Abstract
Gamma-Tocotrienol (γ-T3) is a member of the vitamin E family. Tocotrienols (T3) are powerful antioxidants and possess anti-cancer, neuroprotective and cholesterol lowering properties. T3s inhibit the growth of various cancer cell lines without affecting normal cells. Less is known about the exact mechanisms of action of T3s on cell death and other growth inhibitory pathways. In the present study we demonstrate that γ-T3 induces apoptosis in MDA-MB 231 and MCF-7 breast cancer cells as evident by PARP cleavage and caspase-7 activation. Gene expression analysis of MCF-7 cells treated with γ-T3 revealed alterations in the expression of multiple genes involved in cell growth and proliferation, cell death, cell cycle, cellular development, cellular movement and Gene expression. Further analysis of differentially modulated genes using Ingenuity Pathway Analysis software suggested modulation of canonical signal transduction or metabolic pathways such as NRF-2 mediated oxidative stress response, TGF-β signaling and Endoplasmic Reticulum (ER) stress response. Analysis of ER stress related proteins in MCF-7 and MDA-MB 231 cells treated with γ-T3 demonstrated activation of PERK and pIRE1α pathway to induce ER stress. Activating transcription factor 3 (ATF3) was identified as the most upregulated gene (16.8 folds) in response to γ-T3. ATF3 knockdown using siRNA suggested essential role of ATF3 in γ-T3 induced apoptosis. In summary, we demonstrate that γ-T3 modulates ER stress signaling and have identified ATF3 as a molecular target for γ-T3 in breast cancer cells.
Keywords: Vitamin E, Tocotrienol, Breast Cancer, ER Stress, Unfolded Protein Response (UPR), Apoptosis
Introduction
Vitamin E includes a family of lipophilic micronutrients consisting of four tocopherols and four Tocotrienols (α, β, γ and δ) that consist of a chromanol ring and a side chain. Both tocopherols (Ts) and tocotrienols (T3s) are found in various components of the human diet. Ts are present primarily in nuts and vegetable oils, while T3s are minor plant constituents especially abundant in rice bran, cereal grain, and palm oil. Ts and T3s are well recognized for their antioxidative effects. In general, antioxidants are suggested to reduce cancer by arresting free radical induced DNA damage. Ts have been studied in great detail for their antioxidative property and physiological relevance. However, limited studies have been performed on T3s. It is well documented that T3s possess more powerful anti cancer, neuroprotective and cholesterol lowering properties that are often not exhibited by Ts [1]. The accumulation of T3s in the cells is much greater than tocopherols; this might be one of the reasons that T3s have more significant physiological effects than tocopherols [2]. T3s, particularly γ-T3, suppress the activity of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG CoA), an enzyme involved in cholesterol biosynthesis in the liver, resulting in less cholesterol being manufactured by the liver cells and an overall reduction of plasma cholesterol levels [1, 3]. γ–T3 possesses a hormone like natriuretic function and can potentially prevent hypertension and cardiovascular disease (CVD) caused by high salt intake [4].
T3s have been shown to inhibit the growth of various cancer cells without affecting the growth of normal cells [5–6]. In breast cancer cells, γ-T3s induces apoptosis in irrespective of their estrogen response status [7–9]. Although T3s are likely to be one of the powerful cancer chemotherapeutic/preventive agents in the human diet, their exact mechanisms of action on cell death and other inhibitory pathways are unknown. Gene expression profiling in estrogen receptor (ER) positive, p53 wild type MCF-7 and ER negative, p53 mutant MDA-MB 231 cells treated with tocotrienol rich fraction (TRF) of palm oil suggested different mechanisms in the two cell lines [10]. Lipid peroxidation is one of the mechanisms suggested for its anti-proliferative action [11]. Other mechanisms involve modulation of various signaling pathways including apoptosis by caspase-8 activation and mitochondrial dependency [12–13], inhibition of cell proliferation [6], down-regulation of cyclins [14], reduction in the Pl3K/PDK-1/Akt signaling and NFκb activity [15–16] and modulation of p53, Bax/Bcl2 [17]. Estrogens are known to be involved in breast carcinogenesis. T3s have been demonstrated as antioxidants against the E2 epoxide induced breast cancer carcinogenesis whereas α-tocopherol was found to be least effective [18]. The effects of γ-T3 has also been studied in other cancers such as colon and prostate cancer where γ-T3 has been found to modulate multiple signaling pathways and induce apoptotic cell death. Activation of p53 has been reported in RKO human colon carcinoma cells in response to TRF [17]. Recently, Yap and colleagues reported the modulation of ID family proteins and mesenchymal markers in prostate and breast cancer cells in response to γ-T3 [19–20].
In the present study we examined the whole genome transcription in MCF-7 breast cancer cells, when exposed to γ-T3. We demonstrate that γ-T3 induced apoptosis is associated with induction of early response genes and ER stress transcriptional response in MCF-7 cells. We have characterized the ER stress response induced by γ-T3 in MCF-7 and MDA-MB 231 breast cancer cells.
1. Materials and Methods
2.1. Cell Culture and Media
Human breast cancer cells (MCF-7 and MDA-MB 231) and immortalized normal human breast mammary MCF-10A cells were obtained from Lombardi Comprehensive Cancer Center cell repository and grown in Dulbecco’s modified Eagle medium (DMEM), supplemented with 5% heat-inactivated fetal bovine serum and 25 μg/ml gentamicin (Invitrogen, Carlsbad, CA).
2.2. Chemicals, Reagents and Antibodies
T3s (>95% pure) were from EISAI Corporation (Woodcliff Lake, NJ) and Carotech (Edison, NJ) and dissolved in DMSO. WST-1 reagent used for cytotoxicity assay and protease inhibitor cocktail tablets (Roche Applied Science, Indianapolis, IN); ECL Plus Western blotting detection system (GE Healthcare, Piscataway, NJ); and Coomassie protein assay reagent (BioRad, Hercules, CA).
The following antibodies were obtained commercially: Cleaved PARP, GRP78/Bip, Chop, PERK, Cyclin D-1, p-IRE1α, IRE1α, p-eIF2α, eIF2α, β-Tubulin, and GAPDH antibodies (Cell Signaling Technology Inc, Danvers, MA); ATF3, ATF6 and p-PERK (Santa Cruz Biotechnology, Santa Cruz, CA); β-Actin from Sigma, and ATF4 (Abcam, Cambridge, MA). Secondary antibodies conjugated with horseradish peroxidase included goat anti-mouse IgG, goat anti-rabbit IgG and rabbit anti-goat IgG (Jackson ImmunoResearch, West Grove, PA).
2.3. Cell Viability and Proliferation Assay
Effects of T3s on cell viability and proliferation of breast cancer cells were determined using a cell viability detection kit (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1, 3-benzene disulfonate, WST-1) according to the manufacturer’s instructions (Roche Applied Science, Indianapolis, IN). Briefly, the cells (MCF-10A, MCF-7 and MDA-MB 231) were seeded onto 96-well plates at a density of 3,000cells per well in six replicates, and maintained overnight in 100 μl of 5% fetal bovine serum-containing medium. The following day, cells were treated for 24, 48 and 72 h with different concentrations of α and γ-T3 ranging from 10 to 40 μM made in complete medium using six wells per treatment condition. Control cells were treated with DMSO at a final concentration of 0.1%. At the end of each treatment after 24, 48 and 72 h, 10 μl of the WST-1 reagent was added to each well. Plates were incubated for 2h at 37°C and analyzed at A= 450/600 using a Bio-Rad Model 680 micro plate reader. Cell viability was assessed by trypan blue dye exclusion assay.
2.4. Cell Cycle Analysis
The effects of γ-T3 on cell cycle were determined by flow cytometric analysis of MCF-7 cells treated with γ-T3. The cells were treated with 40 μM of γ-T3 for 24h in triplicates. The floating cells were pooled with the adherent cells, washed and fixed with 70% ethanol followed by FACS analysis. Briefly, the cells were centrifuged and suspended in 1ml of phosphate-buffered saline containing 50μg/ml each of RNase A and propidium iodide (both from Sigma-Aldrich, St. Louis, MO). The stained cells were analyzed using the FACsort (Becton Dickinson, Franklin Lakes, NJ), and Reproman computer software. The percentage of cells containing sub-G1 DNA content was used as an index of apoptosis as described previously [21]. Gating was set to exclude cell debris, doublets, and clumps when determining cells in different stages.
2.5. RNA isolation and Gene Expression Microarray
Gene expression pattern in MCF-7 cells treated with 40 μM γ-T3 were analyzed by microarray studies. Four independent experiments were performed with cells grown on different days and from different stocks. The RNA from cells treated with DMSO control and γ-T3 treated cells was isolated using Trizol (Invitrogen) and further purified using Qiagen RNAeasy kit. The RNA concentrations were determined spectrophotometrically using a Beckman DU640 Spectrophotometer (Beckman Coulter, Brea, CA). RNA quality was assessed using the Agilent 2100 Bioanalyzer. High quality RNA was labeled and hybridized to U133A2 Affymetrix GeneChips following manufacturer’s recommendations (Affymetrix, Santa Clara, CA). Microarray data quality was assessed using various tools including those recommended by Affymetrix. All array data presented here passed the quality control measures. Our data analysis began with preprocessing of the probe-level Affymetrix data (.cel files). We used RMA for background adjustment, quantile method for normalization, and a robust multi-array average for summarization. These methods have provided better performance than MAS 5.0 and MBEI in detecting known levels of differential expression using spike-in Affymetrix data [22–23]. They are implemented in several statistical tools including Bioconductor and BRB-ArrayTools (NCI, Bethesda, MD).
We analyzed the preprocessed data to identify the genes that were differentially expressed between the untreated and treated groups using random variance model implemented in BRB-ArrayTools. The random-variance t-test is an improvement over the standard separate t-test as it permits sharing information among genes within-class variation without assuming that all genes have the same variance [24]. The false discovery rate (FDR) was estimated using the method of Benjami and Hochberg [25]. Probesets were considered statistically significant if their p-value was less than 0.001. With this threshold, 969 probe sets were found statistically significant (p<0.001 and FDR<0.023). These probe sets had a fold change > 1.2. The probe sets with >1.5 fold change were selected for analysis to identify canonical pathways, physiological functions and interaction networks using the Ingenuity Pathway Analysis software (IPA) (Ingenuity Systems, Redwood City, CA).
2.6. Real Time RT-PCR
For Real Time PCR, complementary DNA (cDNA) was synthesized from 1 μg of total RNA using Transcription First Strand cDNA Synthesis Kit (Roche) following the manufacturer’s instructions. The primers were purchased from RealTimePrimers.com (Elkins Park, PA). For each primer set, the meltcurves were performed to verify that the primers amplified a single product and ensure that there were no primer dimers or amplification in the no-template controls. Three independent experiments were performed in triplicate with RNA isolated with different cell stocks, treatments and different days.
PCR amplification was performed with the 7300 Real-Time PCR System (Applied Biosystems, Carlsbad, CA) in 50 μl reactions using 5 μl of cDNA (50 ng of input total RNA), 300 nM each of forward and reverse primer and 1× FastStart SYBR Green PCR Master Mix (Roche). Expression levels of the genes of interest were normalized to GAPDH. The QPCR cycling parameters consisted of 1 cycle of 95°C for 10 minutes, and 40 cycles of (95°C for 15 sec and 60°C for 1 minute). On a given 96- well plate, target and control normalizing genes were run in triplicate. The CT (threshold cycle of amplification) values were determined using the 7300 Real-Time PCR System RQ Study Software (Version 1.3.1) (Applied Biosystems). To determine fold change in expression levels the comparative CT method was used using the formula 2−ΔΔCT.
2.7. Western blotting
Immunoblotting was performed essentially as described previously [26]. After 24 h treatment with γ-T3, adherent and floating cells were collected. Whole cell extracts (total cell homogenates) were prepared by lysing of cells in radioimmune precipitation assay buffer, and proteins were separated on a 4–20% gradient SDS gel (Pierce), followed by transferring of proteins to polyvinylidene difluoride membranes (0.45 μm, Immobilon-P, Millipore, Billerica, MA). Membranes were immunoblotted with the appropriate primary antibody and peroxidase-conjugated secondary antibody. The antigen-antibody complex was determined using the ECL detection assay (Amersham/GE Life Sciences, Piscataway, NJ). Each Western Blot was repeated at least 3 times. Representative Western Blots are shown.
2.8. Statistical Analyses
Cell proliferation experiments were performed in 6 replicates. Cell cycle and RT-PCR experiments were performed in triplicates. Student’s T test was used to analyze treated vs. untreated cells. Results were expressed as averages ± SD. P<0.05 was considered significant. Statistical analysis of microarray data was performed as described above.
3. Results
3.1. Effects of α and γ T3s on the proliferation of human breast cancer cells
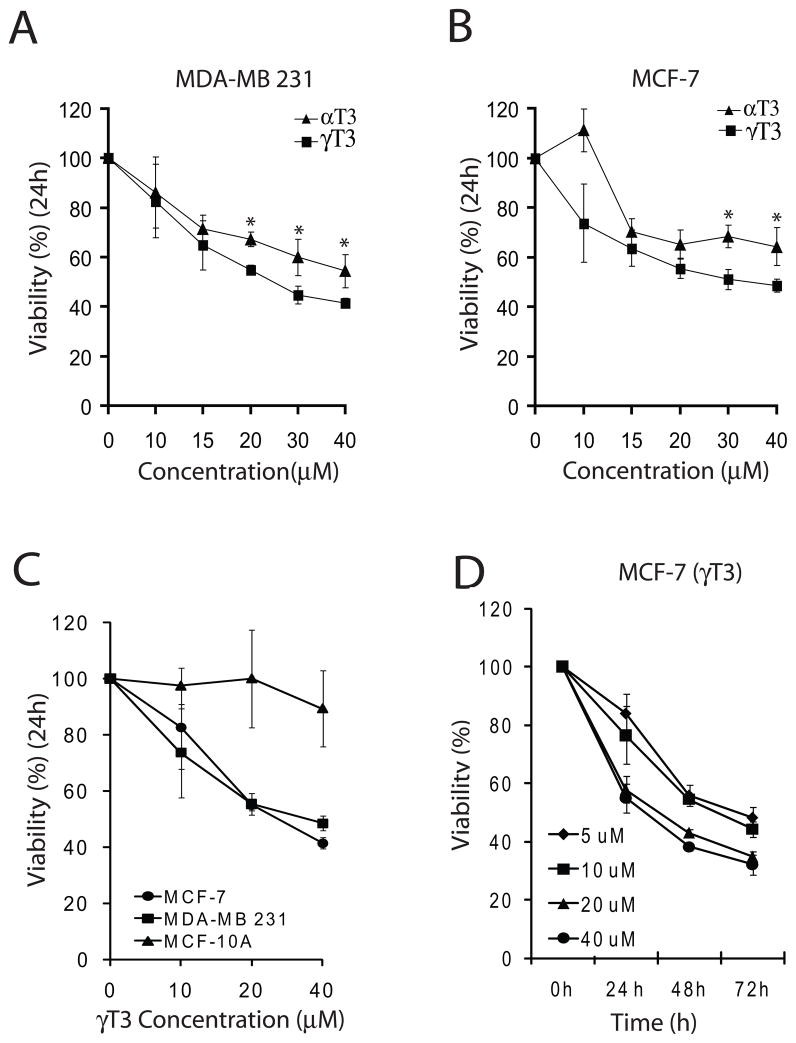
We compared the effects of α and γ T3 on the viability of MCF-7 and MDA-MB 231 cells using WST-1 assay. Both α and γ-T3 inhibited cell proliferation in a dose dependent manner when treated with 10–40 μM of each T3 for 24 h (Fig. 1A, B). γ-T3 exhibited a slightly better cell growth inhibitory effect at 20–40μM as compared with α-T3 in both the cell lines (P<0.05). The inhibitory effect of γ-T3 on MCF-7 and MDA-MB 231 cells was also confirmed by trypan blue dye exclusion assay (data not shown). We concentrated our studies on γ-T3 in subsequent experiments. Next, we compared the effects of γ-T3 on human breast cancer (MCF-7 and MDA-MB 231) with normal breast epithelial cells (MCF-10A). As shown in Fig. 1C, at 24 h, γ-T3 did not inhibit the growth of MCF-10A cells suggesting a cancer specific effect of γ-T3 on cell proliferation. ER status and p53 status have been shown to affect the sensitivity of cancer cell lines to various chemotherapeutic and chemopreventive compounds. We compared the growth inhibitory effects of γ-T3 on ER positive, p53 wild type MCF-7 and ER negative and p53 mutant MDA-MB 231 cells. γ-T3 inhibited the growth of both cell lines in a comparable fashion suggesting that the growth inhibitory properties of γ-T3 are independent of the p53 or ER status (Fig. 1C). To further study the effects of γ-T3 in a time dependent manner, we treated MCF-7 cells with 5–40 μM of tocotrienol for 24, 48 and 72 h. γ-T3 inhibited the growth of MCF-7 cells in a time and dose dependent manner (Fig. 1D).
Fig. 1.
α and γ-T3s inhibit the proliferation of MCF-7 and MDA-MB 231 breast cancer cells without affecting the proliferation of normal immortalized mammary MCF-10A cells. A and B. γ-T3 is slightly more effective in inhibiting cell proliferation of both cells lines. C. γ-T3 inhibits cell proliferation in both MCF-7 and MDA-MB 231 cells with similar efficacy while no affect on the proliferation of normal immortalized mammary MCF-10A cells. D. γ-T3 inhibits the proliferation of MCF-7 cells in a time and dose dependent manner. Cells were seeded at a density of 3000 cells/well in a 96 well plate and treated with DMSO control or different concentrations of α and γ-T3 for indicated time periods. Cell proliferation was determined by WST-1 assay (Roche). A representative of 3 experiments performed in six replicates each is shown. P<0.05 was considered significant.
3.2. Treatment with γ-T3 causes G1 arrest and induction of apoptosis
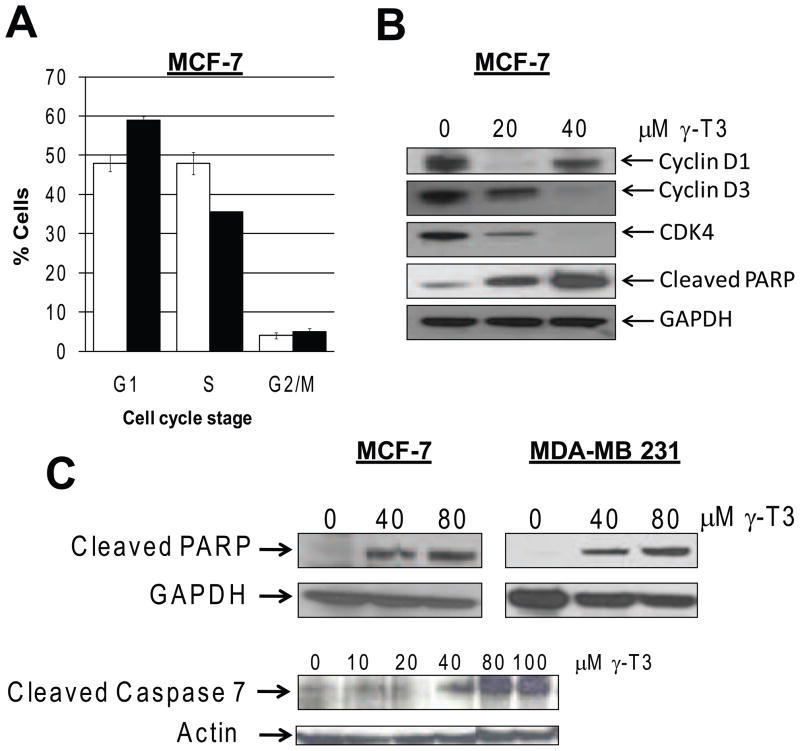
We examined the cell cycle profile of MCF-7 cells treated with γ-T3. Twenty four hour treatment with 40 μM γ-T3 resulted in a modest G1 arrest. Microarray data also revealed cell cycle regulated genes as one of the top 5 categories of altered functions. To study the modulation of cell cycle regulatory proteins, we examined the expression of Cyclin D1/D3 and CDK4 by immunoblotting. D and E-type cyclins and their dependent kinases CDK4/6 play essential roles at the restriction point during G1-S transition of the cell. As shown in Fig. 2B, the G1 arrest caused by γ-T3 treatment was associated with downregulation of cyclin D1/D3 and CDK4.
Fig. 2.
A. γ-T3 treatment of MCF-7 cells causes a modest G-1 arrest. MCF-7 cells were treated with DMSO control (white bars) 40 μM of γ-T3 (black bars) for 24 h and cell cycle distribution was determined by flow cytometry. (representative of 3 experiments, n=3, P<0.05). B. Effects of γ-T3 on cell cycle regulatory proteins. C. γ-T3 induces apoptosis in MCF-7 and MDA-MB 231 cells. Expression of cell cycle regulatory proteins, Cyclin D1, D3, CDK4 and apoptosis indicators, cleaved-PARP and cleaved-caspase 7 was determined by immunoblotting. β-Actin and GAPDH antibodies were used as loading control.
To determine whether, γ-T3 induces apoptotic cell death, we analyzed MCF-7 cells for PARP cleavage and cleaved caspase-7, well established apoptotic markers. Treatment of MCF-7 cells with γ-T3 resulted in increased cleaved PARP and Caspase-7 proteins (Fig. 2B and 2C). As shown in Fig. 2C, γ-T3 also induced apoptosis in MDA-MB231 cells.
3.3. γ-T3 modulates multiple gene functions and signaling pathways in MCF-7 breast cancer cells
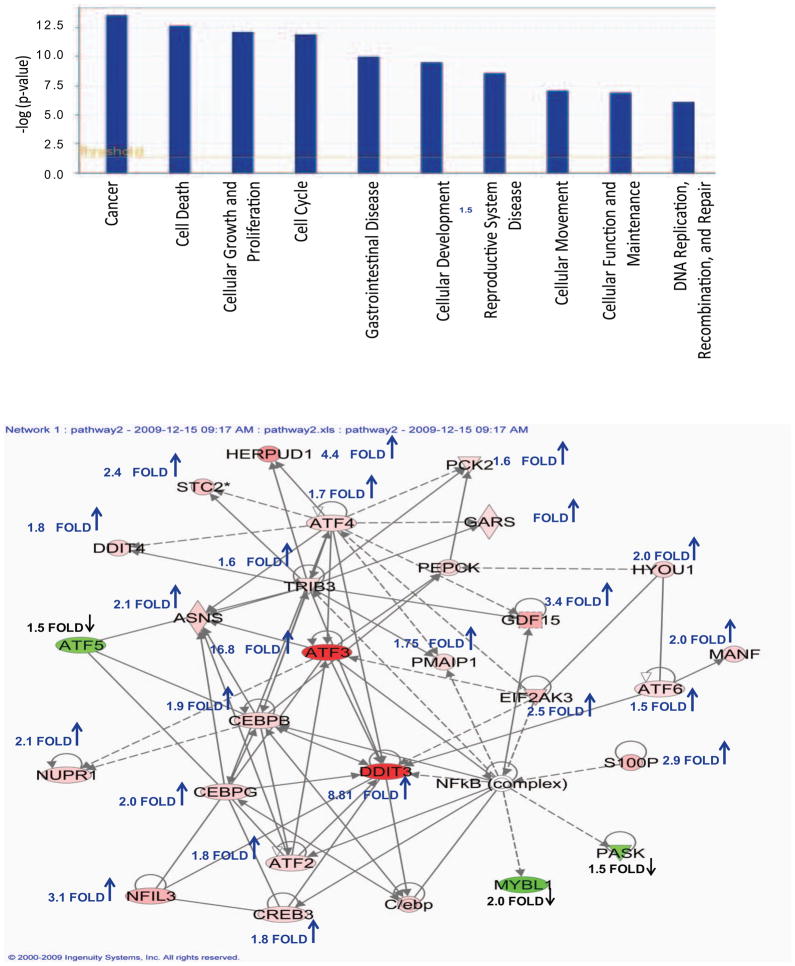
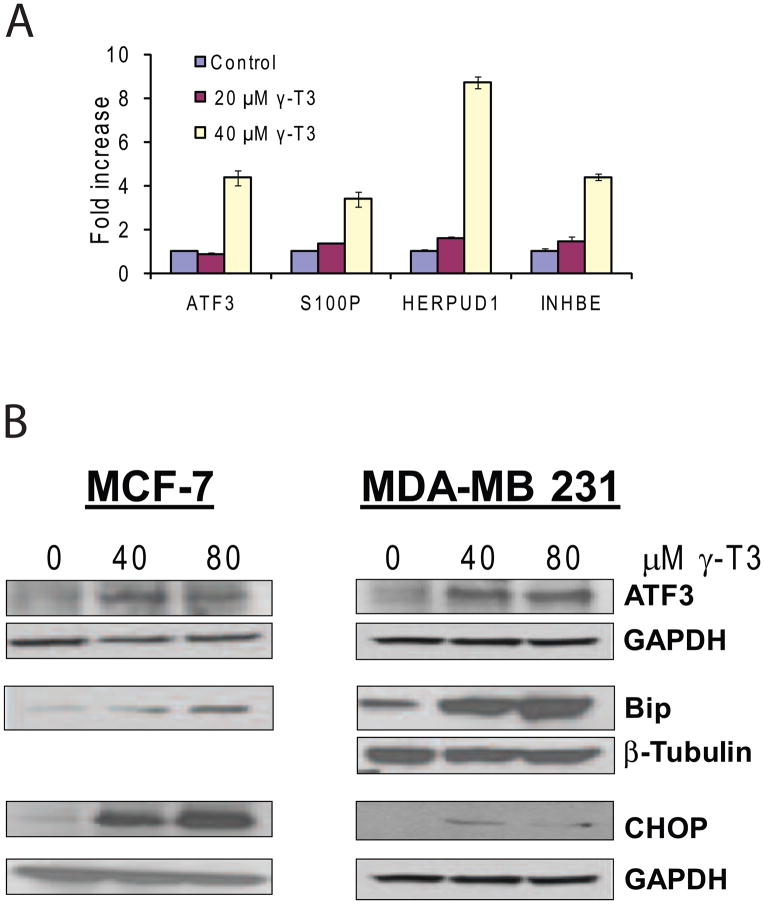
To identify the molecular targets and signaling pathways modulated by γ-T3, MCF-7 cells were treated with DMSO control or 40 μM of γ-T3 and gene expression changes were examined by Affymetrix microarray analysis. Microarray data analysis identified 527 probe sets that were differentially expressed with >1.5 fold change (P<0.001 and FDR<0.023). The complete dataset for microarray data is available in the public database Gene Expression Omnibus (GEO) repository (accession number: GSE21946). Ingenuity analysis of 527 probe sets selected 266 genes in various functional categories and several canonical pathways. Further narrowing down the differentially expressed genes with >2.0 fold change resulted in 127 probe sets that were used in further analysis. 114 probe sets were upregulated and 13 were downregulated by >2.0 fold upon γ-T3 treatment for 24h. The top molecular and cellular functional categories identified were Cellular growth and proliferation, Cell death, Cell cycle, Cellular development, Cellular movement and Gene expression. In order to identify the biological mechanisms involved in response to γ-T3 treatment, 127 probe sets were further analyzed by Ingenuity which suggested modulation of canonical signal transduction or metabolic pathways such as NRF-2 mediated oxidative stress response, TGF-β signaling and Endoplasmic Reticulum (ER) stress response pathway (Fig. 3). A selective list of genes induced or repressed by γ-T3 is shown in Table 1. Activation of ER stress signaling has been demonstrated in anticancer activities of various chemotherapeutic and chemopreventative compounds. The microarray data analysis revealed that γ-T3 treatment modulated several key components of ER stress pathway such as DNA damage-inducible transcript 3 (DDDIT3; also known as GADD153 or CHOP); ATF4, the chaperones HSPA5 (also known as GRP78 or BiP) and HSP90B1 (also known as GRP94); EIF2AK3, PERK, HERPUD1 and PHLDA1. A number of immediate early response genes such as ATF3 (18.5 fold), Fos (2.14 fold), Jun (2.02 fold), and Egr1 (3.86 fold) were also modulated by γ-T3. To validate the gene expression data, we randomly selected 4 genes (ATF3, HERPUD1, S100P and INHBE) with higher fold change values and analyzed their expression in DMSO control and γ-T3 treated MCF-7 cells by real-time RT-PCR. As shown in Fig. 4A, the selected genes were highly induced in γ-T3 treated MCF-7 cells compared to the controls. ATF3 and HERPUD1 have been implicated in ER Stress. S100P is a small calcium binding protein and INHBE is a member of the activin beta family. To further validate our findings at the translational level, we examined the expression of ATF3, GRP78/BIP and CHOP on MCF-7 cells treated with γ-T3. ATF3, GRP78/BiP and CHOP were upregulated in MCF-7 cells upon γ-T3 treatment (Fig. 4B). We asked whether γ-T3 induces a similar response in triple negative MDA-MB 231 cells. Similar to MCF-7 cells, γ-T3 treatment increases the expression of GRP78, ATF3 and CHOP in MDA-MB 231 cells suggesting activation of a common pathway in both cells lines (Fig. 4B).
Fig. 3.
A. Top 10 functional categories that were modulated in 40 μM γ-T3 treated MCF-7 cells as analyzed by Ingenuity Pathway Analysis software. B. Network of the molecular relationship of selected differentially expressed genes in microarray data. Data was analyzed by Ingenuity Pathway Analysis software. The intensity of the node color indicates upregulated (red) and downregulated (green) genes. White node represents a gene that was not identified as a differentially expressed gene and is not part of our dataset, but is included in the network though other genes. Majority of the genes in this network were upregulated as indicated by fold change and upward arrow.
Table 1.
Selected differentially modulated genes in MCF-7 cells after γ-T3 (40μm) treatment for 24h
| Gene symbol | Description | Fold Change |
|---|---|---|
| ER Stress | ||
| ATF3 | activating transcription factor 3 | 16.76104 |
| DDIT3/CHOP/GADD153 | DNA-damage-inducible transcript 3 | 8.294404 |
| DNAJB9/ERDJ4 | DnaJ (Hsp40) homolog, subfamily B, member 9 homocysteine-inducible, endoplasmic reticulum | 7.424944 |
| HERPUD1 | stress-inducible, ubiquitin-like domain member 1 pleckstrin homology-like domain, family A, member | 4.355322 |
| PHLDA1 | 1 protein phosphatase 1, regulatory (inhibitor) subunit | 4.127389 |
| PPP1R15A/GADD34 | 15A | 2.75497 |
| DUSP6 | dual specificity phosphatase 6 | 2.609884 |
| HSPA5 | heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) | 2.544269 |
| EIF2AK3/PERK | eukaryotic translation initiation factor 2-alpha kinase 3 | 2.522865 |
| Others | ||
| INHBE | inhibin, beta E | 14.74111 |
| ANG | angiogenin, ribonuclease, RNase A family, 5 | 5.37427 |
| CLGN | calmegin | 4.113258 |
| ANG | angiogenin, ribonuclease, RNase A family, 5 | 3.72196 |
| EGR1 | early growth response 1 | 3.705403 |
| CAMTA2 | calmodulin binding transcription activator 2 | 3.528805 |
| EGR1 | early growth response 1 | 3.466066 |
| GDF15 | growth differentiation factor 15 | 3.449369 |
| STCH | stress 70 protein chaperone, microsome-associated, 60kDa | 2.836486 |
| HRK | harakiri, BCL2 interacting protein (contains only BH3 domain) | 2.650593 |
| GADD45A | growth arrest and DNA-damage-inducible, alpha | 2.642888 |
| JUN | jun oncogene | 2.37199 |
| TGIF1 | TGFB-induced factor homeobox 1 | 2.216848 |
| GPR87 | G protein-coupled receptor 87 | 2.094095 |
| ARF4 | ADP-ribosylation factor 4 | 2.090843 |
| ANXA1 | annexin A1 | 2.019521 |
| MYBL1 | v-myb myeloblastosis viral oncogene homolog (avian)-like 1 | −2.00495 |
| MEGF9 | multiple EGF-like-domains 9 | −2.01284 |
| SOX11 | SRY (sex determining region Y)-box 11 | −2.02478 |
| RET | ret proto-oncogene | −2.13753 |
Fig. 4.
A. Validation of 4 differentially expressed genes by real-time RT-PCR in control and γ-T3 (20 and 40μM) treated MCF-7 cells. The transcript levels were normalized to GAPDH gene. The data is computed as ratio of indicated genes in γ-T3 treated cells relative to DMSO control treatments (representative of 3 experiments performed in triplicate (P<0.05, mean ± S.E.M. shown), B. Validation of ATF3, BiP and CHOP proteins in MCF-7 and MDA-MB 231 cells by Western blotting. MCF-7 cells: shown are ATF3 and its loading control; GAPDH loading control (lower panel) for Bip and CHOP. MDA-MB 231 cells: ATF3 blot and its loading control GAPDH. Bip and CHOP with their loading controls β-tubulin and GAPDH respectively.
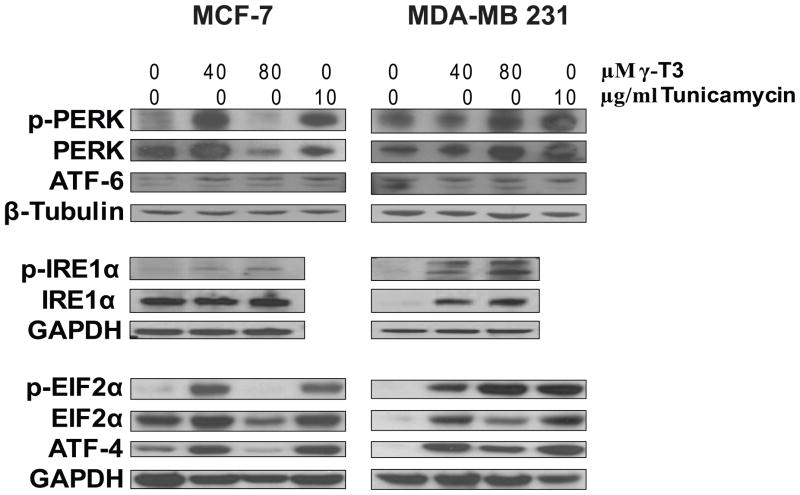
3.4. γ-T3 induces ER Stress and modulates multiple Unfolded Protein Response (UPR) Pathways
Gene expression data suggested modulation of multiple ER stress related pathways in response to γ-T3. To further validate the gene expression data and investigate the ER stress pathways induced by γ-T3, we studied the expression of ER stress related proteins in MCF-7 and MDA-MB 231 cells treated with γ-T3. We used tunicamycin, a known inducer of ER stress as a control treatment. ER stress signaling involves specific unfolded protein response (UPR) that is modulated by three UPR signaling pathways initiated by PERK (PKR-like ER kinase), IRE1α/β and ATF6α/β. Treatment with 40 μM γ-T3 resulted in the activation of PERK and pIRE1α (Fig. 5) in both MCF-7 and MDA-MB 231 cells. No significant change was observed in the expression of ATF6. Next we analyzed the phosphorylation of eukaryotic initiation factor 2α (eIF2α). Activated PERK phosphorylates eIF2α that attenuates mRNA translation and reduces the load of protein synthesis. Phosphorylation of eIF2α is recognized as the most important event during ER stress. As shown in Fig. 5, at 40 μM γ-T3, expression as well as phosphorylation of eIF2α was higher in both the cell lines. Phosphorylation of eIF2α leads to the expression of ATF4 (activating transcription factor 4) that upregulates multiple UPR related genes such as GADD153, ATF3 and GADD34. To analyze the downstream effectors of eIF2α, we analyzed the expression of ATF4 and its downstream targets. Treatment with 40 μM γ-T3 for 24 h induced the expression of ATF4 in both MCF-7 and MDA-MB 231 cells consistent with transcriptional induction of several ATF4 targets such as GADD153/CHOP and ATF3 in MCF-7 cells. As mentioned earlier, γ-T3 treatment did induce the expression of both CHOP and ATF3 at both transcriptional as well as translational level.
Fig. 5.
Modulation of multiple ER stress signaling pathways by γ-T3. MCF-7 and MDA-MB 231 cells were treated with indicated concentrations of γ-T3 for 24 h. Expression of ER stress related proteins was determined by Western blot analysis. GAPDH and β-tubulin antibody were used for loading control. Tunicamycin treatment was used as a positive control for inducing ER stress signaling.
3.5. ATF3 plays an essential role in γ-T3 induced apoptosis in MCF-7 cells
Activating transcription factor 3 (ATF3) is an ER stress related early response gene that is highly elevated (16.8 folds) in gene expression analysis of MCF-7 cells treated with 40 μM γ-T3 for 24 h. ATF3 is a target of ATF4 and is known to be induced by certain chemotherapeutic and chemopreventative compounds. Also, ATF3 has been implicated in the induction of apoptosis by curcumin in breast cancer cells [27]. We asked, whether induction of ATF3 is essential for apoptosis induced by γ–T3. MCF-7 cells were transfected with control siRNA or ATF3 siRNA were treated with 40 μM γ-T3 for 24 h. ATF3 siRNA was able to completely knockdown the expression of ATF3 protein. γ-T3 treatment increased ATF3 protein levels and apoptosis in control siRNA cells as evident by PARP cleavage. PARP cleavage was non-detectable in ATF3siRNA cells upon γ-T3 treatment (Fig. 6). Our data suggest that ATF3 plays an important role in γ-T3 induced apoptosis in human breast cancer cells.
Fig. 6.
ATF3 is essential for γ-T3 induced apoptosis in MCF-7 cells. MCF-7 cells were transfected with control siRNA or ATF3 siRNA for 24h. The cells were treated 24 h post-transfection with 40 μM γ-T3 and PARP cleavage and Cyclin D1 was determined by Western blotting.
4. Discussion
Tocotrienols (T3s) have gained attention due to their powerful anti cancer, neuroprotective and cholesterol lowering properties not exhibited by tocopherols [1]. The accumulation of T3s in the cells is much greater than tocopherols; this might be one of the reasons that T3s have more significant effects than tocopherols [2]. T3s inhibit the growth of various cancer cell lines without affecting the growth of normal cells [5–6]; our results confirmed that at higher concentrations of γ-T3, only the growth of breast cancer cell lines MCF-7 and MDA-MB 231 are inhibited while the normal breast epithelial cells, MCF-10A remain unaffected. Molecular mechanisms involved in the growth inhibitory effects of γ-T3 are poorly understood. Multiple mechanisms of cell growth inhibition by γ-T3 have been suggested including cell cycle arrest, inhibition of NF-kB and AKT pathways, induction of mitochondria dependent apoptosis and reduced angiogenesis [28]. This study was undertaken to identify molecular targets of γ-T3 by gene expression microarray analysis. Effects of α and γ-T3s were studied on MCF-7 and MDA-MB 231 breast cancer cells. Both compounds exhibited a time and dose dependent inhibition in the two cell lines with γ-T3 being slightly more effective than α-T3. In accordance with previous studies we have demonstrated that γ-T3 induces apoptotic cell death in both MCF-7 and MDA-MB 231 cell lines suggesting that the mechanisms involved may be independent of ER/PR/HER status [7–9]. T3s induce cell cycle arrest in G1/S and G2/M phase depending on the cancer cell type [29–33]. We observed a modest G1 arrest in MCF-7 cells treated with 40 μM γ-T3 along with downregulation of cyclin D1, its regulating kinase CDK4 and Cyclin D3, the regulating subunit of CDK4, suggesting a possible mechanism for G1 arrest. Loss of cyclin D1/CDK4 and reduced phosphorylation of Rb has been reported in MDA-MB 231 cells treated with δ-T3 [33]. We noticed that cyclin D1 downregulation was more pronounced at 20 μM than 40 μM γ-T3. Further investigations are being carried out in our laboratory to address this phenomenon. Proteasomal degradation of cyclin D1 during cell cycle regulation has been reported and could explain the loss of cyclin D1 [34]. Cyclin D1 loss was not dose dependent and may be due the proteasomal inhibitory effects of γ-T3 [35]. Redundancy and compensation mechanisms have been reported between cell cycle proteins that may also explain the modest G1 arrest when treated with 40 μM γ-T3 [36]. To further study the molecular targets of γ-T3, we performed a genome wide gene expression study in MCF-7 cells treated with 40 μM γ-T3. Previous studies reporting gene expression data were performed using tocotrienol rich fraction of palm oil and not with the purified T3s [37]. Gene expression studies using purified γ-T3 on breast cancer cells are likely to discover novel targets and further validate the existing targets. Microarray data analysis revealed the modulation of genes involved in immediate early response and ER stress response. One of the prominent genes that we found was ATF3 gene which is the target gene for ATF4 involved in ER stress response. We also demonstrate that similar to previous studies on other chemopreventive agents, ATF3 is essential for γ-T3 induced apoptosis in MCF-7 cells (Fig. 6). Our data supports previous observation that the effectors of ER stress such as CHOP have been shown to be modulated by tocotrienol rich fractions of palm oil [37]. Three different pathways initiated by PERK (PKR-like ER kinase), IRE1α/β and ATF6α/β have been suggested for ER stress. We investigated the key molecules involved in the three pathways to determine the pathway(s) that are active in γ-T3 induced apoptosis. We observed an up-regulation of ER chaperon GRP78/Bip, ATF-4, PERK protein kinase and type I ER transmembrane protein IRE1α in both MCF-7 and MDA-MB 231 cells treated with 40 μM γ-T3. These ER-stress related proteins were greatly diminished in MCF-7 cells at 80 μM and may be due to increased cell death. In response to ER stress, ER chaperone GRP78/Bip releases the ER proteins and binds to unfolded proteins causing the activation of PERK. Once activated, PERK phosphorylates eIF2α, which in turn causes the suppression of general protein synthesis and reduces the protein load on the ER. However, this mechanism allows up-regulated synthesis of selective proteins such as ATF4 transcription factor that modulates other key ER stress effectors such as ATF3 and CHOP to induce apoptosis; one of the possible mechanisms in γ-T3 induced apoptosis.
IRE1α is another core component of the UPR that is induced upon ER stress. It is an endonuclease that splices mRNA for XBP-1 transcription factor transcribing genes important in protein folding and degradation to restore the ER function. In contrast, ATF6 activation is triggered by translocating to the Golgi where it is cleaved resulting in an active transcription factor that induces pro-survival genes. We did not observe any ATF6 modulation in response to γ-T3 treatment. Our study suggests that γ-T3 activates at least two UPR pathways: PERK and IRE1α. While this manuscript was in preparation, two independent groups have also demonstrated recently that γ-T3 induces ER stress in breast cancer cells [38–39], however our approach is still unique as we studied genome wide expression to γ-T3 treatment in MCF-7 breast cancer cells that identified several putative novel mediators of the anticancer effect of γ-T3 that can be exploited in future studies. Further we demonstrated that ATF3 is an essential contributor to the pro-apoptotic response of γ-T3.
Acknowledgments
The studies were conducted using the Tissue Culture, Flow Cytometry and Cell Sorting and Macromolecular Analysis Shared Resources of the Lombardi Comprehensive Cancer Center. We thank Karen Creswell, Xiaojun Zou and James Li for providing excellent technical assistance.
Financial Support (DK): NCI UDC-LCCC U56, UDC Agricultural Experiment Station, MBRS SCORE, NSF HBCU-UP program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sen CK, Khanna S, Roy S. Tocotrienols: Vitamin E beyond tocopherols. Life Sci. 2006;78:2088–98. doi: 10.1016/j.lfs.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noguchi N, Hanyu R, Nonaka A, Okimoto Y, Kodama T. Inhibition of THP-1 cell adhesion to endothelial cells by alpha-tocopherol and alpha-tocotrienol is dependent on intracellular concentration of the antioxidants. Free Radic Biol Med. 2003;34:1614–20. doi: 10.1016/s0891-5849(03)00216-8. [DOI] [PubMed] [Google Scholar]
- 3.Khor HT, Ng TT. Effects of administration of alpha-tocopherol and tocotrienols on serum lipids and liver HMG CoA reductase activity. Int J Food Sci Nutr. 2000;51 (Suppl):S3–11. [PubMed] [Google Scholar]
- 4.Saito H, Kiyose C, Yoshimura H, Ueda T, Kondo K, Igarashi O. Gamma-tocotrienol, a vitamin E homolog, is a natriuretic hormone precursor. J Lipid Res. 2003;44:1530–5. doi: 10.1194/jlr.M300061-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Srivastava JK, Gupta S. Tocotrienol-rich fraction of palm oil induces cell cycle arrest and apoptosis selectively in human prostate cancer cells. Biochem Biophys Res Commun. 2006;346:447–53. doi: 10.1016/j.bbrc.2006.05.147. [DOI] [PubMed] [Google Scholar]
- 6.Sakai M, Okabe M, Yamasaki M, Tachibana H, Yamada K. Induction of apoptosis by tocotrienol in rat hepatoma dRLh-84 cells. Anticancer Res. 2004;24:1683–8. [PubMed] [Google Scholar]
- 7.Nesaretnam K, Stephen R, Dils R, Darbre P. Tocotrienols inhibit the growth of human breast cancer cells irrespective of estrogen receptor status. Lipids. 1998;33:461–9. doi: 10.1007/s11745-998-0229-3. [DOI] [PubMed] [Google Scholar]
- 8.Nesaretnam K, Dorasamy S, Darbre PD. Tocotrienols inhibit growth of ZR-75-1 breast cancer cells. Int J Food Sci Nutr. 2000;51 (Suppl):S95–103. [PubMed] [Google Scholar]
- 9.Birringer M, Pfluger P, Kluth D, Landes N, Brigelius-Flohe R. Identities and differences in the metabolism of tocotrienols and tocopherols in HepG2 cells. J Nutr. 2002;132:3113–8. doi: 10.1093/jn/131.10.3113. [DOI] [PubMed] [Google Scholar]
- 10.Nesaretnam K, Ambra R, Selvaduray KR, Radhakrishnan A, Reimann K, Razak G, et al. Tocotrienol-rich fraction from palm oil affects gene expression in tumors resulting from MCF-7 cell inoculation in athymic mice. Lipids. 2004;39:459–67. doi: 10.1007/s11745-004-1251-1. [DOI] [PubMed] [Google Scholar]
- 11.Komiyama K, Iizuka K, Yamaoka M, Watanabe H, Tsuchiya N, Umezawa I. Studies on the biological activity of tocotrienols. Chem Pharm Bull (Tokyo) 1989;37:1369–71. doi: 10.1248/cpb.37.1369. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Loo G. Disruption of mitochondria during tocotrienol-induced apoptosis in MDA-MB-231 human breast cancer cells. Biochem Pharmacol. 2004;67:315–24. doi: 10.1016/j.bcp.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Shah S, Sylvester PW. Tocotrienol-induced caspase-8 activation is unrelated to death receptor apoptotic signaling in neoplastic mammary epithelial cells. Exp Biol Med (Maywood) 2004;229:745–55. doi: 10.1177/153537020422900806. [DOI] [PubMed] [Google Scholar]
- 14.Galli F, Stabile AM, Betti M, Conte C, Pistilli A, Rende M, et al. The effect of alpha- and gamma-tocopherol and their carboxyethyl hydroxychroman metabolites on prostate cancer cell proliferation. Arch Biochem Biophys. 2004;423:97–102. doi: 10.1016/j.abb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Sylvester PW, Shah S. Intracellular mechanisms mediating tocotrienol-induced apoptosis in neoplastic mammary epithelial cells. Asia Pac J Clin Nutr. 2005;14:366–73. [PubMed] [Google Scholar]
- 16.Shah SJ, Sylvester PW. Gamma-tocotrienol inhibits neoplastic mammary epithelial cell proliferation by decreasing Akt and nuclear factor kappaB activity. Exp Biol Med (Maywood) 2005;230:235–41. doi: 10.1177/153537020523000402. [DOI] [PubMed] [Google Scholar]
- 17.Agarwal MK, Agarwal ML, Athar M, Gupta S. Tocotrienol-rich fraction of palm oil activates p53, modulates Bax/Bcl2 ratio and induces apoptosis independent of cell cycle association. Cell Cycle. 2004;3:205–11. doi: 10.4161/cc.3.2.654. [DOI] [PubMed] [Google Scholar]
- 18.Yu FL, Gapor A, Bender W. Evidence for the preventive effect of the polyunsaturated phytol side chain in tocotrienols on 17beta-estradiol epoxidation. Cancer Detect Prev. 2005;29:383–8. doi: 10.1016/j.cdp.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Yap WN, Zaiden N, Luk SY, Lee DT, Ling MT, Wong YC, et al. In vivo evidence of gamma-tocotrienol as a chemosensitizer in the treatment of hormone-refractory prostate cancer. Pharmacology. 2010;85:248–58. doi: 10.1159/000278205. [DOI] [PubMed] [Google Scholar]
- 20.Yap WN, Chang PN, Han HY, Lee DT, Ling MT, Wong YC, et al. Gamma-tocotrienol suppresses prostate cancer cell proliferation and invasion through multiple-signalling pathways. Br J Cancer. 2008;99:1832–41. doi: 10.1038/sj.bjc.6604763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar D, Whiteside TL, Kasid U. Identification of a novel tumor necrosis factor-alpha-inducible gene, SCC-S2, containing the consensus sequence of a death effector domain of fas-associated death domain-like interleukin- 1beta-converting enzyme-inhibitory protein. J Biol Chem. 2000;275:2973–8. doi: 10.1074/jbc.275.4.2973. [DOI] [PubMed] [Google Scholar]
- 22.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix Gene Chip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–55. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 1995:57, 289–300. [Google Scholar]
- 26.Kumar D, Sakabe I, Patel S, Zhang Y, Ahmad I, Gehan EA, et al. SCC-112, a novel cell cycle-regulated molecule, exhibits reduced expression in human renal carcinomas. Gene. 2004;328:187–96. doi: 10.1016/j.gene.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Yan C, Jamaluddin MS, Aggarwal B, Myers J, Boyd DD. Gene expression profiling identifies activating transcription factor 3 as a novel contributor to the proapoptotic effect of curcumin. Mol Cancer Ther. 2005;4:233–41. [PubMed] [Google Scholar]
- 28.Sen CK, Khanna S, Rink C, Roy S. Tocotrienols: the emerging face of natural vitamin E. Vitam Horm. 2007;76:203–61. doi: 10.1016/S0083-6729(07)76008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu SJ, Ng LT. Tocotrienols Inhibited Growth and Induced Apoptosis in Human HeLa Cells Through the Cell Cycle Signaling Pathway. Integr Cancer Ther. 2010 doi: 10.1177/1534735409357757. [DOI] [PubMed] [Google Scholar]
- 30.Wada S. Chemoprevention of tocotrienols: the mechanism of antiproliferative effects. Forum Nutr. 2009;61:204–16. doi: 10.1159/000212752. [DOI] [PubMed] [Google Scholar]
- 31.Hussein D, Mo H. d-Dlta-tocotrienol-mediated suppression of the proliferation of human PANC-1, MIA PaCa-2, and BxPC-3 pancreatic carcinoma cells. Pancreas. 2009;38:e124–e36. doi: 10.1097/MPA.0b013e3181a20f9c. [DOI] [PubMed] [Google Scholar]
- 32.Sun W, Xu W, Liu H, Liu J, Wang Q, Zhou J, et al. gamma-Tocotrienol induces mitochondria-mediated apoptosis in human gastric adenocarcinoma SGC-7901 cells. J Nutr Biochem. 2009;20:276–84. doi: 10.1016/j.jnutbio.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Elangovan S, Hsieh TC, Wu JM. Growth inhibition of human MDA-mB-231 breast cancer cells by delta-tocotrienol is associated with loss of cyclin D1/CDK4 expression and accompanying changes in the state of phosphorylation of the retinoblastoma tumor suppressor gene product. Anticancer Res. 2008;28:2641–7. [PubMed] [Google Scholar]
- 34.Bai J, Nakamura H, Ueda S, Kwon YW, Tanaka T, Ban S, et al. Proteasome-dependent degradation of cyclin D1 in 1-methyl-4-phenylpyridinium ion (MPP+)-induced cell cycle arrest. J Biol Chem. 2004;279:38710–4. doi: 10.1074/jbc.M403329200. [DOI] [PubMed] [Google Scholar]
- 35.Das M, Das S, Wang P, Powell SR, Das DK. Caveolin and proteasome in tocotrienol mediated myocardial protection. Cell Physiol Biochem. 2008;22:287–94. doi: 10.1159/000149807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–39. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 37.Nesaretnam K, Ambra R, Selvaduray KR, Radhakrishnan A, Canali R, Virgili F. Tocotrienol-rich fraction from palm oil and gene expression in human breast cancer cells. Ann NY Acad Sci. 2004;1031:143–57. doi: 10.1196/annals.1331.014. [DOI] [PubMed] [Google Scholar]
- 38.Wali VB, Bachawal SV, Sylvester PW. Endoplasmic reticulum stress mediates gamma-tocotrienol-induced apoptosis in mammary tumor cells. Apoptosis. 2009;14:1366–77. doi: 10.1007/s10495-009-0406-y. [DOI] [PubMed] [Google Scholar]
- 39.Park SK, Sanders BG, Kline K. Tocotrienols induce apoptosis in breast cancer cell lines via an endoplasmic reticulum stress-dependent increase in extrinsic death receptor signaling. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-0786-2. [DOI] [PubMed] [Google Scholar]