Abstract
Background
CD4-binding site (CD4bs) alterations in gp120 contribute to HIV-1 envelope (Env) mediated fusogenicity and the ability of gp120 to utilize low levels of cell-surface CD4. In a recent study, we constructed three-dimensional models of gp120 to illustrate CD4bs conformations associated with enhanced fusogenicity and enhanced CD4-usage of a modestly-sized panel of blood-derived HIV-1 Envs (n = 16). These conformations were characterized by a wider aperture of the CD4bs cavity, as constrained by the inner-most atoms at the gp120 V1V2 stem and the V5 loop. Here, we sought to provide further validation of the utility of these models for understanding mechanisms that influence Env function, by characterizing the structure-function relationships of a larger panel of Envs derived from brain and other tissues (n = 81).
Findings
Three-dimensional models of gp120 were generated by our recently validated homology modelling protocol. Analysis of predicted CD4bs structures showed correlations between the aperture width of the CD4bs cavity and ability of the Envs to mediate cell-cell fusion, scavenge low-levels of cell-surface CD4, bind directly to soluble CD4, and bind to the Env mAb IgG1b12 whose epitope overlaps the gp120 CD4bs. These structural alterations in the CD4bs cavity were associated with repositioning of the V5 loop.
Conclusions
Using a large, independent panel of Envs, we can confirm the utility of three-dimensional gp120 structural models for illustrating CD4bs alterations that can affect Env function. Furthermore, we now provide new evidence that these CD4bs alterations augment the ability of gp120 to interact with CD4 by increasing the exposure of the CD4bs.
Findings
The human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins (Env) mediate virus entry into cells and exist as trimers, comprising the surface gp120 glycoproteins noncovalently linked to transmembrane gp41 glycoproteins that embed the complex into the viral membrane [1-3]. HIV-1 entry is initiated by gp120 binding to cellular CD4, which facilitates the initial attachment of virus to the target cell [4]. The binding of gp120 to CD4 results in dramatic conformational changes in gp120 that expose the binding site for a secondary coreceptor, which is either of the chemokine receptors CCR5 or CXCR4 (reviewed in [5-7]).
Crystallographic and biochemical studies of gp120 have provided valuable insights into mechanisms involved in CD4 binding and CD4-induced conformational changes [3,8-12]. The unliganded gp120 core of simian immunodeficiency virus (SIV) consists of a highly conserved inner domain which faces the trimer axis and a heavily glycosylated, globular outer domain which is mostly exposed on the surface of the trimer [8]. However, thermodynamic and structural analysis of the gp120-CD4 interaction demonstrated little evidence of a structured CD4 binding pocket on the unliganded gp120, and that CD4bs elements which influence gp120-CD4 affinity are formed from conformational alterations that occur after gp120 has encountered CD4 [2,10]. CD4 interacts with gp120 via surface-exposed residues within three separate regions distributed over six segments of gp120. These regions include the α-helices of the inner domain, the CD4 binding loop of outer domain, and the β20-β21 ribbon which becomes part of the gp120 bridging sheet, which is a structural element of gp120 formed after CD4 binding that is involved in coreceptor binding [3,11].
Changes in CD4 binding to gp120 contribute to different pathophysiological phenotypes of HIV-1, including the fusogenic properties of the Env [13,14]. Env mediates most of the acute cytopathic effects of HIV-1 infection in cultured cells [15], and membrane fusion appears to be an important factor contributing to HIV-1 cytopathicity in vitro [16,17]. In addition, enhancement of pathogenicity of chimeric simian-HIV (SHIV) strains in macaques frequently results from increased Env-mediated fusogenicity [18-22]. Moreover, the cytopathic effects of Env-mediated HIV-1 fusogenicity are evident in humans. For example, the presence of multinucleated giant cells in brain, formed by Env-mediated fusion between infected and uninfected macrophage lineage cells, is characteristic of HIV-1 encephalitis and a neuropathological hallmark of HIV-associated dementia [23].
To better understand the molecular mechanisms contributing to alterations in CD4 binding by primary gp120 proteins and the subsequent influence on Env function, we recently developed and validated a protocol to produce and utilize three-dimensional structural models of gp120 to deduce CD4bs alterations that influence CD4 binding and Env-mediated fusogenicity [13]. Using a modestly-sized panel of blood derived Envs (n = 16), we showed that a wider aperture of the predicted CD4bs cavity, as constrained by the inner-most atoms at the gp120 V1V2 stem and the V5 loop, contributed to increased fusogenicity and ability of gp120 to bind CD4. In the present study, we sought to provide further validation of the utility of these molecular models for understanding mechanisms that influence Env function, by characterizing, for the first time, the structure-function relationships of a larger panel of Envs derived from brain and other tissues (n = 81).
Production and characterization of a panel of primary Env clones
Primary HIV-1 viruses isolated from autopsy brain and/or cerebrospinal fluid, spinal cord, lymph node, spleen or PBMC from subjects CB1, CB3, MACS1, MACS2, MACS3, UK1 and UK7 have been described in detail previously [14,24-27]. The clinical characteristics of the subjects and coreceptor usage profiles of the primary viruses are summarized in Table 1. A 2.1 kb fragment spanning the KpnI to BamHI restriction sites in HIV-1 env (corresponding to nucleotides 6348 to 8478 in HXB2) was amplified from viral cDNA by PCR and cloned into the pSVIII-HXB2 Env expression vector [28], as described previously [29-33]. Between 4 and 6 functional Envs from each virus were identified by entry assays in JC53 cells with Env-pseudotyped GFP reporter viruses, as described previously [30,33-35] (Table 1). The coreceptor specificity of the cloned Envs was determined by entry assays in Cf2th-CD4/CCR5 and Cf2th-CD4/CXCR4 cells [35,36] with Env-pseudotyped luciferase reporter viruses, as described previously [29,35], which recapitulated the coreceptor usage of the primary viruses (Table 1). The Envs were sequenced in their entirety and subjected to multiple sequence alignments (data not shown) and phylogenetic analysis (Figure 1), which together showed that the Envs were independent and compartmentalized according to their tissue of origin. Thus, we established and characterized a new panel of Envs (n = 81) derived from autopsy brain and other tissues of 7 subjects who died from AIDS.
Table 1.
Study subjects, HIV-1 isolates, and summary of Env phenotypes
| Subject | Risk factor | Last CD4 count (cells/μl) | Antiretroviral(s) | HIV-1 encephalitis | Tissues yielding HIV-1 isolates | Name of virus isolate | Coreceptor usage of virus isolate | Envs cloned from virus isolate (n) | Functional | Coreceptor usage of cloned Envs |
|---|---|---|---|---|---|---|---|---|---|---|
| CB1 | MH | 10 | ddI (prior AZT) | Severe | Brain | CB1-BR | X4 | 6 | Yes | All X4 |
| CSF | CB1-CSF | R5 | 6 | Yes | All R5 | |||||
| PBMC | CB1-PBMC | R5 | 6 | Yes | All R5 | |||||
| CB3 | MH | 5 | ddI (prior AZT and ddC) | Severe | S.Cord | CB3-SC | R5 | 6 | Yes | All R5 |
| CSF | CB3-CSF | R5 | 6 | Yes | All R5 | |||||
| PBMC | CB3-PBMC | R5 | 6 | Yes | All R5 | |||||
| MACS1 | MH | 2 | None | Severe | Brain | Macs1-BR | R5X4 | 6 | Yes | All R5X4 |
| Spleen | Macs1-Spln | R5X4 | 6 | Yes | All R5X4 | |||||
| MACS2 | MH | 52 | AZT | Moderate | Brain | Macs2-BR | R5 | 5 | Yes | All R5 |
| L.Node | Macs2-LN | R5 | 6 | Yes | All R5 | |||||
| MACS3 | MH | 95 | None | Moderate | Brain | Macs3-BR | R5 | 6 | Yes | All R5 |
| L.Node | Macs3-LN | R5 | 6 | Yes | All R5 | |||||
| UK1 | IVDU | 87 | ddC (1 mo) | Moderate | Brain | UK1-BR | R5 | 4 | Yes | All R5 |
| UK7 | IVDU | 90 | AZT | Severe | Brain | UK7-BR | R5 | 6 | Yes | All R5 |
The clinical and neuropathological details of the study subjects, and the derivation and characterization of the primary tissue derived HIV-1 isolates have been published previously [14,24,27], and are summarized again here to assist in the interpretation of the data derived from the cloned Envs. Envs were amplified from primary virus isolates by PCR and cloned into the pSVIII-Env expression vector as described previously [29,30,33,35]. Functional Envs were identified by pseudotyping onto Env-deficient GFP reporter virus and entry assays in JC53 cells, as described previously [14,29,30,34,35]. Coreceptor usage of cloned Envs was determined by pseudotyping onto Env-deficient luciferase reporter virus that were generated in 293T cells, and entry assays in Cf2th-CD4 cells expressing CCR5 or CXCR4, as described previously [30,35]. The coreceptor usage of Envs derived from brain and spleen of subject MACS1 has been reported recently [30]. The additional Envs described here have been assigned Genbank accession numbers JN001990 to JN002061. Six functional Envs were cloned from Macs2-BR and UK1-BR viruses, but sequencing and phylogenetic analysis revealed that only 5 and 4 clones, respectively, were independent with unique nucleotide sequences. Thus, only independent Envs are listed here and included for the subsequent structural and functional analyses. MH, male homosexual; IVDU, intravenous drug user; mo, month; ddI, didanosine; AZT, zidovudine; ddC, zalcitabine; CSF, cerebrospinal fluid; PBMC, peripheral blood mononuclear cells; S. Cord, spinal cord; L. Node, lymph node.
Figure 1.
Phylogenetic analysis of env nucleotide sequences. The phylogenetic tree was constructed from an env nucleotide multiple sequence alignment using a maximum likelihood algorithm, as described previously [39]. The nucleotide sequences of HIV-1 AD8, 89.6, JRCSF, YU2, NL4-3 and HXB2 env genes were included for comparison. Numbers associated with each branch are bootstrap values obtained from 1000 replicates. Only values above 700 for the major branches are shown. Branch lengths are proportional to the amount of sequence divergence.
Production of three-dimensional gp120 models and characterization of the CD4bs cavity
We next produced three-dimensional structural models of each of the 81 Envs using a protocol that we described recently [13,32]. Briefly, homology models of CD4-bound gp120 sequences were prepared using the Build Model protocol of the Discovery Studio suite, version 1.6 (Accelrys, San Diego, CA, USA). This approach used the Modeller algorithm to generate an atomic model of the target protein from a template molecule and a sequence alignment. The template-based models were optimized by iterative cycles of conjugate-gradient minimisation against a probability density function that included spatial restraints derived from the template and residue specific properties [37]. The crystal structure of JRFL gp120 containing the V3 variable loop and bound to CD4 and the X5 Fab antibody fragment was used as the template for CD4-bound models [9] (Protein Data Bank ID: 2B4C). The X5 antibody fragment was deleted from the CD4-bound template prior to modeling. The coordinates for gp120 and CD4 were extracted from the 2B4C crystal structure. Sequence alignments were generated between JRFL gp120 and the primary gp120 Env clones. The sequence for CD4 was included as a second polypeptide chain such that the models of gp120 were constructed as complexes with CD4. The V1V2 variable loops were replaced with a GAG linker sequence and the N- and C- termini overhangs were cut using the modeling software.
Similarities in three-dimensional structure were measured by the root mean square deviation (RMSD) of the distances between main-chain atoms (N, Cα, C and O atoms) from crystal and model structures after rigid body superposition, where an RMSD of < 1Å signifies a high level of three-dimensional structural similarity between overlayed proteins. The overall quality of the geometry of gp120 models generated was verified using PROCHECK [38].
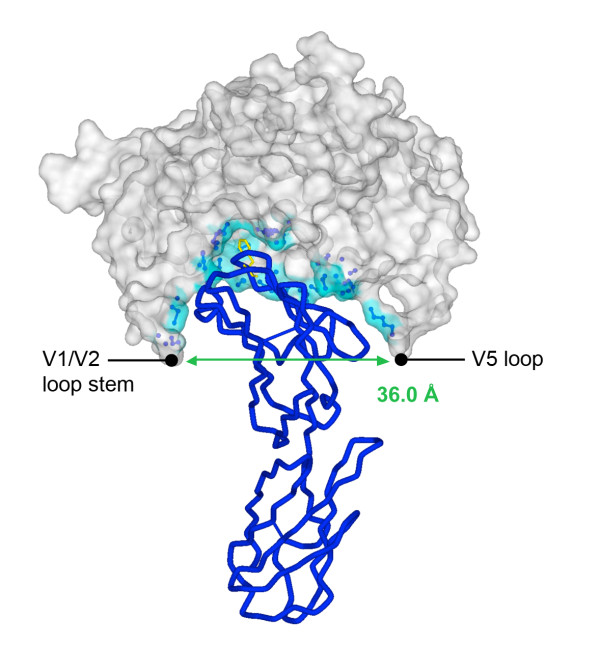
The three-dimensional structural similarity between the 2B4C JRFL crystal structure and the 81 predicted structures of the primary gp120 proteins was < 1.0 Å for all the primary gp120 models (data not shown), indicating a high overall degree of structural similarity. Identical RMSD values for each gp120 model were obtained upon repeated, independent modeling operations (data not shown). The aperture width of the CD4bs cavity was deduced from each of the three-dimensional gp120 structural models by measuring the distance between the inner most atoms present at the stem of the V1V2 loops and the V5 loop, which constrain the CD4 binding pocket of gp120 [9]. Figure 2 shows the derivation of the CD4bs aperture width for the Macs2ln5 gp120 model as an example. Analysis of structural models generated for all the 81 Envs showed that the aperture width of the predicted CD4bs cavity ranges from 30 to 36 Å in this panel of Envs.
Figure 2.
Predicted alterations in the CD4bs cavity from three-dimensional gp120 models. The gp120 model of Macs2ln5 Env is shown in molecular surface representation, and the CD4 molecule is shown in blue Cα wire, with Phe43 of CD4 highlighted in yellow stick representation to show the "Phe43 cavity" of gp120. gp120 residues in the CD4 binding pocket located within 4 Å of the CD4 molecule are shown in ball and stick representation and their molecular surface is highlighted in blue. The width of the CD4bs aperture, as constrained by the inner-most atoms at the gp120 V1V2 stem and the V5 loop, was deduced as described previously [13].
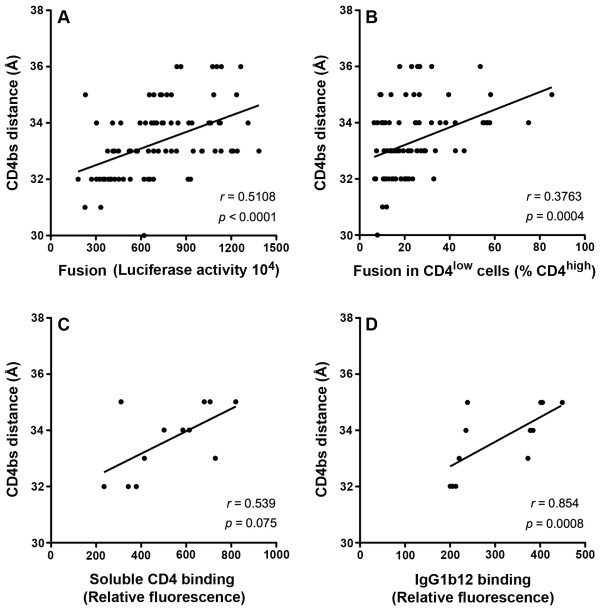
An increased aperture width of the gp120 CD4bs cavity is associated with increased fusogenicity, increased CD4-usage, and increased IgG1b12 Env mAb binding
To determine whether alterations in the width of the gp120 CD4bs aperture may influence Env function, we first performed quantitative cell-cell fusion assays with 293T effector cells expressing equivalent levels of Env on the cell surface, and target cells expressing coreceptor and either relatively high or relatively low levels of CD4 as described previously [13]. In these assays, we observed positive correlations between the width of the gp120 CD4bs cavity and the overall level of cell-cell fusion (Figure 3A), and also with the ability of Env to utilize low levels of CD4 to mediate cell-cell fusion (Figure 3B). Next, to better understand the influence of changes in the gp120 CD4bs aperture on CD4 binding and CD4bs exposure, we measured the ability of Env to bind sCD4 and the Env mAb IgG1b12, whose epitope overlaps the gp120 CD4bs, using a subset of Envs (n = 12, Macs1br2-8 and Macs1sp3-15; see Figure 1), as described previously [13,35]. In these assays, we observed a near significant association between the width of the CD4bs cavity and ability of Env expressed on 293T cells to bind sCD4 (Figure 3C), and a significant correlation between this parameter and the ability of Env to bind IgG1b12 (Figure 3D). Together, these studies, using a large and newly-described panel of primary Envs, demonstrate the utility of three-dimensional modeling of the gp120 CD4bs cavity for better understanding the structural basis of Env-CD4 interactions, confirming the results of our recent study of a different and much smaller panel of Envs [13]. Furthermore, our results provide new evidence suggesting that these predicted CD4bs conformational alterations augment the ability of gp120 to interact with CD4 by increasing the exposure of the CD4bs.
Figure 3.
The effect of gp120 CD4bs cavity alterations on fusogenicity, CD4-dependence, sCD4 binding and CD4bs exposure. The CD4bs aperture widths for each gp120 model were plotted against the ability of Env to mediate cell-cell fusion (A), the ability of Env to utilize low levels of CD4 for cell-cell fusion (B), and the ability of Env to bind sCD4 (C) or the Env mAb IgG1b12 (D), using Prism version 5.0c (GraphPad Software). The methods for these functional and biochemical assays have been described in detail previously, including the extensive use of controls to ensure equivalent expression of Env on the cell surface, protocols for generating (and measuring CD4 expression on) CD4low and CD4high cells, and the empirical determination of sCD4 and IgG1b12 concentrations used that we showed were within the linear range of Env binding [13,32,35]. The Spearman correlation coefficient (r) and P values are shown. P values < 0.05 were considered statistically significant. The data shown are representative of 3 independent experiments.
Repositioning of the V5 loop is associated with conformational alterations in the gp120 CD4bs
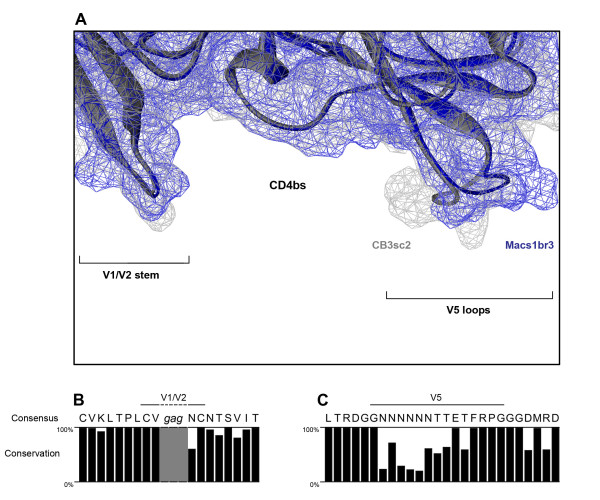
To elucidate the gp120 determinants which may contribute to structural alterations in the CD4bs and which subsequently influence CD4 interactions and fusogenicity, we next compared the structural similarity between CB3sc2 and Macs1br3 gp120 models which are predicted to have the narrowest and widest of the CD4bs apertures (30 and 36 Å, respectively). Overlays of these molecular models revealed a high degree of structural similarity within the V1V2 stem region, but notable structural variation within the V5 loop region (Figure 4A). Furthermore, sequence analysis of all the primary Env clones showed a relatively high degree of sequence homology within the V1V2 stem region (Figure 4B), but a relatively high degree of sequence variation within the V5 loop (Figure 4C). Together, these results suggest that, in this new panel of Envs, alteration in the width of the CD4bs cavity is likely to be due to sequence variability within the V5 region of gp120 which repositions the V5 loop.
Figure 4.
Repositioning of the V5 loop is associated with structural alterations in the CD4bs cavity. The gp120 models of CB3sc2 and Macs1br3 Envs (grey and blue ribbon representation, respectively) were superimposed, and their molecular surfaces were presented as blue or grey wire mesh (A). The consensus V1V2 stem (B) and V5 loop sequence (C) was deduced for all 81 primary Envs, and the degree of conservation at each amino acid position was calculated. The GAG linker sequence, which replaced the V1V2 loops in the crystal and model structures, is shown and highlighted as a grey box in panel (B).
Conclusions
Using a large, independent panel of Envs, we confirmed that structural alterations in the gp120 CD4bs can be deduced using optimized three-dimensional gp120 molecular models, and that these alterations may influence fusogenicity and the ability of gp120 to interact with CD4. We further show, for the first time, that these alterations appear to increase the exposure of the CD4bs. Thus, our study provides new insights into structural mechanisms that contribute to altered interactions between gp120 and CD4. These insights contribute to a better understanding of HIV-1 entry and in addition, may inform the design of Env vaccine immunogens where enhanced exposure of the CD4bs may be desirable to elicit effective neutralizing antibody responses. Furthermore, our modeling approach may be informative for better understanding structural mechanisms contributing to HIV-1 disease progression. Here, we finally describe and characterize a new and relatively large panel of functional Envs from brain and other tissues, which will enhance the capacity of investigators to undertake NeuroAIDS research.
List of abbreviations used
HIV-1: Human immunodeficiency virus type 1; SIV: Simian immunodeficiency virus; SHIV: Simian-human immunodeficiency virus; Env: HIV-1 envelope glycoproteins; CD4bs: CD4 binding site; GFP: Green fluorescent protein; RMSD: Room mean squared deviation; sCD4: soluble CD4; mAb: monoclonal antibody; BR: Brain; CSF: Cerebrospinal fluid; PBMC: Peripheral blood mononuclear cells; SC: Spinal cord; Spln: Spleen; LN: Lymph node
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
LG, JS and PRG designed the experiments. LG and JS performed the experiments. JS and PAR designed the molecular models and interpreted the modeling data. MJC assisted with Env cloning and sequencing, and helped interpret the results. PRG supervised the project and helped interpret the results. LG and PRG wrote the manuscript. All authors helped edit the manuscript and have read and approved the final version.
Contributor Information
Lachlan Gray, Email: lachlang@burnet.edu.au.
Jasminka Sterjovski, Email: Jasminka@burnet.edu.au.
Paul A Ramsland, Email: pramsland@burnet.edu.au.
Melissa J Churchill, Email: churchil@burnet.edu.au.
Paul R Gorry, Email: gorry@burnet.edu.au.
Acknowledgements and funding
We thank D. Gabuzda for providing primary HIV-1 isolates, J. Sodroski for providing pSVIII-HXB2 Env plasmid and Cf2th-CD4/CCR5 cells, J. Sodroski and B. Etemad-Gilbertson for providing Cf2-Luc cells, D. Kabat for providing JC53 cells, and H. Gottlinger for providing pSLV-Tat plasmid.
This study was supported by grants from the Australian National Health and Medical Research Council (NHMRC) to PRG and MJC (433915 & 433920). LG is the recipient of a NHMRC Peter Doherty Fellowship. PRG is the recipient of a NHMRC Level 2 Biomedical Career Development Award. The authors gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program received by the Burnet Institute.
References
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/S0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Doms RW. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- Doms RW, Trono D. The plasma membrane as a combat zone in the HIV battlefield. Genes Dev. 2000;14:2677–2688. doi: 10.1101/gad.833300. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Ancuta P. Coreceptors and HIV-1 pathogenesis. Current HIV/AIDS Reports. 2011;8:45–53. doi: 10.1007/s11904-010-0069-x. [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA. et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- Sterjovski J, Churchill MJ, Roche M, Ellett A, Farrugia W, Wesselingh SL, Cunningham AL, Ramsland PA, Gorry PR. CD4-binding site alterations in CCR5-using HIV-1 envelopes influencing gp120-CD4 interactions and fusogenicity. Virology. 2011;410:418–428. doi: 10.1016/j.virol.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, Bell JE, Wolinsky SM, Gabuzda D. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology. 2007;360:105–119. doi: 10.1016/j.virol.2006.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J, Goh WC, Rosen C, Campbell K, Haseltine WA. Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. Nature. 1986;322:470–474. doi: 10.1038/322470a0. [DOI] [PubMed] [Google Scholar]
- LaBonte JA, Patel T, Hofmann W, Sodroski J. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J Virol. 2000;74:10690–10698. doi: 10.1128/JVI.74.22.10690-10698.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J, Sterjovski J, Gray L, Roche M, Chiavaroli L, Ellett A, Jakobsen MR, Cowley D, da Fonseca Pereira C, Saksena N. et al. Enhanced CD4+ cellular apoptosis by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with progressive HIV-1 infection. Virology. 2010;396:246–255. doi: 10.1016/j.virol.2009.10.029. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Rhone D, Steenbeke T, Sun Y, Manola J, Gelman R, Fanton JW, Racz P, Tenner-Racz K, Axthelm MK. et al. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J Virol. 2001;75:5646–5655. doi: 10.1128/JVI.75.12.5646-5655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Sun Y, Nicholson EK, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J Virol. 2000;74:4433–4440. doi: 10.1128/JVI.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson GB, Halloran M, Schenten D, Lee J, Racz P, Tenner-Racz K, Manola J, Gelman R, Etemad-Moghadam B, Desjardins E. et al. The envelope glycoprotein ectodomains determine the efficiency of CD4+ T lymphocyte depletion in simian-human immunodeficiency virus-infected macaques. J Exp Med. 1998;188:1159–1171. doi: 10.1084/jem.188.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZQ, Muhkerjee S, Sahni M, McCormick-Davis C, Leung K, Li Z, Gattone VH, Tian C, Doms RW, Hoffman TL. et al. Derivation and biological characterization of a molecular clone of SHIV(KU-2) that causes AIDS, neurological disease, and renal disease in rhesus macaques. Virology. 1999;260:295–307. doi: 10.1006/viro.1999.9812. [DOI] [PubMed] [Google Scholar]
- Si Z, Gorry P, Babcock G, Owens CM, Cayabyab M, Phan N, Sodroski J. Envelope glycoprotein determinants of increased entry in a pathogenic simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) passaged in monkeys. AIDS Res Hum Retroviruses. 2004;20:163–173. doi: 10.1089/088922204773004888. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Scarano F, Martin-Garcia J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H. et al. Macrophage Tropism of Human Immunodeficiency Virus Type 1 Isolates from Brain and Lymphoid Tissues Predicts Neurotropism Independent of Coreceptor Specificity. J Virol. 2001;75:10073–10089. doi: 10.1128/JVI.75.21.10073-10089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K. et al. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol. 2002;76:6277–6292. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley D, Gray LR, Wesselingh SL, Gorry PR, Churchill MJ. Genetic and functional heterogeneity of CNS-derived tat alleles from patients with HIV-associated dementia. J Neurovirol. 2011;17:70–81. doi: 10.1007/s13365-010-0002-5. [DOI] [PubMed] [Google Scholar]
- Gray LR, Gabuzda D, Cowley D, Ellett A, Chiavaroli L, Wesselingh SL, Churchill MJ, Gorry PR. CD4 and MHC class 1 down-modulation activities of nef alleles from brain- and lymphoid tissue-derived primary HIV-1 isolates. J Neurovirol. 2011;17:82–91. doi: 10.1007/s13365-010-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H. et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray L, Churchill MJ, Keane N, Sterjovski J, Ellett AM, Purcell DFJ, Poumbourios P, Kol C, Wang B, Saksena N. et al. Genetic and functional analysis of R5X4 human immunodeficiency virus type 1 envelope glycoprotiens derived from two individuals homozygous for the CCR5delta32 allele. J Virol. 2006;80:3684–3691. doi: 10.1128/JVI.80.7.3684-3691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray L, Roche M, Churchill MJ, Sterjovski J, Ellett A, Poumbourios P, Sheffief S, Wang B, Saksena N, Purcell DF. et al. Tissue-specific sequence alterations in the human immunodeficiency virus type 1 envelope favoring CCR5 usage contribute to persistence of dual-tropic virus in the brain. J Virol. 2009;83:5430–5441. doi: 10.1128/JVI.02648-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohagen A, Devitt A, Kunstman KJ, Gorry PR, Rose PP, Korber B, Taylor J, Levy R, Murphy RL, Wolinsky SM, Gabuzda D. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J Virol. 2003;77:12336–12345. doi: 10.1128/JVI.77.22.12336-12345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterjovski J, Roche M, Churchill MJ, Ellett A, Farrugia W, Gray LR, Cowley D, Poumbourios P, Lee B, Wesselingh S. et al. An altered and more efficient mechanism of CCR5 engagement contributes to macrophage tropism of CCR5-using HIV-1 envelopes. Virology. 2010;404:269–278. doi: 10.1016/j.virol.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche M, Jakobsen MR, Sterjovski J, Ellett A, Posta F, Lee B, Jubb B, Westby M, Lewin SR, Ramsland PA. et al. HIV-1 escape from the CCR5 antagonist maraviroc associated with an altered and less efficient mechanism of gp120-CCR5 engagement that attenuates macrophage-tropism. J Virol. 2011;85:4330–4342. doi: 10.1128/JVI.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci USA. 2006;103:15160–15165. doi: 10.1073/pnas.0605513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterjovski J, Churchill MJ, Ellett A, Gray LR, Roche MJ, Dunfee RL, Purcell DF, Saksena N, Wang B, Sonza S. et al. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology. 2007;4:89. doi: 10.1186/1742-4690-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol. 2004;78:12975–12986. doi: 10.1128/JVI.78.23.12975-12986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Gray L, Churchill MJ, Sterjovski J, Witlox K, Learmont J, Sullivan JS, Wesselingh SL, Gabuzda D, Cunningham AL, McPhee DA, Gorry PR. Phenotype and envelope gene diversity of nef-deleted HIV-1 isolated from long term survivors infected from a single source. Virology J. 2007;4:75. doi: 10.1186/1743-422X-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]