey | Gene disruption | Gene loss | Various | Active | Bekpen et al., 2009 [148] | |
| Altered malaria resistance? | HBA2 | Oxygen transport | Alu | >Ape | Duplication | Novel gene | Erythrocytes | Passive | Hess et al., 1983 [156] |
a > = Maximum known distribution.
bERV = endogenous retrovirus
cLTR = long terminal repeat
Table 6.
Specific Examples of transposable elements (TEs) implicated in primate-specific traits: metabolic and other
| TE generated trait | Gene affected | Gene function | TE responsible | Distributiona | Type of event | Effect | Tissue expression | Type of TE-Thrust | Reference |
|---|---|---|---|---|---|---|---|---|---|
| RNF19A | Ubiquitin ligase | Alu | > Human | Exonization | Novel isoform | Various | Active | Huh et al., 2008 [157] | |
| BCL2L11 | Pro-apoptotic | Alu | > Human | Exonization | Novel isoform | Various | Active | Wu et al., 2007 [158] | |
| BCL2L13 | Pro-apoptotic | Alu | > Human | Exonization | Novel isoform | Various (cytosolic instead of mitochondrial) | Active | Yi et al., 2003 [159] | |
| SFTPB | Pulmonary surfactant | Alu/ERVb | Primate | Exonization | Novel isoform | Various | Active | Lee et al., 2009 [160] | |
| Efficiency of ZNF177 transcription and translation | ZNF177 | Transcriptional regulator | Alu/LINE-1c/ERV | > Human | Exonization | Novel isoform | Various | Active | Landry et al., 2001 [161] |
| Production of salivary amylase | AMY1s | Starch digestion | ERV | Old World primate | Regulatory | Major promoter | Salivary gland | Active | Ting et al., 1992 [99] |
| BAAT | Bile metabolism | ERV | > Human | Regulatory | Major promoter | Liver | Active | van de Lagemaat et al., 2003 [73] | |
| CETP | Cholesterol metabolism | Alu | > Human | Regulatory | Negative regulation | Liver | Active | Le Goff et al., 2003 [162] | |
| Absence of FMO1 in adult liver/altered drug metabolism? | FMO1 | Xenobiotic metabolism | LINE-1 | > Human | Regulatory | Negative regulation in liver | Kidney | Active | Shephard et al., 2007 [163] |
| RNF19A | Ubiquitin ligase | LTRd | > Human | Regulatory | Alternative promoter | Various | Active | Huh et al., 2008 [157] | |
| APOC1 | Lipid metabolism | ERV | Ape | Regulatory | Alternative promoter | Various | Active | Medstrand et al., 2001 [134] | |
| KRT18 | Epithelial keratin | Alu | > Human | Regulatory | Retinoic acid responsiveness | Various | Active | Vansant and Reynolds, 1995 [77] | |
| PTH | Parathyroid hormone | Alu | > Old World primate | Regulatory | Negative calcium responsiveness | Parathyroid gland | Active | McHaffie and Ralston, 1995 [164] | |
| PRKACG | cAMP signalling/regulation of metabolism | Unknown | > Old World primate | Retrotransposition | Novel gene | Various | Active | Reinton et al., 1998 [165] | |
| NBR2 | Unknown | Alu | Old World primate | Duplication | Novel gene | Various | Passive | Jin et al., 2004 [166] | |
| LRRC37A | Unknown | Alu | Old World primate | Duplication | Novel genes | Various | Passive | Jin et al., 2004 [166] | |
| ARF2 | GTPase/vesicle trafficking | Alu | Great ape | Inversion | Novel fusion gene | Various | Passive | Jin et al., 2004 [166] | |
| Altered arterial wall function? | ELN | Elastin | Alu | > Old World primate/human | Deletion | Exon losses | Various | Passive | Szabo et al., 1999 [167] |
| Low body mass? | ASIP | Energy metabolism/pigmentation | Alu | Lesser ape (gibbon) | Deletion | Gene loss | Various | Passive | Nakayama and Ishida, 2006 [101] |
a > = Maximum known distribution.
bERV = endogenous retrovirus
cLINE = long interspersed nuclear element
dLTR = long terminal repeat
L1 and the primate-specific Alu predominate in simians [21,40,41], and thus strongly contribute to TE-Thrust in this lineage (Figure 1A). The autonomous L1 is almost universal in mammals, whereas the non-autonomous Alu, like most SINEs, is conspicuously lineage-specific, having been synthesized de novo, extremely unusually, from a 7SL RNA-encoding gene. The confinement of Alu to a single mammalian order is typical of younger SINEs, whereas ancient SINEs, or exapted remnants of them, may be detectable across multiple vertebrate classes [43]. Alu possesses additional unusual characteristics: extreme abundance (1.1 million copies, occurring every 3 kb on average in the human genome), frequent location in gene-rich regions, and a lack of evolutionary divergence [21,44]. Their relatively high homology is most easily explained as being the result of functional selection helping to prevent mutational drift. Thus, Alus have been hypothesized to serve biological functions in their own right, leading to their selection and maintenance in the primate genome [22]. For example, A-to-I RNA editing, which has a very high prevalence in the human genome, mainly occurs within Alu elements [45], which would seem to provide primates with a genetic sophistication beyond that of other mammals. Alus may therefore not represent a peculiar, evolutionary neutral invasion, but rather positively selected functional elements that are resistant to mutational degradation [46]. This has significance for TE-Thrust, as it would greatly prolong the usefulness of Alus as facilitators of evolution within primate lineages.
Other human retro-TEs include the fossil tRNA mammalian-wide intespersed repeat (MIR) SINE, which amplified approximately 130 Mya [21,47] and the much younger SVA, a non-autonomous composite element partly derived from ERV and Alu sequences, which is specific to the great apes and humans [48]. Like Alus, SVAs are mobilised by L1-encoded enzymes and, similar to Alu, a typical full-length SVA is GC-rich, and thus constitutes a potential mobile CpG island. Importantly, ERVs are genome builders/modifiers of exogenous origin [49]. Invasion of ERVs seems to be particularly associated with a key mammalian innovation, the placenta (Table 4). The endogenisation of retroviruses and the horizontal transfer of DNA-TEs into germlines clearly show that the Weismann Barrier is permeable, contrary to traditional theory.
The DNA-TEs, which comprise just 3% of the human genome, are extremely diverse, but are now completely inactive [21,50]. Although some have been exapted within the simian lineage as functional coding sequences (Table 3; Table 4; Table 5; Table 6), DNA-TEs, it seems, cannot now be a significant factor for TE-Thrust in primates, unless there are new infiltrations.
TE-Thrust influences evolutionary trajectories
A key proposal of our TE-Thrust hypothesis is that TEs can promote the origin of new lineages and drive lineage divergence through the engineering of specific traits. Ancestral TEs shared across very many lineages can, by chance, lead to the delayed generation of traits in one lineage but not in another. For example, more than 100 copies of the ancient amniote-distributed AmnSINE1 are conserved as non-coding elements specifically among mammals [51]. However, as they often show a narrow lineage specificity, we hypothesize that younger SINEs (with their partner LINEs) may have a large influence upon the trajectory and the outcomes of the evolution within clades, as is apparent with the Alu/L1 pair in primates (Figure 1A). Probably not all SINEs are equal in this ability; it seems that some SINEs are more readily mobilised than others, and when mobilised, some SINEs are more effective than others at facilitating evolution by TE-Thrust. The extremely abundant primate Alu dimer seems to illustrate this. Whereas the overwhelming majority of SINEs are derived from tRNAs, Alus may have proliferated so successfully because they are derived from the 7SL RNA gene [52], which is part of the signal recognition particle (SRP) that localises to ribosomes. Alu RNAs can therefore bind proteins on the SRP and thus be retained on the ribosome, in position to be retrotransposed by newly synthesized proteins encoded by their partner L1 LINEs [53].
Among the primates, the simians have undergone the greatest evolutionary transitions and radiation. Of the approximately 367 extant primate species, 85% are simians, with the remainder being prosimians, which diverged about 63 Mya. Significantly, large amplifications of L1, and thus of Alus and other sequences confined to simians, offer a plausible explanation for the lack of innovation in the trajectory of evolution in the prosimian lineages, compared with the innovation in the simian lineages. Since their divergence from the basal primates, the simians have experienced repeated periods of intense L1 activity that occurred from about 40 Mya to about 12 Mya [54]. The highly active simian L1s were responsible for the very large amplification of younger Alus and of many gene retrocopies [55]. Possibly, differential activity of the L1/Alu pair may have driven the trajectory and divergence of the simians, compared with the prosimians. The greater endogenization of some retroviruses in simians compared with prosimians [56] may also have played a part. These events may also explain the larger genome size of the simians compared with prosimians [57].
A significant feature of Alus is their dimeric structure, involving a fusion of two slightly dissimilar arms [58]. This added length and complexity seems to increase their effectiveness as a reservoir of evolutionarily useful DNA sequence or as an inducer of ectopic recombination. It may therefore be no coincidence that simian genomes are well endowed with dimeric Alus. Viable SINEs in the less fecund and less evolutionary innovative prosimians are heterogeneous, and include the conventional dimeric Alu, Alu-like monomers, Alu/tRNA dimers and tRNA SINEs [59]. This distinctly contrasts with simian SINEs; in simians, viable SINEs are almost entirely dimeric Alus. Thus, both qualitatively and quantitatively, the Alu dimer seems to represent a key example of the power of a SINE to strongly influence evolutionary trajectory.
Although these coincident events cannot, by themselves, be a clear indication of cause and effect, distinct Alu subfamilies (AluJ, AluS, AluY) correlate with the divergence of simian lineages [38,39]. Whereas the AluJ subfamily was active about 65 Mya when the separation and divergence between the simians and the prosimians occurred, the AluS subfamily was active beginning at about 45 Mya, when the Old World monkey proliferation occurred, followed by a surge in AluY activity and expansion beginning about 30 Mya, contemporaneous with the split between apes and Old World monkeys [38,39]. Thus, periodic expansions of Alu subfamilies in particular seem to correspond temporally with major divergence points in primate evolution. More recent Alu activity may be a factor in the divergence of the human and chimpanzee lineages, with Alus having been three times more active in humans than in chimpanzees [40,60]. Moreover, at least two new Alu subfamilies (AluYa5 and AluYb8) have amplified specifically within the human genome since the human-chimpanzee split [40,60,61].
Passive TE-Thrust mediated by the Alu/L1 pair has also been evident as a force contributing to lineage divergence in the primates. Ectopic recombinations between Alus, in particular, are a frequent cause of lineage-specific deletion, duplication or rearrangement. Comparisons between the human and chimpanzee genomes have revealed the extent to which they have passively exerted their effects in the relatively recent evolutionary history of primates. An examination of human-specific Alu recombination-mediated deletion (ARMD) identified 492 ARMD events responsible for the loss of about 400 kb of sequence in the human genome [62]. Likewise, Han et al. [63] reported 663 chimpanzee-specific ARMD events, deleting about 771 kb of genomic sequence, including exonic sequences in six genes. Both studies suggested that ARMD events may have contributed to the genomic and phenotypic diversity between chimpanzees and humans. L1-mediated recombination also seems to be a factor in primate evolution, with Han et al. [64] reporting 50 L1-mediated deletion events in the human and chimpanzee genomes. The observed high enrichment of TEs such as Alu at low-copy-repeat junctions indicates that TEs have been an important factor in the generation of segmental duplications that are uniquely abundant in primate genomes [39]. Such genomic duplications provide a major avenue for genetic innovation by allowing the functional specialization of coding or regulatory sequences. Karyotypic changes are thought to be an important factor in speciation [65]. Major differences between the human and chimpanzee genomes include nine pericentric inversions, and these have also been linked to TE-mediated recombination events [66]. It thus seems that both the active and passive effects of Alu and L1 have greatly facilitated and influenced the trajectory of simian evolution by TE-Thrust. Transfer RNA-type SINEs, with suitable partner LINEs, probably perform this role in other lineages.
TE-Thrust affects evolutionary trajectory by engineering lineage-specific traits
TEs can act to generate genetic novelties and thus specific phenotypic traits in numerous ways. Besides passively promoting exon, gene or segmental duplications (or deletions) by unequal recombination, or by disruption of genes via insertion, TEs can actively contribute to gene structure or regulation via exaptation. On multiple occasions, TEs have been domesticated to provide the raw material for entire genes or novel gene fusions [11]. More frequently, TEs have contributed partially to individual genes through exonization after acquisition of splice sites [67,68]. Independent exons generated by TEs are often alternatively spliced, and thereby result in novel expressed isoforms that increase the size of the transcriptome [69]. The generation of novel gene sequences during evolution seems to be heavily outweighed by genetic or epigenetic changes in the transcriptional regulation of pre-existing genes [34,70]. Consistent with this, much evidence indicates that a major way in which TEs have acted to functionally modify primate genomes is by actively inserting novel regulatory elements adjacent to genes, thus silencing or enhancing expression levels or changing expression patterns, often in a tissue-specific manner [71-73]. Moreover, because they are highly repetitious and scattered, TEs have the capacity to affect gene expression on a genome-wide scale by acting as distributors of regulatory sequences or CpG islands in a modular form [74]. Many functional binding sites of developmentally important transcription factors have been found to reside on Alu repeats [75]. These include oestrogen receptor-dependent enhancer elements [76] and retinoic acid response elements, which seem to have been seeded next to retinoic acid target genes throughout the primate genome by the AluS subfamily [77]. As a consequence, TEs are able to contribute significantly to the species-specific rewiring of mammalian transcriptional regulatory networks during pre-implantation embryonic development [78]. Similarly, primate-specific ERVs have been implicated in shaping the human p53 transcriptional network [79] and rewiring the core regulatory network of human embryonic stem cells [80].
Certain classes of retro-TEs can actively generate genetic novelty using their retrotranspositional mechanism to partially or fully duplicate existing cellular genes. Duplication is a crucial aspect of evolution, which has been particularly important in vertebrates, and constitutes the primary means by which organisms evolve new genes [81]. LINEs and SVAs have a propensity to transduce host DNA due to their weak transcriptional termination sites, so that 3' flanking regions are often included in their transcripts. This can lead to gene duplication, exon shuffling or regulatory-element seeding, depending on the nature of the sequence involved [37,82,83]. Duplication of genes can also occur via the retrotransposition of mRNA transcripts by LINEs. Such genes are termed retrocopies, which, after subsequent useful mutation, can sometimes evolve into retrogenes, with a new, related function. There are reportedly over one thousand transcribed retrogenes in the human genome [84], with about one new retrogene per million years having emerged in the human lineage during the past 63 Myr [26]. Some primate retrogenes seem to have evolved highly beneficial functions, such as GLUD2 [37].
Specific evidence for TE-Thrust: examples of traits engineered by TEs in the higher primates
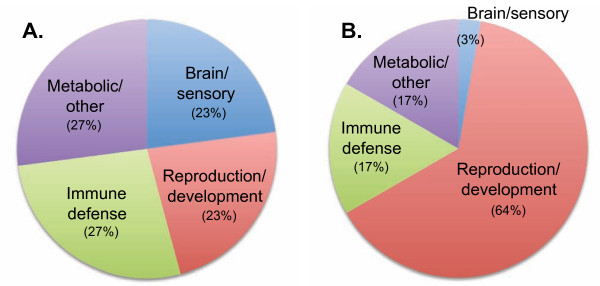
TEs seem to have heavily influenced the trajectories of primate evolution and contributed to primate characteristics, as the simians in particular have undergone major evolutionary advancements in cognitive ability and physiology (especially reproductive physiology). The advancement and radiation of the simians seems to be due, in part and all else being equal, to exceptionally powerful TE-Thrust, owing to its especially effective Alu dimer, partnered by very active novel L1 families, supplemented by ERVs and LTRs. These have engineered major changes in the genomes of the lineage(s) leading to the simian radiations and major transitions. We identified more than 100 documented instances in which TEs affected individual genes and thus were apparently implicated at a molecular level in the origin of higher primate-specific traits (Table 3; Table 4; Table 5; Table 6). The Alu SINE dominated, being responsible for nearly half of these cases, with ERVs/sLTRs being responsible for a third, followed by L1-LINEs at 15% (Figure 1A). Just 2% were due to the young SVAs, and 1% each to ancient MIR SINEs and DNA-TEs. More than half the observed changes wrought by TEs were regulatory (Figure 1B). As discussed below, TEs seem to have influenced four main aspects of the primate phenotype: brain and sensory function, reproductive physiology, immune defence, and metabolic/other (Figure 1C and Table 3; Table 4; Table 5; Table 6). Notably, ERVs, which are often highly transcribed in the germline and placenta [85], were strongly associated with reproductive traits, whereas Alus influenced these four aspects almost equally (Figure 2).
Figure 2.
Comparison of aspects of primate phenotype affected by (A) Alu elements and (B) LTR/ERVs. Based on the published data shown in Tables 3 to 6.
Brain and sensory function
The large brain, advanced cognition and enhanced colour vision of higher primates are distinct from those of other mammals. The molecular basis of these characteristics remains to be fully defined, but from evidence already available, TEs (particularly Alus) seem to have contributed substantially via the origination of novel genes and gene isoforms, or via altered gene transcription (Table 3). Most of the neuronal genes affected by TEs are restricted to the apes, and they seem to have roles in synaptic function and plasticity, and hence learning and memory. These genes include multiple neurotransmitter receptor genes and glutamate dehydrogenase 2 (GLUD2), a retrocopy of GLUD1 that has acquired crucial point mutations. GLUD2 encodes glutamate dehydrogenase, an enzyme that seems to have increased the cognitive powers of the apes through the enhancement of neurotransmitter recycling [37]. The cell cycle-related kinase (CCRK) gene represents a good example of how the epigenetic modification of TEs can be mechanistically linked to the transcriptional regulation of nearby genes [86]. In simians, this gene possesses regulatory CpGs contained within a repressor Alu element, and these CpGs are more methylated in the cerebral cortex of human compared with chimpanzee. Concordantly, CCRK is expressed at higher levels in the human brain [86]. TEs may also affect the brain at a somatic level, because embryonic neural progenitor cells have been found to be permissive to L1 activity in humans [87]. This potentially provides a mechanism for increasing neural diversity and individuality. As our human lineage benefits from a diversity of additional individual talents, as well as shared talents, this phenomenon, if confirmed, could increase the 'fitness' of the human lineage, and is entirely consistent with the concept of differential survival of lineages, as stated in our TE-Thrust hypothesis.
The trichromatic vision of Old World monkeys and apes immensely enhanced their ability to find fruits and other foods, and probably aided them in group identity. This trait evidently had its origin in an Alu-mediated gene-duplication event that occurred about 40 Mya, and subsequently resulted in two separate cone photoreceptor (opsin) genes [36], the tandem OPN1LW and OPN1MW, which are sensitive to long- and medium-wave light respectively. Other mammals possess only dichromatic vision.
Reproductive physiology
Compared with other mammals, simian reproduction is characterized by relatively long gestation periods and by the existence of a hemochorial-type placenta that has evolved additional refinements to ensure efficient fetal nourishment. Available data suggests that TE-Thrust has contributed much of the uniqueness of the higher primate placenta, which seems to be more invasive than that of other mammals, and releases a large number of factors that modify maternal metabolism during pregnancy. These characteristics appear to be due to the generation of novel placenta genes and to various TEs having been exapted as regulatory elements to expand or enhance the expression of pre-existing mammalian genes in the primate placenta (Table 4). The growth hormone (GH) gene locus is particularly notable for having undergone rapid evolution in the higher primates compared with most other mammals. A crucial aspect of this evolutionary advance was a burst of gene-duplication events in which Alu-mediated recombination is implicated as a driving force [88]. The simians thus possess between five and eight GH gene copies, and these show functional specialization, being expressed in the placenta, in which they are thought to influence fetal access to maternal resources during pregnancy [88,89]. Longer gestation periods in simians were accompanied by adaptations to ensure an adequate oxygen supply. One key event was an L1-mediated duplication of the HBG globin gene in the lineage leading to the higher primates, which generated HBG1 and HBG2 [90]. HBG2 subsequently acquired expression specifically in the simian fetus, in which it ensures the high oxygen affinity of fetal blood for more efficient oxygen transfer across the placenta. Old World primates additionally express HBG1 in the fetus, owing to an independent LINE insertion at the beta globin locus [91]. Thus, the important process of placental gas exchange has been extensively improved by TEs in simians, in contrast to that of many mammals, including prosimians, in which fetal and adult haemoglobins are the same.
Two prominent examples of functionally exapted genes whose sequences are entirely TE-derived are syncytin-1 (ERVWE1) and syncytin-2 (ERVWE2). Both of these primate-specific genes are derived from ERV envelope (env) genes [92,93]. The syncytins play a crucial role in simian placental morphogenesis by mediating the development of the fetomaternal interface, which has a fundamental role in allowing the adequate exchange of nutrients and other factors between the maternal bloodstream and the fetus. In a remarkable example of convergent evolution, which attests to the importance of this innovation, two ERV env genes, syncytin-A and syncytin-B, independently emerged in the rodent lineage about 20 Mya [94], as did syncytin-Ory1 within the lagomorphs 12-30 Mya, and these exhibit functional characteristics analogous to the primate syncytin genes [95]. This example, as well as many others (Table 3; Table 4; Table 5; Table 6) suggests the possibility that TE-Thrust may be an important factor in convergent evolution, a phenomenon that can be difficult to explain by traditional theories.
Immune defence
Immune-related genes were probably crucial to the primate lineage by affording protection from potentially lethal infectious diseases. TEs have been reported to contribute to higher primate-restricted transcripts, or to the expression of a wide variety of immunologically relevant genes (Table 5). One example is the insertion of an AluY element into intron 1 of the fucosyltransferase (FUT)1 gene in an ancestor of humans and apes. This enabled erythrocytic expression of FUT1, and thus the ABO blood antigens [96], an adaptation linked to the selective pressure by malarial infection [97]. A particularly good example of a primate-specific adaptation that can be accounted for by a TE is the regulation of the cathelicidin antimicrobial peptide (CAMP) gene by the vitamin D pathway. Only simians possess a functional vitamin D response element in the promoter of this gene, which is derived from the insertion of an AluSx element. This genetic alteration enhances the innate immune response of simians to infection, and potentially counteracts the anti-inflammatory properties of vitamin D [98].
Metabolic/other
TEs seem to underlie a variety of other primate adaptations, particularly those associated with metabolism (Table 6). A striking example, related to dietary change, was the switching of the expression of certain α-amylase genes (AMY1A, AMY1B and AMY1C) from the pancreas to the salivary glands of Old World primates. This event, which was caused by the genomic insertion of an ERV acting as a tissue-specific promoter [99], facilitated the utilization of a higher starch diet in some Old World primates. This included the human lineage, in which consumption of starch became increasingly important, as evidenced by the average human having about three times more AMY1 gene copies than chimpanzees [100]. Another example was the loss of a 100 kb genomic region in the gibbons, due to homologous recombination between AluSx sites [101], resulting in gibbons lacking the ASIP gene involved in the regulation of energy metabolism and pigmentation, which may help to account for their distinctive low body mass, so beneficial for these highly active arboreal primates.
TE-Thrust and divergence of the human lineage
Human and chimpanzee genomes exhibit discernable differences in terms of TE repertoire, TE activity and TE-mediated recombination events [21,40,54,60-64]. Thus, although nucleotide substitutions to crucial genes are important [31], TE-Thrust is likely to have made a significant contribution to the relatively recent divergence of the human lineage [102,103]. In support of this, at least eight of the examples listed (Table 3; Table 4; Table 5; Table 6) are unique to humans. A notable example of a human-specific TE-mediated genomic mutation was the disruption of the CMAH gene, which is involved in the synthesis of a common sialic acid (Neu5Gc), by an AluY element over 2 Mya [104]. This may have conferred on human ancestors a survival advantage by decreasing infectious risk from microbial pathogens known to prefer Neu5Gc as a receptor.
Conclusions
A role for TEs in evolution has long been recognized by many, yet its importance has probably been underestimated. Using primates as exemplar lineages, we have assessed specific evidence, and conclude that it points strongly to an instrumental role for TEs, via TE-Thrust, in engineering the divergence of the simian lineage from other mammalian lineages. TEs, particularly Alu SINEs, have essentially acted as a huge primate-restricted stockpile of potential exons and regulatory regions, and thereby have provided the raw material for these evolutionary transitions. TEs, including Alu SINEs, L1 LINEs, ERVs and LTRs have, through active TE-Thrust, contributed directly to the primate transcriptome, and even more significantly by providing regulatory elements to alter gene expression patterns. Via passive TE-Thrust, homologous Alu and L1 elements scattered throughout the simian genome have led to both genomic gain, in the form of segmental and gene duplications, and genomic loss, by promoting unequal recombination events. Collectively, these events seem to have heavily influenced the trajectories of primate evolution and contributed to characteristic primate traits, as the simian clades especially have undergone major evolutionary advancements in cognitive ability and physiology. Although as yet incompletely documented, the evidence presented here supports the hypothesis that TE-Thrust may be a pushing force for numerous advantageous features of higher primates. These very beneficial features apparently include enhanced brain function, superior fetal nourishment, valuable trichromatic colour vision, improved metabolism, and resistance to infectious-disease agents. Such large evolutionary benefits to various primate clades, brought about by various TE repertories, powerfully demonstrate that if TEs are 'junk' DNA then there is indeed much treasure in the junkyard, and that the TE-Thrust hypothesis could become an important part of some future paradigm shift in evolutionary theory.
Abbreviations
ARMD: Alu recombination-mediated deletion; DNA-TE: DNA transposon; ERV: endogenous retrovirus; L1: LINE-1; LINE: long interspersed nuclear element; LTR: long terminal repeat; MIR: mammalian-wide interspersed repeat; Mya: million years ago; Myr: million years; retro-TE: retrotransposable element; RT: reverse transcriptase; SINE: short interspersed nuclear element; SVA: SINE-VNTR-Alu; TE: transposable element.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
KRO and WKG contributed equally to the writing and the research for this article. Both authors approved the final manuscript.
Contributor Information
Keith R Oliver, Email: K.Oliver@murdoch.edu.au.
Wayne K Greene, Email: W.Greene@murdoch.edu.au.
Acknowledgements
We are grateful to Professor Jen McComb of Murdoch University for critical assessment of the manuscript.
References
- McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- Georgiev GP. Mobile genetic elements in animal cells and their biological significance. Eur J Biochem. 1984;145:203–220. doi: 10.1111/j.1432-1033.1984.tb08541.x. [DOI] [PubMed] [Google Scholar]
- Brosius J. Retroposons - seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. Transposable elements as a molecular evolutionary force. Ann NY Acad Sci. 1999;870:251–264. doi: 10.1111/j.1749-6632.1999.tb08886.x. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Jordan IK. Transposable elements and the evolution of eukaryotic complexity. Curr Issues Mol Biol. 2002;4:65–76. [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Kazazian HH Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Wessler SR. In: The Implicit Genome. Caporale LH, editor. New York: Oxford University Press; 2006. Eukaryotic transposable elements: teaching old genomes new tricks; pp. 138–165. [Google Scholar]
- Biémont C, Vieira C. Genetics: junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Gagen FH. The necessary junk: new functions for transposable elements. Hum Mol Genet. 2007;16:R159–R167. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]
- Böhne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res. 2008;16:203–215. doi: 10.1007/s10577-007-1202-6. [DOI] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. Transposable elements: powerful facilitators of evolution. BioEssays. 2009;31:703–714. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mette MF, Aufsatz W, Jakowitsch J, Matzke AJ. Host defenses to parasitic sequences and the evolution of epigenetic control mechanisms. Genetica. 1999;107:271–287. doi: 10.1023/A:1003921710672. [DOI] [PubMed] [Google Scholar]
- Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Heidmann T. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol Cell Biol. 1996;16:4495–4503. doi: 10.1128/mcb.16.8.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH Jr. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am J Hum Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk JA, Meij IC, Seleme MC, Kano H, Nikopoulos K, Hoefsloot LH, Sistermans EA, de Wijs IJ, Mukhopadhyay A, Plomp AS, de Jong PT, Kazazian HH, Cremers FP. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Walters RD, Kugel JF, Goodrich JA. InvAluable junk: the cellular impact and function of Alu and B2 RNAs. IUBMB Life. 2009;61:831–837. doi: 10.1002/iub.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring E, Hagemann S, Pinsker W. Ancient and recent horizontal invasions of Drosophilids by P elements. J Mol Evol. 2000;51:577–586. doi: 10.1007/s002390010121. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Matjunina LV, Mizrokhi LJ, Georgiev GP. Successive transposition explosions in Drosophila melanogaster and reverse transpositions of mobile dispersed genetic elements. EMBO J. 1985;4:3773–3779. doi: 10.1002/j.1460-2075.1985.tb04147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TM, Hong SJ, Rhyu MG. Periodic explosive expansion of human retroelements associated with the evolution of the hominoid primate. J Korean Med Sci. 2004;19:177–185. doi: 10.3346/jkms.2004.19.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Feschotte C, Pagan HJ, Smith JD, Pritham EJ, Arensburger P, Atkinson PW, Craig NL. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008;18:717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeh DW, Zeh JA, Ishida Y. Transposable elements and an epigenetic basis for punctuated equilibria. BioEssays. 2009;31:715–726. doi: 10.1002/bies.200900026. [DOI] [PubMed] [Google Scholar]
- Gould SJ. The Structure of Evolutionary Theory. Cambridge: The Belknap Press of Harvard University Press; 2002. [Google Scholar]
- Ridley M. Evolution. Oxford: Blackwell Science; 2004. [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M Jr, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Margulis L, Chapman MJ. Endosymbioses: cyclical and permanent in evolution. Trends Microbiol. 1998;6:342–346. doi: 10.1016/S0966-842X(98)01325-0. [DOI] [PubMed] [Google Scholar]
- Monk M. Epigenetic programming of differential gene expression in development and evolution. Dev Genet. 1995;17:188–197. doi: 10.1002/dvg.1020170303. [DOI] [PubMed] [Google Scholar]
- McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- Dulai KS, von Dornum M, Mollon JD, Hunt DM. The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates. Genome Res. 1999;9:629–638. [PubMed] [Google Scholar]
- Burki F, Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hillier LW, Eichler EE, Zody MC, Jaffe DB, Yang SP, Enard W, Hellmann I, Lindblad-Toh K, Altheide TK, Archidiacono N, Bork P, Butler J, Chang JL, Cheng Z, Chinwalla AT, de Jong P, Delehaunty KD, Fronick CC, Fulton LL, Gilad Y, Glusman G, Gnerre S, Graves TA, Hayakawa T, Hayden KE, Huang XQ, Ji HK, Kent WJ, King MC. et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y. et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Labuda D. CORE-SINEs: Eukaryotic short interspersed retroposing elements with common sequence motifs. Proc Natl Acad Sci USA. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda D, Striker G. Sequence conservation in Alu evolution. Nucleic Acids Res. 1989;17:2477–2491. doi: 10.1093/nar/17.7.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, Olshansky M, Rechavi G, Jantsch MF. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Mehler MF. RNA editing, DNA recoding and the evolution of human cognition. Trends Neurosci. 2008;31:227–233. doi: 10.1016/j.tins.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Krull M, Petrusma M, Makalowski W, Brosius J, Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007;17:1139–1145. doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HH Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am J Hum Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Meese E. Human endogenous retroviruses in the primate lineage and their influence on host genomes. Cytogenet Genome Res. 2005;110:448–456. doi: 10.1159/000084977. [DOI] [PubMed] [Google Scholar]
- Pace JK, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Smit AF, Okada N. Functional non-coding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bénit L, Lallemand JB, Casella JF, Philippe H, Heidmann T. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J Virol. 1999;73:3301–3308. doi: 10.1128/jvi.73.4.3301-3308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhao S, Bailey JA, Sahinalp SC, Alkan C, Tuzun E, Green ED, Eichler EE. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–368. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res. 1992;20:487–493. doi: 10.1093/nar/20.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CW. Does SINE evolution preclude Alu function? Nucleic Acids Res. 1998;26:4541–4550. doi: 10.1093/nar/26.20.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, Devine SE. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA. Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen SK, Han K, Wang J, Lee J, Wang H, Callinan PA, Dyer M, Cordaux R, Liang P, Batzer MA. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Lee J, Meyer TJ, Wang J, Sen SK, Srikanta D, Liang P, Batzer MA. Alu recombination-mediated structural deletions in the chimpanzee genome. PLoS Genet. 2007;3:1939–1949. doi: 10.1371/journal.pgen.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Sen SK, Wang J, Callinan PA, Lee J, Cordaux R, Liang P, Batzer MA. Genomic rearrangements by LINE-1 insertion-mediated deletion in the human and chimpanzee lineages. Nucleic Acids Res. 2005;33:4040–4052. doi: 10.1093/nar/gki718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/S0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Sandig C, Chuzhanova N, Goidts V, Szamalek JM, Tänzer S, Müller S, Platzer M, Cooper DN, Hameister H. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes) Hum Mutat. 2005;25:45–55. doi: 10.1002/humu.20116. [DOI] [PubMed] [Google Scholar]
- Sela N, Mersch B, Hotz-Wagenblatt A, Ast G. Characteristics of transposable element exonization within human and mouse. PLoS One. 2010;5:e10907. doi: 10.1371/journal.pone.0010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, Sato S, Davidson BL, Xing Y. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci USA. in press . [DOI] [PMC free article] [PubMed]
- Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Mätlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79:628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68. doi: 10.1016/S0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris J, Fan D, Aleman C, Marks JR, Futreal PA, Wiseman RW, Iglehart JD, Deininger PL, McDonnell DP. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J Biol Chem. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- Vansant G, Reynolds WF. The consensus sequence of a major Alu subfamily contains a functional retinoic acid response element. Proc Natl Acad Sci USA. 1995;92:8229–8233. doi: 10.1073/pnas.92.18.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res. 2010;20:804–815. doi: 10.1101/gr.100594.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA. 2007;104:18613–18618. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH Jr. Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH Jr. Transduction of 3'-flanking sequences is common in L1 retrotransposition. Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci USA. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme S, Bonnaud B, Mallet F. Endogenous retroviruses and animal reproduction. Cytogenet Genome Res. 2005;110:353–364. doi: 10.1159/000084967. [DOI] [PubMed] [Google Scholar]
- Farcas R, Schneider E, Frauenknecht K, Kondova I, Bontrop R, Bohl J, Navarro B, Metzler M, Zischler H, Zechner U, Daser A, Haaf T. Differences in DNA methylation patterns and expression of the CCRK gene in human and nonhuman primate cortices. Mol Biol Evol. 2009;26:1379–1389. doi: 10.1093/molbev/msp046. [DOI] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mendoza A, Escobedo DE, Dávila IM, Saldaña H. Expansion and divergence of the GH locus between spider monkey and chimpanzee. Gene. 2004;336:185–193. doi: 10.1016/j.gene.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Lacroix MC, Guibourdenche J, Frendo JL, Muller F, Evain-Brion D. Human placental growth hormone-a review. Placenta. 2002;23:S87–94. doi: 10.1053/plac.2002.0811. [DOI] [PubMed] [Google Scholar]
- Fitch DH, Bailey WJ, Tagle DA, Goodman M, Sieu L, Slightom JL. Duplication of the gamma-globin gene mediated by L1 long interspersed repetitive elements in an early ancestor of simian primates. Proc Natl Acad Sci USA. 1991;88:7396–7400. doi: 10.1073/pnas.88.16.7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RM, Prychitko T, Gumucio D, Wildman DE, Uddin M, Goodman M. Phylogenetic comparisons suggest that distance from the locus control region guides developmental expression of primate beta-type globin genes. Proc Natl Acad Sci USA. 2006;103:3186–3191. doi: 10.1073/pnas.0511347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Marceau G, Vernochet C, Bénit L, Kanellopoulos C, Sapin V, Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new "syncytin" in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apoil PA, Roubinet F, Despiau S, Mollicone R, Oriol R, Blancher A. Evolution of alpha 2-fucosyltransferase genes in primates: relation between an intronic Alu-Y element and red cell expression of ABH antigens. Mol Biol Evol. 2000;17:337–351. doi: 10.1093/oxfordjournals.molbev.a026314. [DOI] [PubMed] [Google Scholar]
- Cserti CM, Dzik WH. The ABO blood group system and Plasmodium falciparum malaria. Blood. 2007;110:2250–2258. doi: 10.1182/blood-2007-03-077602. [DOI] [PubMed] [Google Scholar]
- Gombart AF, Saito T, Koeffler HP. Exaptation of an ancient Alu short interspersed element provides a highly conserved vitamin D-mediated innate immune response in humans and primates. BMC Genomics. 2009;10:321. doi: 10.1186/1471-2164-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992;6:1457–1465. doi: 10.1101/gad.6.8.1457. [DOI] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, Carter NP, Lee C, Stone AC. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Ishida T. Alu-mediated 100-kb deletion in the primate genome: the loss of the agouti signaling protein gene in the lesser apes. Genome Res. 2006;16:485–490. doi: 10.1101/gr.4763906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ. Transposable element insertions have strongly affected human evolution. Proc Natl Acad Sci USA. 2010;107:19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Satta Y, Gagneux P, Varki A, Takahata N. Alu-mediated inactivation of the human CMP- N-acetylneuraminic acid hydroxylase gene. Proc Natl Acad Sci USA. 2001;98:11399–11404. doi: 10.1073/pnas.191268198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott AM, Mathews MB. snaR genes: recent descendants of Alu involved in the evolution of chorionic gonadotropins. Cold Spring Harb Symp Quant Biol. 2009;74:363–373. doi: 10.1101/sqb.2009.74.038. [DOI] [PubMed] [Google Scholar]
- Watson JB, Sutcliffe JG. Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol Cell Biol. 1987;7:3324–3327. doi: 10.1128/mcb.7.9.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Zhang Y, Wang Z, Zhang Y, Cao C, Zhang PW, Lu SJ, Li XM, Yu Q, Zheng X, Du Q, Uhl GR, Liu QR, Wei L. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput Biol. 2010;6:e1000734. doi: 10.1371/journal.pcbi.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci USA. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mola G, Vela E, Fernández-Figueras MT, Isamat M, Muñoz-Mármol AM. Exonization of Alu-generated splice variants in the survivin gene of human and non-human primates. J Mol Biol. 2007;366:1055–1063. doi: 10.1016/j.jmb.2006.11.089. [DOI] [PubMed] [Google Scholar]
- Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez IR, Mazuruk K, Schoen TJ, Chader GJ. Structural analysis of the human hydroxyindole-O-methyltransferase gene. Presence of two distinct promoters. J Biol Chem. 1994;269:31969–31977. [PubMed] [Google Scholar]
- Fornasari D, Battaglioli E, Flora A, Terzano S, Clementi F. Structural and functional characterization of the human alpha3 nicotinic subunit gene promoter. Mol Pharmacol. 1997;51:250–261. doi: 10.1124/mol.51.2.250. [DOI] [PubMed] [Google Scholar]
- Ebihara M, Ohba H, Ohno SI, Yoshikawa T. Genomic organization and promoter analysis of the human nicotinic acetylcholine receptor alpha6 subunit (CHNRA6) gene: Alu and other elements direct transcriptional repression. Gene. 2002;298:101–108. doi: 10.1016/S0378-1119(02)00925-3. [DOI] [PubMed] [Google Scholar]
- Romanish MT, Nakamura H, Lai CB, Wang Y, Mager DL. A novel protein isoform of the multicopy human NAIP gene derives from intragenic Alu SINE promoters. PLoS One. 2009;4:e5761. doi: 10.1371/journal.pone.0005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjeldbjerg AL, Villesen P, Aagaard L, Pedersen FS. Gene conversion and purifying selection of a placenta-specific ERV-V envelope gene during simian evolution. BMC Evol Biol. 2008;8:266. doi: 10.1186/1471-2148-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Andersson AC, Nilsson BO. Expression of an endogenous retrovirus (ERV3 HERV-R) in human reproductive and embryonic tissues - evidence for a function for envelope gene products. Ups J Med Sci. 1994;99:113–120. doi: 10.3109/03009739409179354. [DOI] [PubMed] [Google Scholar]
- Hsu DW, Lin MJ, Lee TL, Wen SC, Chen X, Shen CK. Two major forms of DNA (cytosine-5) methyltransferase in human somatic tissues. Proc Natl Acad Sci USA. 1999;96:9751–9756. doi: 10.1073/pnas.96.17.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damert A, Löwer J, Löwer R. Leptin receptor isoform 219.1: an example of protein evolution by LINE-1-mediated human-specific retrotransposition of a coding SVA element. Mol Biol Evol. 2004;21:647–651. doi: 10.1093/molbev/msh056. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Polavarapu N, Borodovsky M, McDonald J. Exonization of the LTR transposable elements in human genome. BMC Genomics. 2007;8:291. doi: 10.1186/1471-2164-8-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JW, Kim TH, Yi JM, Park ES, Kim WY, Sin HS, Kim DS, Min DS, Kim SS, Kim CB, Hyun BH, Kang SK, Jung JS, Lee WH, Takenaka O, Kim HS. Molecular evolution of the periphilin gene in relation to human endogenous retrovirus m element. J Mol Evol. 2006;62:730–737. doi: 10.1007/s00239-005-0109-0. [DOI] [PubMed] [Google Scholar]
- Komiyama H, Aoki A, Tanaka S, Maekawa H, Kato Y, Wada R, Maekawa T, Tamura M, Shiroishi T. Alu-derived cis-element regulates tumorigenesis-dependent gastric expression of GASDERMIN B (GSDMB) Genes Genet Syst. 2010;85:75–83. doi: 10.1266/ggs.85.75. [DOI] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Bièche I, Laurent A, Laurendeau I, Duret L, Giovangrandi Y, Frendo JL, Olivi M, Fausser JL, Evain-Brion D, Vidaud M. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod. 2003;68:1422–1429. doi: 10.1095/biolreprod.102.010322. [DOI] [PubMed] [Google Scholar]
- Dunn CA, Romanish MT, Gutierrez LE, van de Lagemaat LN, Mager DL. Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene. 2006;366:335–342. doi: 10.1016/j.gene.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Scofield MA, Xiong W, Haas MJ, Zeng Y, Cox GS. Sequence analysis of the human glycoprotein hormone alpha-subunit gene 5'-flanking DNA and identification of a potential regulatory element as an Alu repetitive sequence. Biochim Biophys Acta. 2000;1493:302–318. doi: 10.1016/s0167-4781(00)00192-5. [DOI] [PubMed] [Google Scholar]
- Wu J, Grindlay GJ, Bushel P, Mendelsohn L, Allan M. Negative regulation of the human epsilon-globin gene by transcriptional interference: role of an Alu repetitive element. Mol Cell Biol. 1990;10:1209–1216. doi: 10.1128/mcb.10.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo MA, Sakagashira M, Eberhardt NL. The human growth hormone gene contains a silencer embedded within an Alu repeat in the 3'-flanking region. Mol Endocrinol. 2006;20:2559–2575. doi: 10.1210/me.2006-0147. [DOI] [PubMed] [Google Scholar]
- Hewitt SM, Fraizer GC, Saunders GF. Transcriptional silencer of the Wilms' tumor gene WT1 contains an Alu repeat. J Biol Chem. 1995;270:17908–17912. doi: 10.1074/jbc.270.30.17908. [DOI] [PubMed] [Google Scholar]
- Bi S, Gavrilova O, Gong DW, Mason MM, Reitman M. Identification of a placental enhancer for the human leptin gene. J Biol Chem. 1997;272:30583–30588. doi: 10.1074/jbc.272.48.30583. [DOI] [PubMed] [Google Scholar]
- Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JW, Kim YH, Lee SR, Kim H, Kim DS, Kim HS, Kang HS, Chang KT. Gain of new exons and promoters by lineage-specific transposable elements-integration and conservation event on CHRM3 gene. Mol Cells. 2009;28:111–117. doi: 10.1007/s10059-009-0106-z. [DOI] [PubMed] [Google Scholar]
- Mätlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006:71753. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanish MT, Lock WM, van de Lagemaat LN, Dunn CA, Mager DL. Repeated recruitment of LTR retrotransposons as promoters by the anti-apoptotic locus NAIP during mammalian evolution. PLoS Genet. 2007;3:e10. doi: 10.1371/journal.pgen.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J Biol Chem. 2001;276:1896–1903. doi: 10.1074/jbc.M006557200. [DOI] [PubMed] [Google Scholar]
- Schulte AM, Lai S, Kurtz A, Czubayko F, Riegel AT, Wellstein A. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc Natl Acad Sci USA. 1996;93:14759–14764. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry JR, Rouhi A, Medstrand P, Mager DL. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol Biol Evol. 2002;19:1934–1942. doi: 10.1093/oxfordjournals.molbev.a004017. [DOI] [PubMed] [Google Scholar]
- Huh JW, Ha HS, Kim DS, Kim HS. Placenta-restricted expression of LTR-derived NOS3. Placenta. 2008;29:602–608. doi: 10.1016/j.placenta.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Sin HS, Huh JW, Kim DS, Kang DW, Min DS, Kim TH, Ha HS, Kim HH, Lee SY, Kim HS. Transcriptional control of the HERV-H LTR element of the GSDML gene in human tissues and cancer cells. Arch Virol. 2006;151:1985–1994. doi: 10.1007/s00705-006-0764-5. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci USA. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ise T, Ha D, Saint Fleur A, Hahn Y, Liu XF, Nagata S, Lee B, Bera TK, Pastan I. Evolution and expression of chimeric POTE-actin genes in the human genome. Proc Natl Acad Sci USA. 2006;103:17885–17890. doi: 10.1073/pnas.0608344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babushok DV, Ohshima K, Ostertag EM, Chen X, Wang Y, Mandal PK, Okada N, Abrams CS, Kazazian HH Jr. A novel testis ubiquitin-binding protein gene arose by exon shuffling in hominoids. Genome Res. 2007;17:1129–1138. doi: 10.1101/gr.6252107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat Genet. 1999;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- Betrán E, Long M. Expansion of genome coding regions by acquisition of new genes. Genetica. 2002;115:65–80. doi: 10.1023/A:1016024131097. [DOI] [PubMed] [Google Scholar]
- Zhang R, Wang YQ, Su B. Molecular evolution of a primate-specific microRNA family. Mol Biol Evol. 2008;25:1493–1502. doi: 10.1093/molbev/msn094. [DOI] [PubMed] [Google Scholar]
- Than NG, Romero R, Goodman M, Weckle A, Xing J, Dong Z, Xu Y, Tarquini F, Szilagyi A, Gal P, Hou Z, Tarca AL, Kim CJ, Kim JS, Haidarian S, Uddin M, Bohn H, Benirschke K, Santolaya-Forgas J, Grossman LI, Erez O, Hassan SS, Zavodszky P, Papp Z, Wildman DE. A primate subfamily of galectins expressed at the maternal-fetal interface that promote immune cell death. Proc Natl Acad Sci USA. 2009;106:9731–9736. doi: 10.1073/pnas.0903568106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caras IW, Davitz MA, Rhee L, Weddell G, Martin DW Jr, Nussenzweig V. Cloning of decay-accelerating factor suggests novel use of splicing to generate two proteins. Nature. 1987;325:545–549. doi: 10.1038/325545a0. [DOI] [PubMed] [Google Scholar]
- Singer SS, Männel DN, Hehlgans T, Brosius J, Schmitz J. From "junk" to gene: curriculum vitae of a primate receptor isoform gene. J Mol Biol. 2004;341:883–886. doi: 10.1016/j.jmb.2004.06.070. [DOI] [PubMed] [Google Scholar]
- Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, Ventura M, Kidd JM, Siswara P, Howard JC, Eichler EE. Death and resurrection of the human IRGM gene. PLoS Genet. 2009;5:e1000403. doi: 10.1371/journal.pgen.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson SJ, Goh FG, Banks H, Krausgruber T, Kotenko SV, Foxwell BM, Udalova IA. The role of transposable elements in the regulation of IFN-lambda1 gene expression. Proc Natl Acad Sci USA. 2009;106:11564–11569. doi: 10.1073/pnas.0904477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini AT, Lee GM, Kinet JP. Involvement of Alu sequences in the cell-specific regulation of transcription of the gamma chain of Fc and T cell receptors. J Biol Chem. 1993;268:1355–1361. [PubMed] [Google Scholar]
- Hambor JE, Mennone J, Coon ME, Hanke JH, Kavathas P. Identification and characterization of an Alu-containing, T-cell-specific enhancer located in the last intron of the human CD8 alpha gene. Mol Cell Biol. 1993;13:7056–7070. doi: 10.1128/mcb.13.11.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CA, Medstrand P, Mager DL. An endogenous retroviral long terminal repeat is the dominant promoter for human beta 1,3-galactosyltransferase 5 in the colon. Proc Natl Acad Sci USA. 2003;100:12841–12846. doi: 10.1073/pnas.2134464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlo S, Davis JR, Mager DL, Kooijman R. Prolactin in man: a tale of two promoters. Bioessays. 2006;28:1051–1055. doi: 10.1002/bies.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedrafita FJ, Molander RB, Vansant G, Orlova EA, Pfahl M, Reynolds WF. An Alu element in the myeloperoxidase promoter contains a composite SP1-thyroid hormone-retinoic acid response element. J Biol Chem. 1996;271:14412–14420. doi: 10.1074/jbc.271.24.14412. [DOI] [PubMed] [Google Scholar]
- Ackerman H, Udalova I, Hull J, Kwiatkowski D. Evolution of a polymorphic regulatory element in interferon-gamma through transposition and mutation. Mol Biol Evol. 2002;19:884–890. doi: 10.1093/oxfordjournals.molbev.a004145. [DOI] [PubMed] [Google Scholar]
- Hess JF, Fox M, Schmid C, Shen CK. Molecular evolution of the human adult alpha-globin-like gene region: insertion and deletion of Alu family repeats and non-Alu DNA sequences. Proc Natl Acad Sci USA. 1983;80:5970–5974. doi: 10.1073/pnas.80.19.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JW, Kim DS, Ha HS, Lee JR, Kim YJ, Ahn K, Lee SR, Chang KT, Kim HS. Cooperative exonization of MaLR and AluJo elements contributed an alternative promoter and novel splice variants of RNF19. Gene. 2008;424:63–70. doi: 10.1016/j.gene.2008.07.030. [DOI] [PubMed] [Google Scholar]
- Wu M, Li L, Sun Z. Transposable element fragments in protein-coding regions and their contributions to human functional proteins. Gene. 2007;401:165–171. doi: 10.1016/j.gene.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Yi P, Zhang W, Zhai Z, Miao L, Wang Y, Wu M. Bcl-rambo beta, a special splicing variant with an insertion of an Alu-like cassette, promotes etoposide- and Taxol-induced cell death. FEBS Lett. 2003;534:61–68. doi: 10.1016/S0014-5793(02)03778-X. [DOI] [PubMed] [Google Scholar]
- Lee JR, Huh JW, Kim DS, Ha HS, Ahn K, Kim YJ, Chang KT, Kim HS. Lineage specific evolutionary events on SFTPB gene: Alu recombination-mediated deletion (ARMD), exonization, and alternative splicing events. Gene. 2009;435:29–35. doi: 10.1016/j.gene.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Landry JR, Medstrand P, Mager DL. Repetitive elements in the 5' untranslated region of a human zinc-finger gene modulate transcription and translation efficiency. Genomics. 2001;76:110–116. doi: 10.1006/geno.2001.6604. [DOI] [PubMed] [Google Scholar]
- Le Goff W, Guerin M, Chapman MJ, Thillet J. A CYP7A promoter binding factor site and Alu repeat in the distal promoter region are implicated in regulation of human CETP gene expression. J Lipid Res. 2003;44:902–910. doi: 10.1194/jlr.M200423-JLR200. [DOI] [PubMed] [Google Scholar]
- Shephard EA, Chandan P, Stevanovic-Walker M, Edwards M, Phillips IR. Alternative promoters and repetitive DNA elements define the species-dependent tissue-specific expression of the FMO1 genes of human and mouse. Biochem J. 2007;406:491–499. doi: 10.1042/BJ20070523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHaffie GS, Ralston SH. Origin of a negative calcium response element in an ALU-repeat: implications for regulation of gene expression by extracellular calcium. Bone. 1995;17:11–14. doi: 10.1016/8756-3282(95)00131-V. [DOI] [PubMed] [Google Scholar]
- Reinton N, Haugen TB, Orstavik S, Skålhegg BS, Hansson V, Jahnsen T, Taskén K. The gene encoding the C gamma catalytic subunit of cAMP-dependent protein kinase is a transcribed retroposon. Genomics. 1998;49:290–297. doi: 10.1006/geno.1998.5240. [DOI] [PubMed] [Google Scholar]
- Jin H, Selfe J, Whitehouse C, Morris JR, Solomon E, Roberts RG. Structural evolution of the BRCA1 genomic region in primates. Genomics. 2004;84:1071–1082. doi: 10.1016/j.ygeno.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Szabó Z, Levi-Minzi SA, Christiano AM, Struminger C, Stoneking M, Batzer MA, Boyd CD. Sequential loss of two neighboring exons of the tropoelastin gene during primate evolution. J Mol Evol. 1999;49:664–671. doi: 10.1007/PL00006587. [DOI] [PubMed] [Google Scholar]