Abstract
Background
Burkholderia cenocepacia is a threatening nosocomial epidemic pathogen in patients with cystic fibrosis (CF) or a compromised immune system. Its high level of antibiotic resistance is an increasing concern in treatments against its infection. Strain B. cenocepacia J2315 is the most infectious isolate from CF patients. There is a strong demand to reconstruct a genome-scale metabolic network of B. cenocepacia J2315 to systematically analyze its metabolic capabilities and its virulence traits, and to search for potential clinical therapy targets.
Results
We reconstructed the genome-scale metabolic network of B. cenocepacia J2315. An iterative reconstruction process led to the establishment of a robust model, iKF1028, which accounts for 1,028 genes, 859 internal reactions, and 834 metabolites. The model iKF1028 captures important metabolic capabilities of B. cenocepacia J2315 with a particular focus on the biosyntheses of key metabolic virulence factors to assist in understanding the mechanism of disease infection and identifying potential drug targets. The model was tested through BIOLOG assays. Based on the model, the genome annotation of B. cenocepacia J2315 was refined and 24 genes were properly re-annotated. Gene and enzyme essentiality were analyzed to provide further insights into the genome function and architecture. A total of 45 essential enzymes were identified as potential therapeutic targets.
Conclusions
As the first genome-scale metabolic network of B. cenocepacia J2315, iKF1028 allows a systematic study of the metabolic properties of B. cenocepacia and its key metabolic virulence factors affecting the CF community. The model can be used as a discovery tool to design novel drugs against diseases caused by this notorious pathogen.
Background
Burkholderia cenocepacia is a Gram-negative opportunistic pathogen and formerly Genomovar III of Burkholderia cepacia complex (Bcc). The Bcc comprises at least 17 taxonomically related species [1-3], which have developed diverse niches from the natural environment [4] and humans as they have emerged as pathogens in patients with cystic fibrosis (CF), chronic granulomatous disease, and in immunocompromised individuals [5]. B. cenocepacia is the dominant Bcc species in patients with CF, accounting for between 50% and 80% of the infection cases [5]. It also causes many instances of non-CF clinical infections, such as for cancer patients [6,7]. As a representative isolate for the spread of an epidemic CF strain, B. cenocepacia J2315 belongs to a clonal lineage known as ET12, which is of increased transmissibility and dominates fatal infections among CF patients in the United Kingdom and Canada [8-12]. B. cenocepacia J2315 is notorious for its high resistance to the majority of clinically useful antimicrobial agents [6,13], including antimicrobial peptides [14,15]. Yet the mechanisms of host infection and drug resistance remain mostly unknown.
The genome of B. cenocepacia J2315 has been sequenced and recently annotated [13]. It is one of the largest Gram-negative genomes consisting of three circular chromosomes with 3.8, 3.2 and 0.8 million base pairs (Mb) respectively and a plasmid. Its complex genome encodes a broad range of metabolic capabilities, and numerous virulence and drug resistance functions that allow it to survive under a variety of conditions and invade immunocompromised individuals. It is vital to develop a systems-level metabolic model for this opportunistic human pathogen to explore and gain insights into its versatile metabolic capability and disease-causing mechanism, and eventually aid in finding potential clinical therapeutic targets. The genome-scale metabolic reconstruction enables integration of genomic information with metabolic activities observed in phenotypic experiments and other "omics" measurements to elicit hidden biological knowledge that would have been otherwise difficult to obtain.
In this study, we presented the manually curated genome-scale metabolic network of B. cenocepacia J2315, named as iKF1028, which accounts for the major metabolic pathways for the synthesis of each component of biomass and for the degradation of common biologically important carbon sources. Syntheses pathways for key virulence factors highly associated with metabolism were particularly emphasized and reconstructed. The in silico model was validated by performing BIOLOG substrate utilization assays, which can test the ability of a microorganism to oxidize various substrates simultaneously [16]. Model-driven analysis and discoveries, including refinement of gene annotation, and gene and enzyme essentiality, were carried out to define the architecture of the genome-wide metabolic and transport network and assist the identification of potential drug targets. Model iKF1028 provides researchers a framework to explore and understand the global metabolism of B. cenocepacia J2315 and its key metabolic virulence factors affecting CF patients upon infection. It allows a broad spectrum of basic and practical applications, especially the application for drug design which may open new doors for anti-infection strategies.
Results and discussion
Characteristics of the genome-scale metabolic network of B. cenocepacia J2315
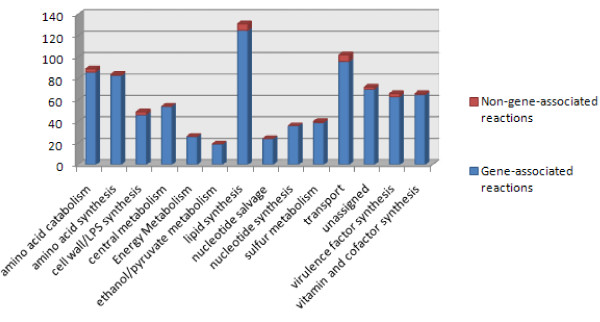
The genome-scale reconstructed metabolic model of B. cenocepacia J2315, referred by the conventional naming rules [17] as iKF1028, consists of 859 internal reactions (including transport) and 834 metabolites. The reconstruction accounts for 1,028 genes, covering 14.4% of the 7,116 protein coding genes identified from whole genome sequencing (http://www.ncbi.nlm.nih.gov/genome?term=burkholderia%20cenocepacia%20J2315). The model iKF1028 includes all major pathways required for cell growth and the degradation of common biologically important carbon sources of B. cenocepacia. Apart from these central metabolic pathways, model iKF1028 also includes pathways associated with key metabolic virulence factors, which provides insights into how the system-level metabolic properties affect pathogenicity. For an overview, the properties of the J2315 genome and the reconstructed model iKF1028 were summarized in Table 1. Genome-scale metabolic models have been successfully used to study many pathogenic bacteria, including Staphylococcus aureus [18-20], Acinetobacter baumannii [21], Mycobacterium tuberculosis [22], Salmonella typhimurium [23], and Pseudomonas aeruginosa [24]. A basic comparison between the model iKF1028 and the above five recently published metabolic reconstructions is also illustrated in Table 1. Schematic representation of the metabolic network of B. cenocepacia J2315 with key metabolic virulence factors is shown in Figure 1. Figure 2 enumerates the metabolic pathways included in iKF1028 and, for each pathway, the number of reactions with assigned and non-assigned genes. The high ratio of gene-associated reactions shows that the reconstructed metabolic model of B. cenocepacia J2315 is reliable. [Additional file 1 for iKF1028 and Additional file 2 is in SBML format]
Table 1.
Comparison of properties of reconstructed metabolic network for selected pathogens
| Model | N.A.* | AbyMBEL891 | iNJ661 | iRR1083 | iMO1056 | iKF1028 |
|---|---|---|---|---|---|---|
| Genome size | 2.8 Mb | 3.93 Mb | 4.4 Mb | 4.8 Mb | 6.3 Mb | 8.1 Mb |
| Included genes | 758 | 650 | 661 | 1,083 | 1,056 | 1,028 |
| Total reactions | 1,497 | 891 | 939 | 1,087 | 883 | 859 |
| Gene-associated reactions (% of total reactions) | 1,278 | 713 (80%) | 723 (77%) | 1,018 (93.7%) | 839 (95%) | 832 (96.9%) |
| Non-gene-associated reactions | 219 | 46 | 216 | 69 | 44 | 27 |
| Transport reactions | 146 | 130 | 93 | 230 | 133 | 102 |
| Metabolites | 1,431 | 778 | 828 | 744 | 760 | 834 |
Properties of metabolic reconstruction of B. cenocepacia J2315 (iKF1028) were compared with other published metabolic reconstructions of pathogenic microbes, S. aureus N315 (2009) [18] (which is the improvement of the N315 reconstruction by Becker and Palsson 2005 [19], and Heinermann et al. 2005 [20]), A. baumannii AYE (AbyMBEL891) (2010) [21], M. tuberculosis H37Rv (iNJ661) (2007) [22], S. typhimurium LT2 (iRR1083) (2009) [23], and P. aeruginosa PAO1 (iMO1056) (2008) [24]. *: Not available for the reconstruction of S. aureus N315.
Figure 1.
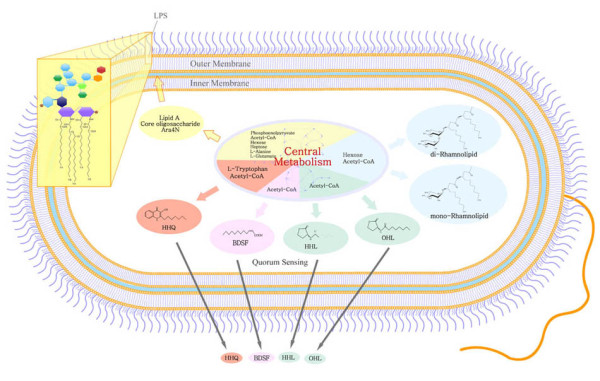
Schematic representation of the metabolic network in B. cenocepacia J2315, referred as model iKF1028.
Figure 2.
Metabolic pathways included in iKF1028 and the distribution of gene-associated and non-gene-associated reactions for each pathway.
Metabolic virulence factors in model iKF1028
The success of B. cenocepacia as a pathogen originates from the ability of its large genome to encode numerous virulence mechanisms [13], including quorum sensing (QS) [25-30], siderophores-based iron uptake systems [31-33], cable pili and adhesion [34-36], motility [37,38], hemolysin [39], ZmpA and ZmpB proteases [40-42], phospholipases [43], secretion systems [44-46], lipopolysaccharides (LPS) [15,47-49], and extracellular capsule [50]. Syntheses of the key metabolic virulence factors of these virulent mechanisms, namely QS, LPS and rhamnolipids, were incorporated and analyzed in iKF1028. Table 2 lists the virulence-associated pathways and the required proteins and precursors for syntheses of virulence factors in each pathway.
Table 2.
Virulence factors incorporated in the metabolic network reconstruction of B. cenocepacia J2315
| Virulence factors | Proteins involved | Major metabolic precursors |
|---|---|---|
| Lipopolysaccharide components | ||
| Lipid A | LpxA, LpxB, LpxC, LpxD, LpxH, LpxK, KdsA, KdsB, KdsC, KdtA, KdoO, HtrB | O2, UDP-N-acetyl-D-glucosamine, (R)-3-Hydroxytetradecanoyl-ACP, (R)-3-Hydroxyhexadecanoyl-ACP, Myristoyl-ACP, D-arabinose 5-phosphate, Phosphoenolpyruvate, |
| Core oligosaccharide | GmhA, RfaE, GmhB, HldD, RmlD, WbiI, WaaC, WaaF, WabP, WabR, WabO, WabS, WaaL | Sedoheptulose 7-phosphate, UDP-glucose, UDP-N-acetyl-D-glucosamine, dTDP-4-dehydro-6-deoxy-L-mannose, L-Alanine |
| Ara4N modification | ArnA1, ArnA2, ArnB, ArnC, ArnT | L-Glutamate, 10-Formyltetrahydrofolate, UDPglucuronate |
| Quorum sensing | ||
| AHLs | CepI, CciI | S-adenosyl-L-methionine, Octanoyl-ACP, Hexanoyl-ACP |
| HHQ | KynA, KynB, KynU, PqsA, PqsB, PqsC, PqsD | L-Tryptophan, 3-oxodecanoyl-ACP |
| BDSF | RpfF, FadA, FadB, FadH | (S)-Hydroxydecanoyl-CoA |
| Rhamnolipids | RhlA, RhlB, RhlC, PhaC | dTDP-4-dehydro-6-deoxy-L-mannose, (R)-3-Hydroxydecanoyl-ACP |
Ara4N, 4-amino-4-deoxy-arabinose; AHLs, N-acylhomoserine lactones; HHQ, 2-heptyl-4-quinolone; BDSF, cis-2-dodecenoic acid
The LPS produced by B. cenocepacia J2315 has an important role in both disease aetiology and antibiotic resistance [51,52]. LPS usually consists of three components: lipid A, core oligosaccharide, and O antigen. Although there were some studies on characterizing the features of LPS in B. cenocepacia, all these studies focused on a certain part/component of LPS. So far, there is no systematic elucidation of the LPS structure and composition specifically for B. cenocepacia strain J2315, nor any global analysis on its biosynthesis process of the LPS. In this study, we depicted the detailed features of the complete LPS structure in B. cenocepacia J2315 by integrating all available reports on LPS. We also reconstructed the LPS-synthesis pathways supplemented with all necessary proteins involved and major metabolic precursors, as illustrated in Figure 3. According to our study, in J2315, each of the three components has a very unique feature. The lipid A portion is modified by an additional Ara4N residue [49,53], which had been shown to reduce the binding of cationic antibiotics and was proposed as a potential drug target [54]. The inner core oligosaccharide contains an unusual KDO-KO-Ara4N residue instead of the typical KDO-KDO residue [15,51,55]. The outer core comprises various polysaccharides including L-glycero-D-manno-heptose, glucose, galactose, quinovosamine, and rhamnose [51]. The O-antigen portion of LPS in J2315 was interrupted by an insertion element in BCAL3125 [47,56]. These differences might indicate the reason why strain J2315 is of remarkably distinct activity.
Figure 3.

Specific structure of Lipopolysaccharide (LPS) in B. cenocepacia J2315 and the synthesis pathways of LPS as well as the proteins involved. The lipid A portion of LPS is composed of two linked glucosamine residues (purple hexagon) with fatty acid side chains (wavy lines), (R)-3-hydroxyhexadecanoic (C16:0 (3-OH)) in an amide linkage and (R)-3-hydroxytetradecanoic (C14:0 (3-OH)) acid and tetradecanoic acid (C14:0) in an ester linkage. There are 4-amino-4-deoxyarabinose (Ara4N, brown sphere) moieties attached to the phosphate residues in the lipid A backbone. The inner core oligosaccharide contains unusual KDO-KO-Ara4N residue linked to the lipid A (KDO: 3-deoxy-D-manno-octulosonic acid, dark blue hexagon; KO: D-glycero-D-talo-octulosonic acid, light blue hexagon). Various polysaccharides comprise the outer core oligosaccharide (L-glycero-D-manno-heptose, blue heptagon; glucose, dark green hexagon; galactose, light green hexagon; quinovosamine, orange hexagon; rhamnose, red hexagon). J2315 cannot make complete LPS O-antigen, owing to an insertion element in BCAL3125 [47].
B. cenocepacia strains possess multiple quorum sensing systems, which regulate the expression of versatile virulence determinants, such as biofilm formation and motility. Strain J2315 owns the ability to synthesize and recognize three types of chemical signals used for cell-to-cell communication: N-acylhomoserine lactones (AHLs), 4-quinolones (4Qs), and the DSF-like molecule cis-2-dodecenoic acid (BDSF). Two AHLs-based QS systems have been found in J2315, namely CciIR and CepIR [25,57], which can both produce N-hexanoyl-L-homoserine lactone (C6-HSL) and N-octanoyl-L-homoserine lactone (C8-HSL) signals using acyl side chain (Hexanoyl-ACP and Octanoyl-ACP, respectively) and S-adenosyl-methionine (SAM) as precursors [58]. The CepIR system is conserved in all species of the Bcc. The CciIR system is encoded within a pathogenicity island, designated as the B. cenocepacia island (cci), which was the first time that cell-signalling genes were found on a genomic island [59]. The 4Qs-based signal, the 2-heptyl-4-quinolone (HHQ), is produced by B. cenocepacia strains [26]. HHQ is the precursor of 2-heptyl-3-hydroxy-4-quinolone (PQS) [60] and its synthesis requires four proteins: PqsA, PqsB, PqsC, and PqsD. It had been reported that the exported HHQ from B. cenocepacia can be recognized by Pseudomonas aeruginosa within which HHQ is converted into PQS which is one of the QS signals for P. aeruginosa [26], highlighting the possibility of inter-species communication during the CF co-infection caused by P. aeruginosa and B. cenocepacia. BDSF is a newly discovered signal molecule produced by B. cenocepacia [28]. The synthesis of BDSF requires the gene BCAM0581 [29].
The synthesis pathway of rhamnolipids was also reconstructed in iKF1028. Although there has not been any report demonstrating that B. cenocepacia can produce rhamnolipid, Dubeau et al demonstrated that Burkholderia thailandensis has the orthologs of rhlA, rhlB, and rhlC, which are responsible for the biosynthesis of rhamnolipids in P. aeruginosa [61]. By protein similarity search against the UniProt database, proteins coded by genes BCAM2340, BCAM2338, and BCAM2336 in B. cenocepacia J2315 were identified as highly similar in sequence to rhlA, rhlB, and rhlC in both B. thailandensis (with BLAST E value of 1E-121, 1E-173, and 1E-108, respectively) and P. aeruginosa (with BLAST E value of 3E-60, 7E-98, and 1E-67, respectively). This facilitates us to hypothesize that B. cenocepacia can potentially generate rhamnolipids. Further experimental investigations are needed.
Model validation and gap-filling using phenotype data
BIOLOG substrates utilization assays for B. cenocepacia J2315 were performed in triplicates in order to validate and refine the model. In silico growth on various substrates was simulated by setting each of them as sole carbon source and its uptake rate to 10 mmol/gcell/h under aerobic conditions based on M9 minimal medium. The simulation was performed on the ToBiN platform by flux balance analysis, as described in Methods. Of the 95 carbon sources tested, 40 could be directly compared with the in silico model of B. cenocepacia J2315, iKF1028. Preliminary disagreement between BIOLOG assays and in silico predictions were probably due to metabolic gaps, improper gene annotations and unacquainted transporters. These discrepancies were checked through gap analysis and literature mining. After continuous gap-filling and network refinement, the overall prediction accuracy was improved to 87.5%, a value that supported iKF1028 as being a proper reconstruction of the B. cenocepacia J2315 core metabolism (comparison results are showed in Table 3).
Table 3.
Comparison with the BIOLOG substrates utilization assays
| Class | Carbon source | BIOLOG results | In silico prediction | Agreement |
|---|---|---|---|---|
| Carbohydrates | N-Acetyl-D-glucosamine | No Growth | No Growth | yes |
| D-Galactose | Growth | Growth | yes | |
| α-D-Glucose | Growth | Growth | yes | |
| m-Inositol | No Growth | No Growth | yes | |
| Sucrose | Growth | Growth | yes | |
| D-Trehalose | Growth | Growth | yes | |
| Carboxylic acids | Acetic acid | Growth | Growth | yes |
| cis-Aconitic acid | Growth | Growth | yes | |
| Citric acid | Growth | Growth | yes | |
| D-Gluconic acid | Growth | Growth | yes | |
| β-Hydroxybutyric acid | Growth | Growth | yes | |
| α-Ketoglutaric acid | Growth | Growth | yes | |
| D,L-Lactic acid | Growth | Growth | yes | |
| Malonic acid | Growth | Growth | yes | |
| Propionic acid | No Growth | No Growth | yes | |
| Quinic acid | Growth | Growth | yes | |
| D-Saccharic acid | Growth | Growth | yes | |
| Succinic acid | Growth | Growth | yes | |
| Amino acids | L-Alanine | Growth | Growth | yes |
| L-Asparagine | Growth | Growth | yes | |
| L-Aspartic acid | No Growth | Growth | no | |
| L-Glutamic acid | Growth | Growth | yes | |
| L-Histidine | Growth | Growth | yes | |
| Hydroxy-L-proline | Growth | Growth | yes | |
| L-Leucine | No Growth | Growth | no | |
| L-Ornithine | No Growth | Growth | no | |
| L-Phenylalanine | Growth | Growth | yes | |
| L-Proline | Growth | Growth | yes | |
| L-Pyroglutamic Acid | Growth | Growth | yes | |
| L-Serine | Growth | Growth | yes | |
| L-Threonine | No Growth | Growth | no | |
| D,L-Carnitine | No Growth | No Growth | yes | |
| γ-Aminobutyric acid | Growth | Growth | yes | |
| Miscellaneous | Succinamic acid | Growth | Growth | yes |
| Uridine | No Growth | No Growth | yes | |
| Thymidine | No Growth | No Growth | yes | |
| Putrescine | No Growth | No Growth | yes | |
| 2,3-Butanediol | No Growth | No Growth | yes | |
| Glycerol | No Growth | Growth | no | |
| D-Glucose-6-Phosphate | Growth | Growth | yes | |
Of the remaining 55 carbon sources tested, 14 were indirectly compared with the model due to the missing knowledge of whether the transport mechanisms of these compounds exist in J2315 or not. Initially, all those 14 carbon sources showed a no-growth phenotype both in silico and in the BIOLOG assays. Then we made the assumption that each of these carbon sources could be transported into the cell (i.e. to function as intracellular compounds) and re-tested whether the in silico model can grow on each of them. The results showed that 11 of the 14 carbon sources enabled iKF1028 to grow after applying the above assumption. This supports the hypothesis that J2315 lacks of transporters for all those 11 carbon sources, even though their catabolic pathways are complete. For the rest 3 carbon sources, the agreement between the in silico results and BIOLOG assays remained.
As the catabolism of the remaining 41 carbon sources out of 55 has not been well studied and information regarding their role in the cell could not be found in public resources, these 41 carbon sources cannot be analyzed in our model. (Complete comparison results with BIOLOG assays are supplied in the Additional file 3)
Model-driven refinement of genome annotation
The reconstruction of metabolic network allowed the identification and refinement of improperly annotated genes of B. cenocepacia J2315 from the public biological databases. Careful effort was made in this work to rectify the current genome annotation based on metabolic gap analysis, BLAST searches, BIOLOG substrate utilization assays, and literature mining. The full list of refinement of genome annotation derived from the genome-scale metabolic reconstruction is shown in Table 4.
Table 4.
Proposed annotation refinements
| Gene Locus | Current Annotation (Burkholderia.com) | Proposed Reannotation | Protein name | Protein ID | Evidence |
|---|---|---|---|---|---|
| BCAL0691, BCAL2945 | Putative cytidylyltransferase, D-beta-D-heptose 7-phosphate kinase | Bifunctional protein RfaE domain II and I, respectively, sugar kinase/adenylyltransferase | RfaE | - | Modelling evidence, RfaE is necessary for biosynthesis of ADP-L-glycero-D-manno-heptose, a precursor for LPS inner core biosynthesis; BLAST search of RfaE from P. aeruginosa gave E values of 9E-35 and 1E-75, respectively |
| BCAL0780 | Putative multiphosphoryl transfer protein | Glucose-specific enzyme IIA component of PTS | Crr | TC-4.A.1.1.1 | BIOLOG assays indicated growth on glucose; BLAST search of Crr from E.coli gave an E value of 1E-28 and Identities of 40% |
| BCAL0781 | Phosphotransferase system, IIbc component | Glucose/N-acetyl glucosamine-specific IIC component | PtsG/NagE | - | Evidence from BIOLOG assays; BLAST search of PtsG, NagE from E.coli gave E values of 3E-107, 7E-151 and Identities of 43%, 56%, respectively |
| BCAL0802 | Gene locus is not assigned in Burkholderia Genome Database and KEGG | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase | IspE | EC-2.7.1.148 | Modelling evidence, IspE is necessary for biosynthesis of polyprenyl-PP, a precursor for ubiquinone biosynthesis; BLAST E value of 4E-172; assigned gene locus of BCAL0802 (from 872938 to 873820) in GeneDB database |
| BCAL1281 | Hypothetical protein | Ornithine N-acyltransferase | OlsB | EC-2.3.1.- | Physiological evidence from Weissenmayer et al. (2002); OlsB is required for the first step of ornithine-derived lipid biosynthesis; BLAST E value of 1E-29 |
| BCAL1431, BCAL1432, BCAL1433 | Putative sugar transport system | Galactose transport | MglB, MglA, MglC | TC-3.A.1.2.3 | BIOLOG assays indicated growth on galactose; and BLAST E values (< 2E-23) |
| BCAL1933, BCAL1934 | Putative formyltransferase, NAD-dependent epimerase/dehydratase family protein | UDP-Ara4N formyltransferase, UDP-4'-keto-5'-carboxypentose decarboxylase | ArnA1, ArnA2 | - | Evidence from Ortega et al. (2006). Unlike other bacteria in which arnA is a single gene encoding a bifunctional enzyme, two distinct genes were found in J2315 (arnA1 and arnA2) and both are required for Ara4N biosynthesis |
| BCAL2388 | Hypothetical protein | Glucose-1-phosphate adenylyltransferase | - | EC-2.7.7.27 | Modelling evidence, a missing protein is required for glycogen biosynthesis; and BLAST search against UNIPROT database gave an E value of 9E-58 |
| BCAL3280 | Putative carbon-nitrogen hydrolase protein | Succinamic acid amidohydrolase | - | EC-3.5.1.3 | BIOLOG assays indicated growth on succinamic acid; modelling showed a missing protein in this pathway; BLAST search against UNIPROT database gave an E value of 3E-46 |
| BCAL3365 | Putative gluconate permease | D-gluconate: H+ symporter | GntP | TC-2.A.8.1.3 | BIOLOG assays indicated growth on D-Gluconic acid; modelling revealed a lack of transporter; BLAST E values of 4E-68 |
| BCAM0469 | Putative aldehyde dehydrogenase | Aldehyde dehydrogenase A, NAD-linked | AldA | EC-1.2.1.21 | Modelling evidence: a gene is missing to synthesize glycolaldehyde which is required for biosynthesis of vitamin B6; BLAST E value of 2E-74 |
| BCAM1404 | Probable exported glycosyl hydrolase | Sucrose-6-phosphate hydrolase | ScrB | EC-3.2.1.26 | BIOLOG assays indicated growth on sucrose; modelling showed missing protein along the pathway; gene locus identified from annotation as 93% similarity from Staphylococcus aureus and E value of 1E-33 |
| BCAM2340, BCAM2338, BCAM2336 | Putative (R)-3-hydroxydecanoyl-ACP: CoA transacylase, putative glycosyltransferase, putative sugar transferase | Rhamnosyltransferase chain A, Rhamnosyltransferase chain B, Rhamnosyltransferase 2 | RhlA, RhlB, RhlC | - | Strong physiological evidence from Dubeau et. al. (2009): Burkholderia cepacia complex (Bcc) can synthesize rhamnolipids; high homologous similarity of RhlA, RhlB, RhlC found in B. cenocepacia J2315 when compared with P. aeruginosa PAO1 and B. thailandensis |
| BCAM2496, BCAM2497, BCAM2498 | Binding-protein-dependent transport system protein, ABC transporter ATP-binding protein, extracellular solute-binding protein | Thiamin transport via ATP-binding protein | ThiP, ThiQ, ThiB | TC-3.A.1.19.1 | Genetic evidence: J2315 is unable to biosynthesize thiamin, which is an important cofactor to grow, by itself and could only obtain it from culture medium; BLAST search of ThiP, ThiQ, ThiB from E.coli got good result |
| BCAM2723 | Putative outer membrane porin protein | Pyroglutatmate porin OpdO | OpdO | TC-1.B.25.1.7 | Evidence from BIOLOG assays; BLAST search of OpdO from P. aeruginosa PAO1 versus the B. cenocepacia J2315 genome gave an E value of 4E-32 |
| BCAM2795 | Hypothetical protein | 1,4-lactonase | - | EC-3.1.1.25 | BIOLOG assays indicated growth on galactose; modelling suggested a protein is missing in this pathway; BLAST search of 1,4-lactonase from Xanthomonas campestris gave an E value of 4E-55 and identities of 42% |
The first type of refinement was to re-annotate genes in iKF1028 - based on literature evidence and BLAST searches - that have been improperly annotated. An example is the gene BCAL1281, which was annotated in both the Burkholderia Genome Database and KEGG as a "hypothetical protein", but that was now reassigned coding for an "ornithine N-acyltransferase". It was reported that the outer membrane of "B. cepacia" [62] possesses unusual polar lipids, ornithine amide lipids (OL) [63,64]. In addition, the protein OlsB is required for the first step of OL biosynthesis [65]. By BLAST searches of OlsB against the UNIPROT database, the gene BCAL1281 of B. cenocepacia J2315 was identified with high similarity.
Another type of annotation refinement was based on gap analyses, which pinpointed reactions for which the gene products involved were missing. For instance, we identified a missing reaction that should be catalyzed by IspE and that takes part in the biosynthesis of polyprenyl-PP, a necessary precursor of the ubiquinone biosynthesis. The IspE encoding genes for other strains of B. cenocepacia (AU1054, HI2424, MC0-3) could be identified in the Burkholderia Genome Database and KEGG. By querying GeneDB, we found that the genomic location from 872938 to 873820, named BCAL0802, was not assigned any function in the above two databases. A BLAST search of BCAL0802 against the UNIPROT database revealed a perfect match with IspE from other B. cenocepacia strains. Consequently, BCAL0802 is annotated as a 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase and this missing reaction was supplemented into the model iKF1028. This example exemplifies how the reconstruction process can drive the reconciliation of isolated data from different biological databases.
BIOLOG substrate utilization assays have already been successfully used on the refinement of metabolic reconstructions [24]. It is an efficient approach for gap analysis and refinement of genome annotation. For example, in our study we can highlight the case for the substrate D-galactose, associated in BIOLOG assays with a growth phenotype. From that result we inferred that J2315 should contain a transport mechanism for D-galactose. By homology searches of MglA, MglB, and MglC, which are galactose-binding proteins conveying galactose into the cell, gene BCAL1431 (mglB), BCAL1432 (mglA), and BCAL1433 (mglC) were identified and they had been annotated as a putative sugar ABC transporter ATP-binding protein, a putative ribose ABC transport system, and a putative sugar transport system permease protein respectively. The ordering of mglB, mglA, and mglC is consistent with a) previous studies indicating that the mgl operon contains three genes and the genes are transcribed in the order of mglB, mglA, and mglC [66] and b) mglA and mglC being located downstream of mglB [67]. As a result, the annotations of BCAL1431-1433 were refined to account for the galactose transport. In total, 7 genes were reannotated based on BIOLOG assays.
Gene essentiality analysis
The term 'essential gene' means a gene for which knockout is lethal (i.e. no biomass yield) under certain conditions (e.g. glucose minimal medium) [68]. Identification of essential genes is helpful to understand the basic functions required for survival and it is an efficient way to discover novel targets for new antimicrobial therapies. Here in this study, iKF1028 was used as a framework to predict computationally identified essential genes in B. cenocepacia J2315 on both M9 minimal medium and synthetic CF medium (SCFM). About 19% (192) and 15% (154) of the 1,028 metabolic genes included in iKF1028 were predicted to be essential on M9 and SCFM media, respectively. There are more genes predicted as essentials on M9 than on SCFM, which indicates the influence of the living environment on the bacterium. 147 overlapping predictions were on both media. These essential genes are unequally located on the three chromosomes and most of the essential genes are located on chromosome 1 (Figure 4a). This result agrees with known features of the J2315 genome: chromosome 1 contains a higher proportion of coding sequence (CDS) involved in central metabolism and other house-keeping functions, whereas chromosomes 2 and 3 contain a greater proportion of CDS encoding accessory functions [13].
Figure 4.
Gene essentiality analysis. (a) Distribution of essential genes predicted on M9 and SCFM respecively; (b) Overlapping essentail genes among in silico prediction on M9, SCFM, and essential genes with in vivo evidence from two P. aeruginosa strains: P. aeruginosa PAO1 and P. aeruginosa PA14.
To assess the predictive potential of the model, we compared the in silico essential genes predicted on SCFM with experimental essentiality data for P. aeruginosa PAO1 and P. aeruginosa PA14 [69,70] since there is no experimental gene essentiality data available for B. cenocepacia. As both P. aeruginosa and B. cenocepacia are CF pathogens, and B. cenocepacia was historically classified under the genus Pseudomonas [71], it is possible that partial similarity exists between them. SCFM was chosen due to its similarity to the nutritional composition of sputum from CF patients. The common set of the essential genes from two P. aeruginosa strains was chosen for comparison in order to reduce the effect of strain-dependent variation. Totally, there are 294 in silico essential metabolic and non-metabolic genes of B. cenocepacia J2315 with high similarity to the common set of P. aeruginosa by BLAST searches, of which 91 in vivo essential genes are present in iKF1028. Genes in the in vivo essential set but not in the iKF1028 were assumed to be involved either in non-metabolic functions or in accessory functions of metabolism. A total of 55% (50) of the in silico predicted essential genes agreed with the in vivo essential genes based on gene homology with P. aeruginosa (Figure 4b, refer to Additional file 4 for detailed information about essential genes).
Based on gene essentiality analysis, the genome-scale metabolic model of B. cenocepacia was further refined. For example, BCAL0660 and BCAL3421, which were homologous genes encoding protein AccC according to their annotation, were originally both included in the model with the gene-protein-reaction (GPR) relationship of "BCAL0660 or BCAL3421". Through in silico gene deletion study, BCAL3421 is identified as non-essential, which is inconsistent with the in vivo essential gene results. Such discrepancy was subject to further analysis. AccA, AccB, AccC, and AccD were four subunits of Acetyl-CoA Carboxylase (ACC) catalyzing the first step of fatty acid biosynthesis [72]. The gene accB, of which the locus in J2315 is BCAL3420, is frequently adjacent to the gene accC and both genes are cotranscribed and form an operon together [73,74]. Furthermore, BCAL3420 and BCAL3421 shows greater than 2-fold expression for J2315 under CF conditions versus soil conditions, yet BCAL0660 shows an opposite result [75]. Taken together, BCAL0660 was excluded from model iKF1028. Further studies are necessary to validate the function of BCAL0660.
Identification of essential enzymes and potential drug targets
Essential enzyme/protein refers to a gene product (catalyzing the relevant reactions) for which individual deletion (i.e. imposing the fluxes through these reactions to zero) is lethal under certain conditions. Through FBA using iKF1028,we could obtain a collection of essential enzymes (protein), based on which 45 essential enzymes were identified as potential drug targets and supported by experimental evidences from literatures. There are 39 of them which were also predicted as drug targets for P. aeruginosa PAO1 [76]. All the 39 targets are nonhomologous to human protein sequences and thus could serve as potential candidate antibiotic drug targets for CF patients infected by both B. cenocepacia J2315 and P. aeruginosa PAO1. Among 39 targets, there are 9 targets, namely AccA, AccB, AccC, AccD, MurA, FolP, PhoA, RibE, and RibH, which have approved drugs in the DrugBank database [77].
The other 6 potential targets, namely ArnT, ArnB, ArnC, ArnA1, ArnA2, and Ugd are unique in B. cenocepacia J2315. ArnT, ArnB, ArnC, ArnA1, and ArnA2 are necessary proteins required for the synthesis of Ara4N, which is an additional moiety of LPS specially presented in B. cenocepacia J2315. Ara4N is essential for the viability of B. cenocepacia J2315 and significantly contributes to high resistance to antimicrobial peptides (AMPs) [54]. AMPs have been proposed as agents for treating CF infections [78,79]. It had also been demonstrated that arnC transposon mutant was survival-defective and attenuated in infected rats [48]. The UDP-glucose dehydrogenase (Ugd), which catalyzes the conversion of UDP-glucose to UDP-glucuronic acid and is the initial step in the synthesis of UDP-Ara4N, is also necessary for the viability of B. cenocepacia and its resistance to polymyxin B [80]. These targets are potentially useful for designing strategies against B. cenocepacia J2315. Further studies are necessary to test their applicability. An overview of the 45 proposed targets is given in Table 5.
Table 5.
Proposed essential enzymes that can be candidate drug targets for B. cenocepacia J2315
| Functional subsystem | EC No. | Protein | Enzyme name |
|---|---|---|---|
| Amino acid metabolism | EC-4.2.3.4 | AroB | 3-dehydroquinate synthase |
| EC-4.2.3.5 | AroC | Chorismate synthase | |
| EC-1.1.1.25 | AroE | Shikimate dehydrogenase | |
| EC-1.3.1.26 | DapB | Dihydrodipicolinate reductase | |
| EC-2.3.1.117 | DapD | Tetrahydrodipicolinate succinylase | |
| EC-5.1.1.7 | DapF | Diaminopimelate epimerase | |
| EC-2.7.2.4 | LysC | Aspartate kinase | |
| Lipid synthesis | EC-6.4.1.2 | AccA* | Acetyl-CoA carboxylase carboxyltransferase subunit-α |
| EC-6.4.1.2 | AccB* | Acetyl-CoA carboxylase biotin carboxyl carrier protein subunit | |
| EC-6.4.1.2 | AccC* | Acetyl-CoA carboxylase biotin carboxylase subunit | |
| EC-6.4.1.2 | AccD* | Acetyl-CoA carboxylase subunit-β | |
| EC-2.7.8.8 | PssA | Phosphatidylserine synthase | |
| Cell wall/LPS synthesis | - | ArnA1# | UDP-Ara4N formyltransferase |
| - | ArnA2# | UDP-4-keto-5-carboxypentose decarboxylase | |
| - | ArnB# | UDP-4-ketopentose aminotransferase | |
| - | ArnC# | Ara4N Und-P transferase | |
| - | ArnT# | Ara4N transferase | |
| EC-3.6.1.27 | BacA | Undecaprenyl pyrophosphate phosphatase | |
| EC-2.5.1.55 | KdsA | 2-dehydro-3-deoxyphosphooctonate aldolase | |
| EC-2.7.7.38 | KdsB | 3-deoxy-manno-octulosonate cytidylyltransferase | |
| EC-2.3.1.129 | LpxA | UDP-N-acetylglucosamine acyltransferase | |
| EC-2.4.1.182 | LpxB | Lipid-A-disaccharide synthase | |
| EC-3.5.1.- | LpxC | UDP-3-O-[3-hydroxymyristoyl]N-acetylglucosamine deacetylase | |
| EC-2.7.1.130 | LpxK | Tetraacyldisaccharide 4'-kinase | |
| EC-2.5.1.7 | MurA* | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | |
| EC-1.1.1.158 | MurB | UDP-N-acetylmuramate dehydrogenase | |
| EC-6.3.2.8 | MurC | UDP-N-acetylmuramate--L-alanine ligase | |
| EC-6.3.2.9 | MurD | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase | |
| EC-6.3.2.13 | MurE | UDP-N-acetylmuramoylalanyl-D-glutamate--2, 6-diaminopimelate ligase | |
| EC-2.4.1.227 | MurG | Undecaprenyldiphospho-muramoylpentapeptide-β-N-acetylglucosaminyltransferase | |
| EC-1.1.1.22 | Udg# | UDP-glucose dehydrogenase | |
| EC-2.4.1.- | WaaF | UDP-glucose:(heptosyl) LPS-α-1,3-glucosyltransferase | |
| EC-5.1.3.13 | RmlC | dTDP-4-dehydrorhamnose 3,5-epimerase | |
| Vitamin and cofactor synthesis | EC-2.7.11.5 | AceK | Bifunctional isocitrate dehydrogenase kinase/ phosphatase protein |
| EC-4.1.2.25 | FolB | Dihydroneopterin aldolase | |
| EC-2.5.1.15 | FolP* | Dihydropteroate synthase | |
| EC-1.2.1.70 | HemA | Glutamyl-tRNA reductase | |
| EC-2.1.2.11 | PanB | 3-methyl-2-oxobutanoate hydroxymethyltransferase | |
| EC-6.3.2.1 | PanC | Pantoate--β-alanine ligase | |
| EC-1.1.1.169 | PanE | 2-dehydropantoate 2-reductase | |
| EC-3.1.3.1 | PhoA* | Alkaline phophatase | |
| EC-3.5.4.25 | RibB | Bifunctional 3,4-dihydroxy-2-butanone 4-phosphate synthase | |
| EC-3.5.4.26 | RibD | Riboflavin-specific deaminase/reductase | |
| EC-2.5.1.9 | RibE* | Riboflavin synthase subunit-α | |
| EC-2.5.1.9 | RibH* | 6,7-dimethyl-8-ribityllumazine synase |
# Essential enzymes unique for B. cenocepacia J2315 (others are shared between B. cenocepacia J2315 and P. aeruginosa PAO1).
* The proteins have had approved drugs from the DrugBank database.
Conclusions
In this study, we reconstructed the first manually curated genome-scale metabolic network of B. cenocepacia J2315, a Gram-negative pathogen for CF patients. An iterative reconstruction process led to the establishment of the model, termed iKF1028, which captures the important metabolic capabilities and biosynthesis of key metabolic virulence factors. The model iKF1028 shows its predictive potential when compared with BIOLOG assays. Model-driven analyses on gene annotation refinement and identification of gene and enzyme essentiality analyses are helpful to understand the genome and discover promising novel drug targets. Through careful investigation, we proposed 45 enzymes that catalyze reactions predicted to be essential for growth with priority to be considered as drug targets. The model will keep being further validated and improved with experimentally determined biomass composition, large-scale gene deletion experimental data, proteome, and metabolome data, as they become available for B. cenocepacia. The model herein developed provides a valuable tool to explore the metabolic space of B. cenocepacia, to describe its metabolic wiring under a range of conditions, to pinpoint possible targets and to generate testable hypotheses. Taken together, our study underlined the value of the model iKF1028 as a framework to systematically study the metabolic capabilities of B. cenocepacia and its metabolic virulence factors affecting the CF community.
Methods
Reconstruction of the metabolic network
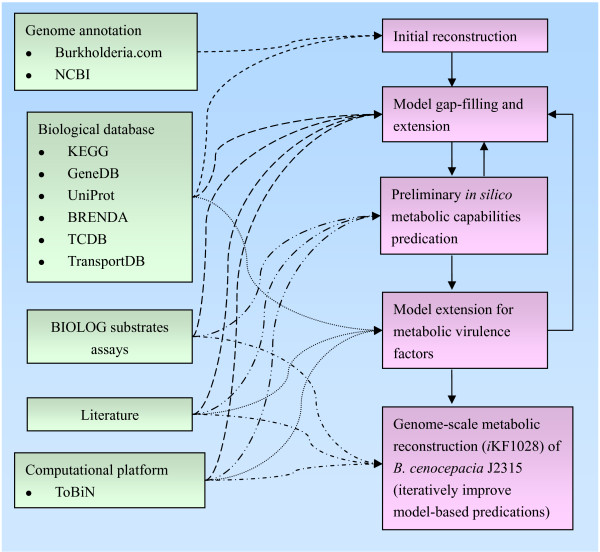
The reconstruction process for B. cenocepacia J2315 is illustrated in Figure 5. The process followed the procedure described previously [81]. The reconstruction was carried out on ToBiN (Toolbox for Biochemical Networks, http://www.lifewizz.com), which was first mentioned in the paper [82]. ToBiN is a modular platform for metabolic modelling and the structural analysis of networks. It consists of a collection of open-source computational tools. Sets of reactions can be uploaded in the platform via a web interface, merged with already existing sets, and the resulting stoichiometric matrix is then processed by the server as a FBA problem. The linear solver that ToBiN used is the Clp (Coin-or linear programming), an open-source linear programming solver written in C++ and is part of the COIN-OR (Computational Infrastructure for Operations Research) project (http://www.coin-or.org). The platform works in a similar way as the COBRA toolbox with the main difference that, by being web-based, it permits users to adopt a more efficient and collaborative workflow.
Figure 5.
The process for genome-scale metabolic reconstruction of B. cenocepacia J2315. The left side indicates resources used for reconstruction, and the right side indicates the reconstruction process. Initial reconstruction started from genome annotation and other biological databases. Gap-filling was a continuous step throughout the reconstruction by probing missing reactions in a pathway which causes in silico growth infeasible, and subsequently closing these gaps by referring to the biological databases, extensive literature mining, and comparison with BIOLOG substrate utilization assays [89,90]. This improved model was then extended by adding key metabolic virulence factors for B. cenocepacia from the literature. The process of model development and validation against experimental data was iteratively repeated until the genome-scale metabolic model was robust.
An initial draft reconstruction was derived from the annotated genome of Burkholderia cenocepacia J2315 available at the Burkholderia Genome Database (http://www.burkholderia.com). To link annotated genes to proteins and proteins to reactions, biological databases such as KEGG, GeneDB, UniProt, BRENDA, Transport Classification Database (TCDB), and TransportDB were used [83-88]. Manual curations were performed to establish gene-protein-reaction (GPR) associations, which connect genetic data to reactions in the metabolic network and allow for subsequent exploration of metabolic phenotypes using genetic perturbations.
After the initial reconstruction was generated, gaps in metabolic pathways necessary to produce biomass components and key virulence factors were filled by cautious literature mining, BIOLOG substrates utilization assays, and BLAST searches on homology and protein sequence similarity analyses [89,90]. The genome annotation was refined as consequence of the gap-filling and model extension process.
Flux Balance Analysis (FBA) was carried out throughout this study to explore the metabolic capabilities of iKF1028 under various environments. In addition to minimal medium, synthetic cystic fibrosis medium (SCFM) representing the physical living environment during CF infection was simulated in silico to investigate the metabolic flux distribution in a CF-like condition.
Biomass composition
The biomass composition in the genome-scale metabolic model of B. cenocepacia J2315 was adapted by selecting the well-studied biomass composition of E. coli as a template [91], since there's no experimental data available about the biomass composition of B. cenocepacia. However, the amount of metabolic precursors to formulate the cellular component was specific to B. cenocepacia according to previous study [20]. Moreover, the relative fatty acid composition of the lipids required for growth was based on data specific to B. cenocepacia [63,64,92,93] and listed in Table 6. Further details are provided in the supplemental material [Additional file 5].
Table 6.
Amino lipid composition of B. cenocepacia J2315
| Fatty acid | PE | PG | CLPN | OL | |
|---|---|---|---|---|---|
| Saturated | 16:0 | + | + | + | ND |
| Unsaturated | 16:1 | + | + | + | ND |
| 18:1 | + | + | + | + | |
| Hydoxy | 14:0 3OH | ND | ND | ND | + |
| 16:0 3OH | ND | ND | ND | + | |
| Cyclopropane | 17:0 CYC | + | + | + | ND |
| 19:0 CYC | + | + | + | ND | |
PE, phosphatidylethanolamine; PG, phosphatidylglycerol; CLPN, cardiolipin; OL, ornithine amide lipid; ND, not determined; plus symbol (+), fatty acid was detected in a significant amount
Flux balance analysis
Flux balance analysis (FBA) is an algorithm based on linear programming (LP) and on the assumption that the represented metabolic network is in steady-state (i.e. all the intracellular metabolite concentrations are constant). Being a LP problem, FBA also requires the selection of an objective function and of whether the value for that same function should be maximized or minimized. FBA is usually used to compute the optimal growth yield (the maximized objective function) based on the assumption that the evolutionary fitness of the organism depends on growth alone and, consequently, the implicit regulatory mechanism are organized to permit the theoretical maximal growth. If the system of equations (stoichiometric matrix which represents the metabolic network) is feasible, the algorithm generates an optimal flux distribution for that same network, taking into account the imposed thermodynamic constraints (reaction directionality) and limits on substrate uptake rates. The mathematical description is as follows:
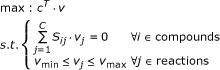 |
Where S is a stoichiometric matrix containing i rows representing metabolites and j columns representing reactions, v is a vector of all reaction fluxes, vmin and vmax are imposed lower and upper bounds on flux vj respectively, and cT is a vector of coefficients for each reaction that is to be maximized.
In silico media composition
Two different living environments were simulated in silico for strain J2315: M9 minimal medium [94], which contains PO43-, SO42-, NH4+, H+, Fe2+, K+, Mg2+, Na+, H2O, and thiamine, with glucose or other BIOLOG substrates as sole carbon source; and synthetic CF sputum medium (SCFM) [95] representing the nutrient conditions inside a host-cell during CF infection. Details of the simulated SCFM composition are provided in the supplemental material [Additional file 6].
BIOLOG assay
To validate the model and estimate the metabolic capabilities of strain J2315, BIOLOG assay was performed by using various carbon sources for strain cultivation [16]. The BIOLOG assay was carried out in triplicates using Biolog GN2 MicroPlates (Biolog, Inc.), which can test the ability of a microorganism to oxidize a panel of 95 different carbon sources simultaneously. The procedure for using the MicroPlates was according to the manufacturer's specification. The strain J2315 was obtained from DSMZ GmbH (DSMZ 16553, equivalent to LMG 16656 as which strain J2315 has been deposited in the BCCM/LMG Bacteria Collection). The strain was cultured overnight in CASO agar plate. Then the bacteria were swabbed from the plate surface and suspended in GN/GP inoculating fluid (Biolog, Inc.) and 150 μl of the suspension was transferred to each well of the GN2 MicroPlate. The MicroPlates were incubated at 30°C for 48 hours and were read by a microplate reader at 24 and 48 hours and analyzed with the Biolog MicroLog3 4.20 software (Biolog, Inc.). A comparison between the BIOLOG results and in silico predictions is provided in the supplemental material. [Additional file 3]
Gene and enzyme essentiality
FBA can be used to interpret genetic modification, such as gene deletion and enzyme inhibition, and subsequently make comprehensive in silico predictions on gene and enzyme essentiality [96]. To assess the essentiality of a gene, its GPR is checked for a unique relation with the associated reaction(s). If the gene is necessary to the reaction, the reaction flux will be constrained to zero and a solution for the maximal growth yield is searched. The deleted gene is predicated to be essential if, as consequence of that added constraint, the value of the objective function (growth yield) changes to zero. The deletion of every gene accounted in the model was simulated for growth on minimal medium with glucose as sole carbon source, and on SCFM. Similarly, an enzyme is considered essential if, by constraining to zero the flux on every associated reaction that has no alternative means of catalysis, the value for the growth yield changes to zero. The essentiality of every enzyme accounted in the model was analysed for growth on SCFM.
Authors' contributions
KCF, HSZ, SCY, SHC, and KLZ carried out the reconstruction of Burkholderia cenocepacia J2315. KCF designed the study and performed the analysis. KCF and CMCL arranged the BIOLOG substrate utilization analysis. JW and VMDS supervised the research. KCF, CMCL, VMDS, and JW drafted the manuscript. GP provided technical support for the reconstruction of B. cenocepacia J2315 in ToBiN. MG was involved in the early stage of the development of iKF1028 in ToBiN. All authors read and approved the final manuscript.
Supplementary Material
Reconstructed metabolic network model iKF1028
Model iKF1028 in SBML format
BIOLOG assays
Gene essentiality comparison results
Biomass detailed information
Synthetic CF medium simulated in silico
Contributor Information
Kechi Fang, Email: fangkc@psych.ac.cn.
Hansheng Zhao, Email: zhaohs@psych.ac.cn.
Changyue Sun, Email: suncy@psych.ac.cn.
Carolyn M C Lam, Email: carolyn.lam@helmholtz-hzi.de.
Suhua Chang, Email: changsh@psych.ac.cn.
Kunlin Zhang, Email: zhangkl@psych.ac.cn.
Gurudutta Panda, Email: gurudutta.panda@helmholtz-hzi.de.
Miguel Godinho, Email: mig@lifewizz.com.
Vítor A P Martins dos Santos, Email: vitor.martinsdossantos@wur.nl.
Jing Wang, Email: wangjing@psych.ac.cn.
Acknowledgements and Funding
We acknowledge funding from the European Union (Projects PROBACTYS: Grant No. 29104 and MICROME: No. 222886), the National Basic Research Program of China (973 Program) (Grant No. 2011CBA00802) from the Ministry of Science and Technology of the People's Republic of China, the Project for Young Scientists Fund, Institute of Psychology, Chinese Academy of Sciences (Grant No. O9CX115011), and Portuguese Fundação para a Ciência e Tecnologia (Project METAGUT; Grant No. ERA-PTG/SAU/0003/2008).
References
- Mahenthiralingam E, Baldwin A, Dowson CG. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol. 2008;104:1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- Vanlaere E, Lipuma JJ, Baldwin A, Henry D, De Brandt E, Mahenthiralingam E, Speert D, Dowson C, Vandamme P. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol. 2008;58:1580–1590. doi: 10.1099/ijs.0.65634-0. [DOI] [PubMed] [Google Scholar]
- Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol. 2009;59:102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- Parke JL, Gurian-Sherman D. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu Rev Phytopathol. 2001;39:225–258. doi: 10.1146/annurev.phyto.39.1.225. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Baldwin A, Vandamme P. Burkholderia cepacia complex infection in patients with cystic fibrosis. J Med Microbiol. 2002;51:533–538. doi: 10.1099/0022-1317-51-7-533. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat Rev Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- Mann T, Ben-David D, Zlotkin A, Shachar D, Keller N, Toren A, Nagler A, Smollan G, Barzilai A, Rahav G. An outbreak of Burkholderia cenocepacia bacteremia in immunocompromised oncology patients. Infection. 2010. [DOI] [PubMed]
- Govan JR, Brown PH, Maddison J, Doherty CJ, Nelson JW, Dodd M, Greening AP, Webb AK. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342:15–19. doi: 10.1016/0140-6736(93)91881-L. [DOI] [PubMed] [Google Scholar]
- Martin DW, Mohr CD. Invasion and intracellular survival of Burkholderia cepacia. Infect Immun. 2000;68:24–29. doi: 10.1128/IAI.68.1.24-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme P, Holmes B, Coenye T, Goris J, Mahenthiralingam E, LiPuma JJ, Govan JR. Burkholderia cenocepacia sp. nov.--a new twist to an old story. Res Microbiol. 2003;154:91–96. doi: 10.1016/S0923-2508(03)00026-3. [DOI] [PubMed] [Google Scholar]
- Drevinek P, Holden MT, Ge Z, Jones AM, Ketchell I, Gill RT, Mahenthiralingam E. Gene expression changes linked to antimicrobial resistance, oxidative stress, iron depletion and retained motility are observed when Burkholderia cenocepacia grows in cystic fibrosis sputum. BMC Infect Dis. 2008;8:121. doi: 10.1186/1471-2334-8-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry N, Du W, Lane D, Pasta F. Improved electrotransformation and decreased antibiotic resistance of the cystic fibrosis pathogen Burkholderia cenocepacia strain J2315. Appl Environ Microbiol. 2010;76:1095–1102. doi: 10.1128/AEM.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S. et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol. 2009;191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Cho Y, Dinh NN, Waring AJ, Lehrer RI. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob Agents Chemother. 1998;42:2206–2214. doi: 10.1128/aac.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet SA, Flannagan RS, Kooi C, Sokol PA, Valvano MA. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J Bacteriol. 2006;188:2073–2080. doi: 10.1128/JB.188.6.2073-2080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JM, Rhoden DL. Preliminary evaluation of Biolog, a carbon source utilization method for bacterial identification. J Clin Microbiol. 1991;29:1143–1147. doi: 10.1128/jcm.29.6.1143-1147.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR) Genome Biology. 2003;4:R45. doi: 10.1186/gb-2003-4-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Burd H, Liu J, Almaas E, Wiest O, Barabasi AL, Oltvai ZN, Kapatral V. Comparative genome-scale metabolic reconstruction and flux balance analysis of multiple Staphylococcus aureus genomes identify novel antimicrobial drug targets. J Bacteriol. 2009;191:4015–4024. doi: 10.1128/JB.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker SA, Palsson BO. Genome-scale reconstruction of the metabolic network in Staphylococcus aureus N315: an initial draft to the two-dimensional annotation. BMC Microbiol. 2005;5:8. doi: 10.1186/1471-2180-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann M, Kummel A, Ruinatscha R, Panke S. In silico genome-scale reconstruction and validation of the Staphylococcus aureus metabolic network. Biotechnol Bioeng. 2005;92:850–864. doi: 10.1002/bit.20663. [DOI] [PubMed] [Google Scholar]
- Kim HU, Kim TY, Lee SY. Genome-scale metabolic network analysis and drug targeting of multi-drug resistant pathogen Acinetobacter baumannii AYE. Mol Biosyst. 2010;6:339–348. doi: 10.1039/b916446d. [DOI] [PubMed] [Google Scholar]
- Jamshidi N, Palsson BO. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol. 2007;1:26. doi: 10.1186/1752-0509-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan A, Reed J, Shin S, Palsson B, Daefler S. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst Biol. 2009;3:38. doi: 10.1186/1752-0509-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt MA, Puchalka J, Fryer KE, Martins dos Santos VA, Papin JA. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol. 2008;190:2790–2803. doi: 10.1128/JB.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl L. Quorum sensing in the genus Burkholderia. Int J Med Microbiol. 2006;296:103–110. doi: 10.1016/j.ijmm.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Eberl L. From a local dialect to a common language. Chem Biol. 2006;13:803–804. doi: 10.1016/j.chembiol.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Sokol PA, Malott RJ, Riedel K, Eberl L. Communication systems in the genus Burkholderia: global regulators and targets for novel antipathogenic drugs. Future Microbiol. 2007;2:555–563. doi: 10.2217/17460913.2.5.555. [DOI] [PubMed] [Google Scholar]
- Boon C, Deng Y, Wang LH, He Y, Xu JL, Fan Y, Pan SQ, Zhang LH. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2008;2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- Deng Y, Boon C, Eberl L, Zhang LH. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J Bacteriol. 2009;191:7270–7278. doi: 10.1128/JB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RP, McCarthy Y, Watt SA, Niehaus K, Dow JM. Intraspecies signaling involving the diffusible signal factor BDSF (cis-2-dodecenoic acid) influences virulence in Burkholderia cenocepacia. J Bacteriol. 2009;191:5013–5019. doi: 10.1128/JB.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol PA, Darling P, Woods DE, Mahenthiralingam E, Kooi C. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding L-ornithine N(5)-oxygenase. Infect Immun. 1999;67:4443–4455. doi: 10.1128/iai.67.9.4443-4455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer KL, Thomas MS. Isolation and characterization of Burkholderia cenocepacia mutants deficient in pyochelin production: pyochelin biosynthesis is sensitive to sulfur availability. J Bacteriol. 2004;186:270–277. doi: 10.1128/JB.186.2.270-277.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser MB, Majumdar S, Hani E, Sokol PA. Importance of the ornibactin and pyochelin siderophore transport systems in Burkholderia cenocepacia lung infections. Infect Immun. 2004;72:2850–2857. doi: 10.1128/IAI.72.5.2850-2857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjan US, Sun L, Goldstein R, Forstner JF. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J Bacteriol. 1995;177:1030–1038. doi: 10.1128/jb.177.4.1030-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjan U, Ackerley C, Forstner J. Interaction of cblA/adhesin-positive Burkholderia cepacia with squamous epithelium. Cell Microbiol. 2002;4:73–86. doi: 10.1046/j.1462-5822.2002.00171.x. [DOI] [PubMed] [Google Scholar]
- Sajjan U, Liu L, Lu A, Spilker T, Forstner J, LiPuma JJ. Lack of cable pili expression by cblA-containing Burkholderia cepacia complex. Microbiology. 2002;148:3477–3484. doi: 10.1099/00221287-148-11-3477. [DOI] [PubMed] [Google Scholar]
- Tomich M, Herfst CA, Golden JW, Mohr CD. Role of flagella in host cell invasion by Burkholderia cepacia. Infect Immun. 2002;70:1799–1806. doi: 10.1128/IAI.70.4.1799-1806.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban TA, Griffith A, Torok AM, Smolkin ME, Burns JL, Goldberg JB. Contribution of Burkholderia cenocepacia flagella to infectivity and inflammation. Infect Immun. 2004;72:5126–5134. doi: 10.1128/IAI.72.9.5126-5134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison ML, Poxton IR, Govan JR. Burkholderia cepacia produces a hemolysin that is capable of inducing apoptosis and degranulation of mammalian phagocytes. Infect Immun. 1998;66:2033–2039. doi: 10.1128/iai.66.5.2033-2039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooi C, Corbett CR, Sokol PA. Functional analysis of the Burkholderia cenocepacia ZmpA metalloprotease. J Bacteriol. 2005;187:4421–4429. doi: 10.1128/JB.187.13.4421-4429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooi C, Subsin B, Chen R, Pohorelic B, Sokol PA. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect Immun. 2006;74:4083–4093. doi: 10.1128/IAI.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooi C, Sokol PA. Burkholderia cenocepacia zinc metalloproteases influence resistance to antimicrobial peptides. Microbiology. 2009;155:2818–2825. doi: 10.1099/mic.0.028969-0. [DOI] [PubMed] [Google Scholar]
- Korbsrisate S, Tomaras AP, Damnin S, Ckumdee J, Srinon V, Lengwehasatit I, Vasil ML, Suparak S. Characterization of two distinct phospholipase C enzymes from Burkholderia pseudomallei. Microbiology. 2007;153:1907–1915. doi: 10.1099/mic.0.2006/003004-0. [DOI] [PubMed] [Google Scholar]
- Tomich M, Griffith A, Herfst CA, Burns JL, Mohr CD. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infect Immun. 2003;71:1405–1415. doi: 10.1128/IAI.71.3.1405-1415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engledow AS, Medrano EG, Mahenthiralingam E, LiPuma JJ, Gonzalez CF. Involvement of a plasmid-encoded type IV secretion system in the plant tissue watersoaking phenotype of Burkholderia cenocepacia. J Bacteriol. 2004;186:6015–6024. doi: 10.1128/JB.186.18.6015-6024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey KM, Glendinning KJ, Morgan JA, Hart CA, Winstanley C. Caenorhabditis elegans killing assay as an infection model to study the role of type III secretion in Burkholderia cenocepacia. J Med Microbiol. 2006;55:967–969. doi: 10.1099/jmm.0.46618-0. [DOI] [PubMed] [Google Scholar]
- Ortega X, Hunt TA, Loutet S, Vinion-Dubiel AD, Datta A, Choudhury B, Goldberg JB, Carlson R, Valvano MA. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J Bacteriol. 2005;187:1324–1333. doi: 10.1128/JB.187.4.1324-1333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt TA, Kooi C, Sokol PA, Valvano MA. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infect Immun. 2004;72:4010–4022. doi: 10.1128/IAI.72.7.4010-4022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Soyza A, Silipo A, Lanzetta R, Govan JR, Molinaro A. Chemical and biological features of Burkholderia cepacia complex lipopolysaccharides. Innate Immun. 2008;14:127–144. doi: 10.1177/1753425908093984. [DOI] [PubMed] [Google Scholar]
- Parsons YN, Banasko R, Detsika MG, Duangsonk K, Rainbow L, Hart CA, Winstanley C. Suppression-subtractive hybridisation reveals variations in gene distribution amongst the Burkholderia cepacia complex, including the presence in some strains of a genomic island containing putative polysaccharide production genes. Arch Microbiol. 2003;179:214–223. doi: 10.1007/s00203-003-0518-7. [DOI] [PubMed] [Google Scholar]
- Silipo A, Molinaro A, Ierano T, De Soyza A, Sturiale L, Garozzo D, Aldridge C, Corris PA, Khan CM, Lanzetta R, Parrilli M. The complete structure and pro-inflammatory activity of the lipooligosaccharide of the highly epidemic and virulent gram-negative bacterium Burkholderia cenocepacia ET-12 (strain J2315) Chemistry. 2007;13:3501–3511. doi: 10.1002/chem.200601406. [DOI] [PubMed] [Google Scholar]
- Ortega X, Silipo A, Saldias MS, Bates CC, Molinaro A, Valvano MA. Biosynthesis and structure of the Burkholderia cenocepacia K56-2 lipopolysaccharide core oligosaccharide: truncation of the core oligosaccharide leads to increased binding and sensitivity to polymyxin B. J Biol Chem. 2009;284:21738–21751. doi: 10.1074/jbc.M109.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinion-Dubiel AD, Goldberg JB. Lipopolysaccharide of Burkholderia cepacia complex. J Endotoxin Res. 2003;9:201–213. doi: 10.1179/096805103225001404. [DOI] [PubMed] [Google Scholar]
- Ortega XP, Cardona ST, Brown AR, Loutet SA, Flannagan RS, Campopiano DJ, Govan JR, Valvano MA. A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J Bacteriol. 2007;189:3639–3644. doi: 10.1128/JB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Raetz CRH. Identification and characterization of a 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) oxidase; KdoO. The FASEB Journal. 2010.
- Kenna DT, Barcus VA, Langley RJ, Vandamme P, Govan JR. Lack of correlation between O-serotype, bacteriophage susceptibility and genomovar status in the Burkholderia cepacia complex. FEMS Immunol Med Microbiol. 2003;35:87–92. doi: 10.1016/S0928-8244(02)00442-X. [DOI] [PubMed] [Google Scholar]
- Malott RJ, Baldwin A, Mahenthiralingam E, Sokol PA. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect Immun. 2005;73:4982–4992. doi: 10.1128/IAI.73.8.4982-4992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P, Winzer K, Chan WC, Camara M. Look who's talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc Lond B Biol Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A, Sokol PA, Parkhill J, Mahenthiralingam E. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia. Infect Immun. 2004;72:1537–1547. doi: 10.1128/IAI.72.3.1537-1547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Dubeau D, Deziel E, Woods DE, Lepine F. Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiol. 2009;9:263. doi: 10.1186/1471-2180-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Vandamme P, Govan JR, LiPuma JJ. Taxonomy and identification of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:3427–3436. doi: 10.1128/JCM.39.10.3427-3436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S, Pitt T. Burkholderia (Pseudomonas) cepacia: Surface chemistry and typing methods. REV MED MICROBIOL. 1995;6:1–9. [Google Scholar]
- Taylor CJ, Anderson AJ, Wilkinson SG. Phenotypic variation of lipid composition in Burkholderia cepacia: a response to increased growth temperature is a greater content of 2-hydroxy acids in phosphatidylethanolamine and ornithine amide lipid. Microbiology. 1998;144(Pt 7):1737–1745. doi: 10.1099/00221287-144-7-1737. [DOI] [PubMed] [Google Scholar]
- Gao JL, Weissenmayer B, Taylor AM, Thomas-Oates J, Lopez-Lara IM, Geiger O. Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol Microbiol. 2004;53:1757–1770. doi: 10.1111/j.1365-2958.2004.04240.x. [DOI] [PubMed] [Google Scholar]
- Harayama S, Bollinger J, Iino T, Hazelbauer GL. Characterization of the mgl operon of Escherichia coli by transposon mutagenesis and molecular cloning. J Bacteriol. 1983;153:408–415. doi: 10.1128/jb.153.1.408-415.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm LV, Young NR, Frye JG, Hardham JM. Identification and sequences of the Treponema pallidum mglA and mglC genes. DNA Seq. 1996;6:293–298. doi: 10.3109/10425179609020876. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R. et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- Cronan JE Jr, Waldrop GL. Multi-subunit acetyl-CoA carboxylases. Prog Lipid Res. 2002;41:407–435. doi: 10.1016/S0163-7827(02)00007-3. [DOI] [PubMed] [Google Scholar]
- Li SJ, Cronan JE Jr. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Hamid AM, Cronan JE. Coordinate expression of the acetyl coenzyme A carboxylase genes, accB and accC, is necessary for normal regulation of biotin synthesis in Escherichia coli. J Bacteriol. 2007;189:369–376. doi: 10.1128/JB.01373-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder-Himes DR, Konstantinidis KT, Tiedje JM. Identification of potential therapeutic targets for Burkholderia cenocepacia by comparative transcriptomics. PLoS One. 2010;5:e8724. doi: 10.1371/journal.pone.0008724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal D, Samal A, Sakharkar KR, Sakharkar MK. Targeting multiple targets in Pseudomonas aeruginosa PAO1 using flux balance analysis of a reconstructed genome-scale metabolic network. J Drug Target. 2010. [DOI] [PubMed]
- Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Parente J, Harris SM, Woods DE, Hancock RE, Falla TJ. Antimicrobial peptide therapeutics for cystic fibrosis. Antimicrob Agents Chemother. 2005;49:2921–2927. doi: 10.1128/AAC.49.7.2921-2927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookherjee N, Hancock RE. Cationic host defence peptides: innate immune regulatory peptides as a novel approach for treating infections. Cell Mol Life Sci. 2007;64:922–933. doi: 10.1007/s00018-007-6475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet SA, Bartholdson SJ, Govan JR, Campopiano DJ, Valvano MA. Contributions of two UDP-glucose dehydrogenases to viability and polymyxin B resistance of Burkholderia cenocepacia. Microbiology. 2009;155:2029–2039. doi: 10.1099/mic.0.027607-0. [DOI] [PubMed] [Google Scholar]
- Edwards JS, Covert M, Palsson BO. Metabolic modelling of microbes:the flux-balance approach. Environmental Microbiology. 2002;4:133–140. doi: 10.1046/j.1462-2920.2002.00282.x. [DOI] [PubMed] [Google Scholar]
- Diez MS, Lam CM, Leprince A, dos Santos VA. (Re-)construction, characterization and modeling of systems for synthetic biology. Biotechnol J. 2009;4:1382–1391. doi: 10.1002/biot.200900173. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K. et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–195. doi: 10.1093/nar/gkn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Palsson BO. Systematizing the generation of missing metabolic knowledge. Biotechnol Bioeng. 2010;107:403–412. doi: 10.1002/bit.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JL, Patel TR, Chen KH, Joyce AR, Applebee MK, Herring CD, Bui OT, Knight EM, Fong SS, Palsson BO. Systems approach to refining genome annotation. Proc Natl Acad Sci USA. 2006;103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Molecular Systems Biology. 2007;3 doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci E, Kroppenstedt RM. Defferentiation of Species Combined into the Burkholderia cepacia Complex and Related Taxa on the Basis of Their Fatty Acid Patterns. Journal of Clinical Microbiology. 2006;44 doi: 10.1128/JCM.44.3.1159-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SA, Ramos CG, Almeida F, Meirinhos-Soares L, Wopperer J, Schwager S, Eberl L, Leitao JH. Burkholderia cenocepacia J2315 acyl carrier protein: a potential target for antimicrobials' development? Microb Pathog. 2008;45:331–336. doi: 10.1016/j.micpath.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BO, Agarwalla S. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol. 2006;188:8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reconstructed metabolic network model iKF1028
Model iKF1028 in SBML format
BIOLOG assays
Gene essentiality comparison results
Biomass detailed information
Synthetic CF medium simulated in silico