Abstract
Fast and sensitive virus detection techniques, which can be rapidly deployed at multiple sites, are essential to prevent and control the future epidemics and bioterrorism threats. In this letter, we demonstrate a label-free optofluidic-nanoplasmonic sensor that can directly detect intact viruses from biological media at clinically relevant concentrations with little to no sample preparation. Our sensing platform is based on extraordinary light transmission effect in plasmonic nanoholes and utilizes group specific antibodies for highly divergent strains of rapidly evolving viruses. So far, the questions remain for the possible limitations of this technique for virus detection, as the penetration depths of the surface plasmon polaritons (SPP) are comparable to the dimensions of the pathogens. Here, we demonstrate detection and recognition of small enveloped RNA viruses (vesicular stomatitis virus and pseudo-typed Ebola) as well as large enveloped DNA viruses (vaccinia virus) within a dynamic range spanning three orders of magnitude. Our platform, by enabling high signal noise measurements without any mechanical or optical isolation, opens up opportunities for detection of a broad range of pathogens in typical biology laboratory settings.
Keywords: Biosensing, Plasmonics, Virus Detection, Vaccinia, Ebola, Vesicular Stomatitis Virus
Early detection of infectious viral diseases is a serious public health, homeland security, and armed forces issue. A number of recent outbreaks of viral diseases (e.g. H1N1 flu, H5N1 flu and SARS) in recent years have raised significant fears that such viruses could rapidly spread and turn into a pandemic similar to 1918 Spanish flu that killed more than 50 million people1. A critical aspect of recognizing and controlling future epidemics will be the development of rapid and sensitive diagnostic techniques that can be rapidly deployed at multiple sites2. Traditional detection methods such as cell culturing, enzyme-linked immunosorbant assays (ELISA), and polymerase chain reaction (PCR). are not readily compatible with point-of-care use without the existence of entensive infrastructure3,4. Cell culturing is a time consuming, highly specialized and labor intensive process. In some cases, viruses cannot be cultured at all5. ELISA technique require multiple steps and agents with a potential to create quenching interactions among each other6. PCR, another powerful diagnostic tool based on detection of nucleic fragments in clinical samples, requires significant sample preparation, and can be confounded by inhibitors within a clinical sample7. PCR also provides only an indirect test of the infection8–10. Viral nucleic acid fragments can be present in the host organism after the infection has been “cleared” or effectively neutralized8–10. In addition, while PCR is a robust and accurate technique in detecting known strains, it may not be adaptable to newly emerged or highly divergent strains of an infections agent. An example of this is the recent description of a new strain of Ebola that was not identified in PCR-based diagnostics11. Therefore, highly sensitive/specific, compact, fast and easy to use virus diagnostics are needed to prevent further spread at the onset of a viral epidemic.
Label free biosensors have recently emerged as promising diagnostic tools for cancer and infectious diseases12–24. These sensors circumvent the need for fluorescence/radio-active tagging or enzymatic detection, and enable compact, simple, inexpensive point-of-care diagnostics. Various sensing platforms based on optical12–17, electrical22,23 and mechanical18–21signal transduction mechanisms have been offered for applications ranging from laboratory research to clinical diagnostics and drug development to combating bioterrorism. Among these sensing platforms, optical detection platforms are particularly promising. Optical biosensors allow remote transduction of the biomolecular binding signal from the sensing volume without any physical connection between the excitation source and the detection channel25,26. Unlike mechanical and electrical sensors, they are also compatible with physiological solutions and are not sensitive to the changes in the ionic strengths of the solutions27,28. However, a drawback of the most currently-used optical biosensors is that they require precise alignment of light coupling to the biodetection volume15–17,24. As a result, these technologies are not particularly suitable for point-of-care applications. Nanoplasmonic biosensors are distinctive among photonic sensors as they allow direct coupling of the perpendicularly incident light and constitute a robust sensing platform minimizing the alignment requirements for light coupling12–14,29–32. This capability also opens up opportunities for multiplexed detection29. In addition, the extraordinary transmission (EOT) signals in plasmonic nanohole arrays create an excellent detection window enabling spectral measurements with minimal background noise and high signal-to-noise ratios33–35. In a recent work, we have demonstrated a novel approach combining nanofluidics and plasmonic sensing in a single platform enabling both the resonant transmission of light and the active transport of fluidics through the holes35. With this newly developed hybrid nanohole sensor, we achieved higher sensitivities and faster sensor response times as a result of our lift-off free nanofabrication technique in combination with the targeted analyte delivery scheme to the sensing surface35–37. To our knowledge, nanohole based sensing platforms have not been used for the detection of intact viruses in the past. Questions remain for the possible limitations of the technique as the penetration depths of the surface plasmon polaritons (SPP) are comparable to the dimensions of the viruses38–40.
In this letter, we demonstrate that the optofluidic-nanoplasmonic sensors enable direct detection of intact viruses from biologically relevant media in a label free fashion with little to no sample preparation. Our interests are mainly focused in detection and recognition of small, enveloped RNA viruses. As a family, viruses that utilize RNA as their genetic material make up almost all of the alarming new infectious diseases (Category A, B, and C biothreats) and are a large component of the existing viral threats (influenza, rhinovirus, etc). Some of these viruses, e.g. the Ebola hemorrhagic fever virus are both emerging infectious and biological threat agent 41,42. Patients presenting with RNA virus infections often show symptoms that are not virus specific43. Thus, there is great interest in developing sensitive and rapid diagnostics for these viruses to help direct proper treatment. Our sensing platform uses antiviral immunoglobulins immobilized at the sensor surface for specific capturing of the virions. Unlike PCR, our scheme allows us to take advantage of group specific antibodies, which have historically been able to identify a broad range of known and even previously unknown pathogens (i.e. novel mutant strains)11,44. In addition, our detection platform is capable of quantifying virus concentrations. Such quantitative detection makes it possible to detect not only the presence of the intact viruses in the analyzed samples, but also the intensity of the infection process. A dynamic range spanning three orders of magnitude from 106 PFU/ml to 109 PFU/ml is shown in experimental measurements proving that our platform enables label free virus detection within a concentration window ranging from clinical testing to drug screening. We also extended our studies to show the suitability of this technology for enveloped DNA viruses (vaccinia virus)45. Another advantage of this platform is that due to the non-destructive nature of the detection scheme, captured virions and their nucleic acid load (genome) can be used in further studies46. In this study, experiments are performed in ordinary biosafety level 1 and 2 laboratory settings without any need for mechanical or light isolation as it is expected from any practical technology. This technology, enabling fast and compact sensing of the intact viruses, could play an important role in early and point-of-care detection of viruses in clinical settings as well as in biodefense contexts.
Device Operation Principles
The detection scheme based on our optofluidic-nanoplasmonic sensor is illustrated in Fig. 1a–b. The device consists of a suspended nanohole array grating that couples the normally incident light to surface plasmons, electromagnetic waves trapped at metal/dielectric interface in coherence with collective electron oscillations35,47–49. The extraordinary light transmission resonances are observed at specific wavelengths, λres approximated by 50–53:
| (1) |
where the grating coupling enables the excitation of the surface plasmons (Fig. 1c–d). Here, a0 is the periodicity of the array and i, j are the grating orders. This resonance wavelength is strongly correlated with the effective dielectric constant of the adjacent medium around the plasmonic sensor51,52. As biomolecules/pathogens bind to the metal surface or to the ligands immobilized on the metal surface, the effective refractive index of the medium increases, and the red-shifting of the plasmonic resonance occurs54. Unlike techniques based on external labeling, such resonance shifting operate as a reporter of the molecular binding phenomena in a label free fashion and enables transduction of the capturing event directly to the far field optical signal 55–57. Exponential decay of the extent of the plasmonic excitation results in subwavelength confinement of the electromagnetic field to the metal/dielectric interface58. As a result, the sensitivity of the biosensor to the refractive index changes decreases drastically with the increasing distance from the surface, thereby minimizing the effects of refractive index variations due to the temperature fluctuations in the bulk medium58.
Fig. 1d, demonstrates a typical set of experimental end-point measurements for selective detection of vesicular stomatitis virus (VSV) at a concentration of 109 PFU/ml. Here, the transmission light spectra are acquired from an optofluidic-nanohole array of 90 µm × 90 µm with a periodicity of 600 nm and an aperture radius of 220 nm. Spectra are given for both before (blue curve) and after (red curve) the incubation of the virus containing sample. The sharp resonance feature observed at 690 nm (blue curve) with 25 nm full width at half maximum (FWHM) is due to the extraordinary light transmission phenomena through the optically thick gold film. This transmission resonance (blue curve) corresponds to the excitation of the (1,0) grating order SPP mode at the metal/dielectric interface of the antibody immobilized detection sensor50. After the incubation process (enabling the diffusive delivery of analytes) with the virus containing sample, a strong red-shifting (~100nm) of the plasmonic resonance peak is observed (red curve) in end point measurements. This red-shifting is related to the accumulated biomass on the functionalized sensing surface. Such a strong resonance shift results in a color change of the transmitted light, which is large enough to discern visually without a spectrometer. For the unfunctionalized control sensors (Fig. 1c), a negligible red-shifting (~1 nm) of the resonances is observed (blue vs red curves), possibly due to the non-specific binding events. This measurement clearly demonstrates that optofluidic biosensors are promising candidates for specific detection of viruses. At lower concentrations of viruses (<108 PFU/ml) spectral shifts are more modest and require spectral measurements. However, considering that concentrations of certain types of viruses in infected samples reaches to the concentrations comparable to our visual detection limit, our platform offers unique opportunities for the development of rapid point-of-care diagnostics59.
Device Fabrication
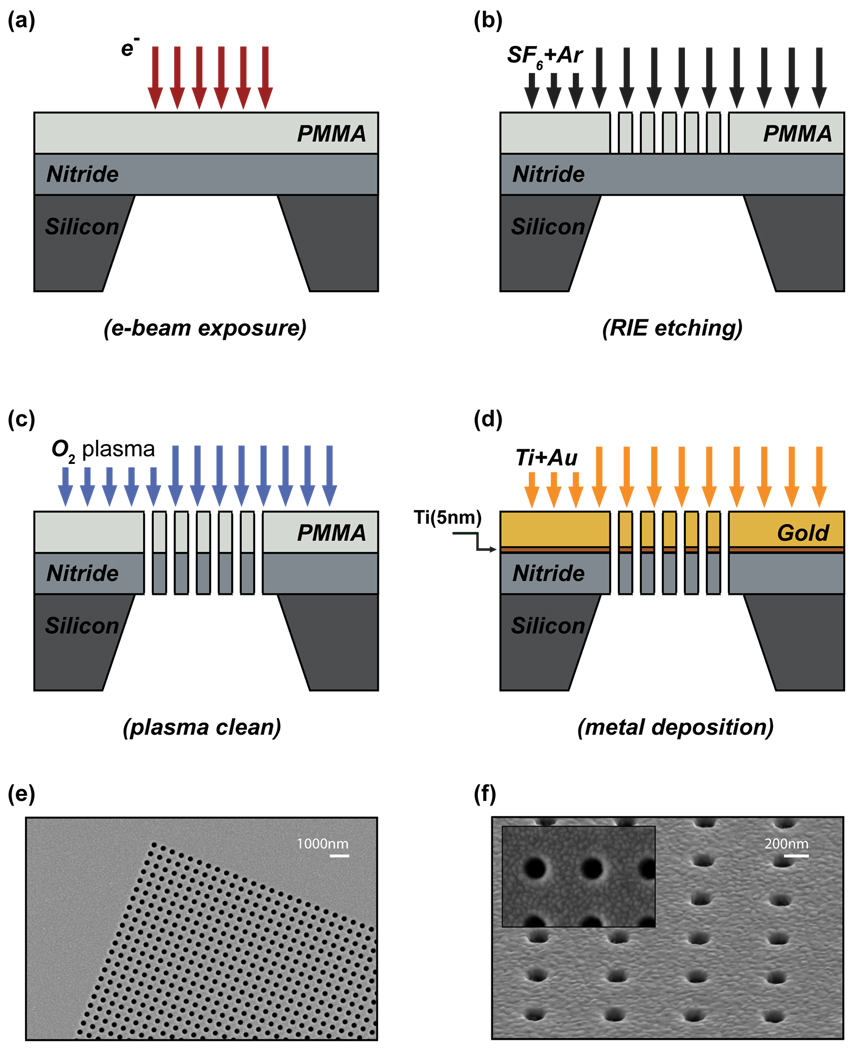
A lift-off free nanofabrication technique, based on positive resist e-beam lithography and direct deposition of metallic layers, was developed to fabricate optofluidic-plasmonic biosensors35. This scheme eliminates the need for lift-off processes as well as operationally slow focused ion beam lithography, which introduces optically active ions. As a result, we achieved high quality plasmonic resonances (15–20 nm FWHM) and high figures of merit (FOM~40) for refractive index sensitivities, which is defined as shift per refractive index unit (RUI) divided by the width of the surface plasmon resonances in energy units35. The fabrication scheme is summarized in Fig. 2. Initially, free standing SiNx membranes are created using a series of photolithographic and chemical wet etching (KOH) processes (not shown)60. The membranes are then covered with positive e-beam resist poly(methyl methacrylate) (PMMA) and e-beam lithography is performed to define the nanohole pattern in the resist (Fig. 2a). This pattern is transferred to the SiNx membrane through a reactive ion etching process (Fig. 2b). After the removal of the resist with an oxygen plasma etching process (Fig. 2c), a photonic crystal like free standing SiNx membrane is defined. Sequential deposition of the metal layers (5 nm Ti, 100 nm Au) results in a free standing plasmonic nanoholes transmitting light at resonance (Fig. 2d) 35. As demonstrated repeatedly in the experiments, this scheme allows fabrication of metallic nanohole arrays without clogging the openings with extremely high yield/reproducibility and with minimal surface roughness (Fig. 2e–f)35.
Virus preparation
VSV and virus pseudotypes. Baby hamster kidney (BHK) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 7% fetal bovine serum and 2 mM glutamine. Cells were grown to 85–95% confluence and then infected with VSV (Indiana serotype, Orsay strain) in DMEM at a low multiplicity of infection (MOI=0.01). 24 hours postinfection (hpi), media was harvested and virus titer was determined by plaque assay. We also used VSV that was engineered to lack the VSV glycoprotein but contain the gene for the Ebola glycoprotein. The resulting virus expresses the glycoprotein from Ebola Zaire and incorporates this protein as its envelope glycoprotein, a process known as pseudotyping. The VSV-pseudotyped with Ebola GP (PT-Ebola) was grown in a similar fashion to that described for wild-type VSV, but media was harvested at 48 hpi. Purified virus was obtained through sedimentation of virus at 100,000XG for 1 hour followed by resuspension in PBS or 10mM Tris pH 8.0. Resuspended virus was checked for purity by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Blue staining, aliquoted and stored at −80°C. Vaccinia virus. A549 cells were cultured in medium described above. Cells were infected with Vaccinia (Western Reserve strain) in DMEM at an MOI=0.01. 24 hpi media was harvested and virus titers were determined via plaque assay. Aliquots were stored at −80°C.
Antibodies
8G5 antibodies targeting the single external VSV glycoprotein were a kind gift of Douglas S. Lyles (Wake Forest). Antibodies were obtained from hybridoma supernatants. Purification of 8G5 antibodies from hybridoma supernatents was accomplished by protein A purification. M-DA01-A5 antibodies targeting the Ebola glycoprotein were a kind gift of Lisa Hensley (The United States Army Medical Research Institute of Infectious Diseases-USAMRIID). A33L antibodies against Vaccinia virus were a kind gift of Jay Hooper (USAMRIID).
Surface Functionalization
Surfaces are immobilized with protein A/G (Pierce, IL) at a concentration of 1mg/ml in PBS (10 mM phosphate buffer, 137 mM NaCl and 2.7 ml KCl)61. Protein A/G is chosen as a template for the immobilization of the virus specific antibodies due to its high affinity to the Fc region of the IgG molecules62,63. Protein-AG is a recombinant fusion protein that contains the four Fc binding domains of protein A and two of the Protein G. Unlike protein A, the binding of chimeric protein A/G is less dependent upon the pH. The elimination of the non-specific binding regions to the serum proteins (including albumin) makes it an excellent choice for immobilization of the immunoglobulins. Proper orientation of the antibodies is imposed by this template (Fig. 3a)63.
Antibody Immobilization
Specific detection of viruses in a label free fashion requires an effective method to distinguish non-specific binding of the viruses to the optofluidic-plasmonic sensor surface. Selectivity is achieved by surface immobilized highly specific antiviral immunoglobulins showing strong affinity to the viral membrane proteins, called glycoproteins (GP)64. GPs are presented on the outside of the assembled virus membrane and bind to receptors on the host cell membrane in order to enter into the cell. Antibodies that recognize the VSV-GP (8G5)65,66, that recognize Ebola-GP (M-DA01-A5) and that recognize Vaccinia-GP (A33L) 67 were spotted on an array of sensors at a concentration of 0.5 mg/ml in PBS (Fig. 3a). The sensitivity of any immunoassay is highly dependent on the spotting of the antibodies. Higher concentrations of antiviral antibodies with respect to the virion concentrations are needed [virion] < [IgG], so that the spectral shift is proportional to the concentration of the virions instead of being limited by the antiviral immunoglobulin concentration68. After a 60 min of incubation, unbound antibody was removed by a three step post-incubation washing process. No blocking agent was needed to block the antibody-free protein A/G surface, since the viruses do not directly bind to the protein A/G functionalized surface61.
The successful functionalization of the sensing surface is monitored with end-point measurements after each incubation and washing processes. As shown in Fig. 3b, the accumulated biomass on the sensing surface results in red-shifting of the air (1,0) resonance (black curve) due to the increasing local refractive index at the metal/dielectric of interface of the nanoplasmonic biosensor. Initially, a red shifting for about 4 nm was observed (blue curve), after the protein A/G funtionalization. Protein A/G template is then used to immobilize the 8G5-VSV specific antibodies at a concentration of 0.5 mg/ml. A spectral shift of 14 nm (red curve) is observed after the antibody immobilization, confirming the successful functionalization of the surface.
Reference sensors
Reference sensors were incorporated into the chip design to correct for any drift and noise signal due to the unexpected changes in the measurement conditions or nonspecific binding events. Two different types of spotting conditions, one functionalized with protein A/G only and one without any functionalized biomolecules, were tested to determine the optimum configuration for the reference sensors. For the control sensors functionalized with protein-A/G, it was observed that after the introduction of the antibodies to the detection sensor, a red-shifting of the resonance is observed. This observation is associated to the relocation of the anti-viral immunoglobulins, during the washing processes, from antibody immobilized detection spots to the protein A/G immobilized control spots as a result of the high affinity of the protein A/G to the IgG antibodies. On the other hand, for the reference sensors with no protein A/G layer, red shifting of the resonance after the introduction of the viruses was minimal. Accordingly, unfunctionalized nanohole sensors were used for reference measurements.
PT-Ebola and Vaccinia Virus Detection
For broad adaptability of our platform, detection of hemorrhagic fever viruses (e.g. Ebola virus) and poxviruses (e.g. monkeypox or variola, the causative agent of smallpox) is an important test. These viruses are of particular interest to public health and national security41,69. Though we were not able to directly test these viruses because of biosafety considerations, we use a genetically derived VSV-pseudotyped Ebola (PT-Ebola), where the Ebola glycoproteins are expressed on the virus membrane instead of the VSV’s own glycoprotein70. PT-Ebola is a viable surrogate to analyze the behavior of Ebola, since the expressed glycoprotein folds properly and is fusion competent. The pseudotyped viruses have been successfully used as vaccine against Ebola in nonhuman primate models and can be used at lower biosafety levels (BSL2 versus BSL4).
For these experiments, antibodies against the Ebola glycoprotein were immobilized on the 9 of 12 sensors on a single chip, while 3 sensors were reserved for reference measurements. Successful functionalization of the protein-A/G and the antibodies were confirmed by spectral measurements (Fig. 4a). Following the immobilization of the antibodies, PT-Ebola (at a concentration of 108 PFU/ml) in a PBS buffer solution) was added onto the chips and incubated for 90 min. After the washing process, transmission spectra were collected (Fig. 4a). Consistent red-shifting of the plasmonic resonances were observed on antibody-coated spots indicating PT-Ebola detection (≥ 14nm red shift), while reference sensors showed no spectral shift (red bars, Fig. 4b). This occurred with high repeatability (9 of 9 sensors) and excellent signal-to-noise ratios. Similarly, we tested our platform for the detection of enveloped DNA poxviruses. To do this, we utilized Vaccinia virus, a poxvirus that is commonly used as a prototype for more pathogenic viruses such as smallpox and monkeypox71. A similar approach (A33L Vaccinia antibody to the external protein immobilized on 9 of 12 sensors, incubation with intact vaccinia virus at the same concentration of 108 PFU/ml) yielded similar positive results to those seen with Ebo-VSV (Fig. 4c). All of the 9 sensors detected the virus, while all of the reference sensors indicated minimal binding (Fig. 4d).
For sensors close to the spotted sample edges, both weaker (8 nm in the case of Vaccinia virus) and stronger (20–21 nm in the case of pseudo-Ebola virus) spectral shifts were observed. This is related to the non-uniform virus concentrations around the spot edge. Measurements obtained from multiple sensors improved the robustness of the assay. Repeatability of the measurements was readily observed; all functionalized nanohole sensors showed a consistent shift ranging from 14–21 nm (Fig. 4b,4d). This observation shows a clear quantitative relation between the spectral shifts and virus concentrations. Such quantification is not possible with techniques based on fluorescent labeling (ELISA). Although Vaccina viruses have relatively larger dimensions than the PT-Ebola viruses, comparable spectral shifts are observed. This observation clearly indicates that the capturing efficiency of the viruses, thus the accumulated biomass, is not only controlled by the concentrations of the virions but also controlled by the affinity of the virus-IgG interactions72. Without doubt, strength of such interactions is strongly affected by the complex mixture of the envelope proteins and the surroundings of the viral subunits72,73. In fact, the structure and the conformation state of the membrane incorporated glycoproteins may strongly differ from those of the purified ones72. Accordingly, techniques based on detection of recombinant and refined virus specific proteins or viral peptides are not suitable for medical studies of in-vivo behavior of live viruses. Instead, techniques enabling direct detection of entire viral particles in medically relevant biological media are needed. While most studies in this field are confined to detection of individual viral components such as glycoproteins and nucleic acids, we demonstrate that our detection platform enables direct detection of intact viruses73,74.
Virus Detection in Biological Media
To demonstrate the applicability of our detection platform in biologically relevant systems, we extended our experiments to the detection of intact viruses directly from biological media (cell growth medium + 7% fetal calf serum). These conditions provide a number of potentially confounding factors (high serum albumin levels, immunoglobulins and growth factors) that could add unwanted background signal, thus this was an important test for the robustness of our detection system. In Fig. 5, it is shown that the initial Pr-AG functionalization (1 mg/ml) resulted in 4 nm red shifting of the resonances. Subsequently, anti-VSV (0.5 mg/ml) immobilization was confirmed with the ~15 nm red shifting of the resonances. Finally, VSV was applied to the chips at a concentration of 106 PFU/ml in a DMEM/FBS medium. Measurements, following an incubation period of 90 min and post washing processes, showed a 4 nm resonance shift for the anti-viral immunoglobin functionalized spots. In reference sensors, red-shifting of the resonances was seen, but was limited to only 1.3 nm due to the non-specific binding of the serum proteins. The specific capturing of the intact viruses at a low concentration of 106 PFU/ml is clearly distinguishable at the antibody functionalized sensors. This observation demonstrates the potential of this platform for clinical applications. Due to our ability to quantify non-specific binding on an individual chip, the presence of a small amount of background does not pose a fundamental bottleneck for the viability of this technology. In fact, this technology is sufficient for microbiology laboratories involving culturing of the viruses. In addition, it is likely that the technology can be adapted “as is” for successful diagnosis of herpesvirus, poxvirus and some gastroenteric infections, since a detection limit of 107–108 PFU/ml is usually sufficient for clinical applications59. Given that the resolution limit of detection system is 0.05 nm, it is likely that much lower concentrations (<105 PFU/ml) can be detected with the current platform. Background shifting due to the non-specific binding could be a problem at lower concentrations of analytes, however this limitation can be considerably reduced and significant improvements in detection limits of the devices can be achieved by optimizing the surface chemistry.
Conclusion
The present study provides proof-of-concept biosensing platform for fast, compact, quantitative and label free sensing of viral particles with minimal sample processing. We demonstrate that the extraordinary light transmission phenomena on plasmonic nanohole array can be adapted for pathogen detection without being confounded by the surrounding biological media. The sensing platform uses antiviral immunoglobulins immobilized at the sensor surface for specific capturing of the intact virions and is capable of quantifying their concentrations. Direct detection of different types of viruses (VSV, PT-Ebola and Vaccinia) are shown. A dynamic range spanning three orders of magnitude from 106 PFU/ml to 109 PFU/ml is shown in experimental measurements corresponding to virion concentration within a window relevant to clinical testing to drug screening. Moreover, projected low detection limits (<105 PFU/ml) of the viruses in biologically relevant media clearly demonstrates the feasibility of the technology for earlier diagnosis of viruses directly from the human blood. It is important to note that the ease of multiplexing afforded by this approach is a crucial aspect of the biosensor design. The optofluidic-plasmonic sensors can be readily expanded into a multiplexed format, where the various viral antibodies are immobilized at different locations to selectively detect the pathogens in an unknown sample. The advantage of the optofluidic-plasmonic sensor is its ability to detect intact virus particles and identify them without damaging the virus structure or the nucleic acid load (gnome), so that the samples could be further studied46.The proposed approach opens up biosensing applications of extraordinary light transmission phenomena for a broad range of pathogens, and can be directly utilized in typical biology laboratory settings.
ACKNOWLEDGMENTS
H. Altug and J. H. Connor would like to acknowledge Peter Paul Career Award for providing support. A. A. Yanik, J. H. Connor and H. Altug also acknowledge Helen Fawcett for fruitful discussions. Authors extend their gratitude to Douglas S. Lyles (Wake Forest) for providing 8G5 antibody to recognize VSV, to Lisa Hensley (The United States Army Medical Research Institute of Infectious Diseases) for providing Ebola M-DA01-A5 glycoprotein antibodies, and Jay Hooper for providing antibody against Vaccinia virus (A33L) (The United States Army Medical Research Institute of Infectious Diseases). H. Altug gratefully acknowledges support from NSF CAREER Award (ECCS-0954790), ONR Young Investigator Award, Massachusetts Life Science Center New Investigator Award, NSF Engineering Research Center on Smart Lighting (EEC-0812056), Boston University Photonics Center and Army Research Laboratory. JHC acknowledges support from the NIH grant AI064606. Authors thank Mike Ekins for the illustrations.
REFERENCES
- 1.Potter CW. J. Appl Microbiol. 2001;91:572–579. doi: 10.1046/j.1365-2672.2001.01492.x. [DOI] [PubMed] [Google Scholar]
- 2.Tseng D, Mudanyali O, Oztoprak C, Isikman SO, Sencan I, Yaglidere O, Ozcan A. Lab Chip. 2010;10:1787–1792. doi: 10.1039/c003477k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henkel JH, Aberle SW, Kundi M, Popow-Kraupp T. J. Med. Virol. 1997;53:366–371. [PubMed] [Google Scholar]
- 4.Bao PD, Huang TQ, Liu XM, Wu TQJ. Raman Spectrosc. 2001;32:227–230. [Google Scholar]
- 5.Karst SM. Viruses. 2010;2:748–781. doi: 10.3390/v2030748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones LJ, Upson RH, Haugland N, Panchuk-Voloshina M, Zhou J, Haugland RP. Anal. Biochem. 1999;271:119–120. doi: 10.1006/abio.1997.2259. [DOI] [PubMed] [Google Scholar]
- 7.Drosten C, Panning M, Guenther S, Schmitz H. J Clin Microbiol. 2002;40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaydos CA, Crotchfelt KA, Howell MR, Kralian S, Hauptman P, Quinn TC. J. Infect. Dis. 1998;117:417. doi: 10.1086/514207. [DOI] [PubMed] [Google Scholar]
- 9.Liebert U. Intervirology. 1997;40:176. doi: 10.1159/000150544. [DOI] [PubMed] [Google Scholar]
- 10.Schneider-Schaulies J, Meulen V, Schneider-Schaulies S. J. Neurovirol. 2003;9:247. doi: 10.1080/13550280390193993. [DOI] [PubMed] [Google Scholar]
- 11.Towner JS, Sealy TK, Khristova ML, Albarino CG, Conlan S, Reeder SA, Quan PL, Lipkin WI, Downing R, Tappero JW, Okware S, Lutwama J, Bakamutumaho B, Kayiwa J, Comer JA, Rollin PE, Ksiazek TG, Nichol ST. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surbhi L, Stephan L, Naomi JH. Nature Photonics. 2007;1:641–648. [Google Scholar]
- 13.Sandra B, Carly SL, Muhammed G, Bruce J, Rozell C, Johnson DH, Naomi JH. Nano Letters. 2006;6:1687–1692. doi: 10.1021/nl060865w. [DOI] [PubMed] [Google Scholar]
- 14.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Nature Mater. 2008;7:442. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 15.Homola J. Anal. Bioanal. Chem. 2003;377:528–539. doi: 10.1007/s00216-003-2101-0. [DOI] [PubMed] [Google Scholar]
- 16.Arnold S, Khoshsima M, Teraoka I, Holler S, Vollmer F. Opt. Lett. 2003;28:272–327. doi: 10.1364/ol.28.000272. [DOI] [PubMed] [Google Scholar]
- 17.Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Science. 2007;317:783–787. doi: 10.1126/science.1145002. [DOI] [PubMed] [Google Scholar]
- 18.Savran CA, Knudsen SM, Ellington AD, Manalis SR. Analytical Chemistry. 2004;76:3194–3198. doi: 10.1021/ac049859f. [DOI] [PubMed] [Google Scholar]
- 19.Joonhyung L, Jaesung J, Demir A, Cagri AS, Bashir R. Appl Phys Lett. 2008;93 013901. [Google Scholar]
- 20.Fritz J, Baller MK, Lang HP, Rothuizen H, Vettiger P, Meyer E, Güntherodt H, Gerber C, Gimzewski JK. Science. 2000;288:316–318. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 21.Savran CA, Knudson SM, Ellington AD, Manalis SR. Analytical Chemistry. 2004;76:3194. doi: 10.1021/ac049859f. [DOI] [PubMed] [Google Scholar]
- 22.Cui Y, Wei QQ, Park HK, Lieber CM. Science. 2001;293:1289–1292. doi: 10.1126/science.1062711. [DOI] [PubMed] [Google Scholar]
- 23.Yuri LB, Young SS, Woon-Seok Y, Michael A, Gabriel K, James RH. J. Am. Chem. Soc. 2006;128:16323–16331. [Google Scholar]
- 24.Terrel M, Digonnet MJF, Fan S. Applied Optics. 2009;48:4874–4879. doi: 10.1364/AO.48.004874. [DOI] [PubMed] [Google Scholar]
- 25.Pineda MF, Chan LL-YChan, Kuhlenschmidt T, Choi CJ, Kuhlenschmidt M, Cunningham BT. IEEE Sensors Journal. 2009;9:470–477. [Google Scholar]
- 26.Lee J, Icoz K, Roberts A, Ellington AD, Savran CA. Analytical Chemistry. 2010;82:197–202. doi: 10.1021/ac901716d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta AK, Nair PR, Akin D, Ladisch MR, Broyles S, Alam MA, Bashir R. Proc Natl Acad Sci USA. 2006;103:13362–13367. doi: 10.1073/pnas.0602022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern E, Wagner R, Sigworth FJ, Breaker R, Fahmy TM, Reed MA. Nano Lett. 2007;7:3405–3409. doi: 10.1021/nl071792z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J-C, Ji J, Hogle JM, Larson DN. Biosensors and Bioelectronics. 2009;24:2334–2338. doi: 10.1016/j.bios.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesuffleur A, Im H, Lindquist NC, Oh SH. Appl. Phys. Lett. 2007;90:243110. [Google Scholar]
- 31.Matthew E, Stewart N, Mack H, Malyarchuk V, Soares JANT, Lee T-W, Gray SK, Nuzzo RG, Rogers JA. Proc Natl Acad Sci USA. 2006;103:17143–17148. doi: 10.1073/pnas.0606216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brolo AG, Gordon R, Leathem B, Kavanagh KL. Langmuir. 2004;20:4813–4815. doi: 10.1021/la0493621. [DOI] [PubMed] [Google Scholar]
- 33.Yanik AA, Wang X, Erramilli S, Hong MK, Altug H. Appl. Phys. Lett. 2008;93 doi: 10.1063/1.4737942. 081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanik AA, Adato R, Erramilli S, Altug H. Optics Express. 2009;17:20900–20910. doi: 10.1364/OE.17.020900. [DOI] [PubMed] [Google Scholar]
- 35.Yanik AA, Huang M, Artar A, Chang T-Y, Altug H. Appl. Phys. Lett. 2010;96 021101. [Google Scholar]
- 36.Sheehan PE, Whitman LJ. Nano Lett. 2005;5:803. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 37.Squires TM, Messinger RJ, Manalis SR. Nat. Biotechnol. 2008;26:417. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 38.Liedberg B, Lundström I, Stenberg E. Sens. Actuators B. 1993;11:63–72. [Google Scholar]
- 39.Snopok BA, Kostyukevych KV, Rengevych OV, Shirshov YM, Venger EF, Kolesnikova IN, Lugovskoi EV. Semicond. Phys., Quantum Electron. Optoelectron. 1998;1:121. [Google Scholar]
- 40.Zia R, Selker MD, Brongersma ML. Phys. Rev. B. 2005;71:165431. [Google Scholar]
- 41.Bray M. Antiviral Res. 2003;57:53–60. doi: 10.1016/s0166-3542(02)00200-0. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki Y, Gojobori T. Molecular Biology and Evolution. 1997;14:800–806. doi: 10.1093/oxfordjournals.molbev.a025820. [DOI] [PubMed] [Google Scholar]
- 43.Drosten C, Kummerer BM, Schmitz H, Gunther S. Antiviral Research. 2003;57:61–87. doi: 10.1016/s0166-3542(02)00201-2. [DOI] [PubMed] [Google Scholar]
- 44.Yu JS, Liao HX, Gerdon AE, Huffman B, Scearce RM, McAdams M, Alam SM, Popernack PM, Sullivan NJ, Wright D, Cliffel DE, Nabel GJ, Haynes BF. J Virol Methods. 2006;137:219–228. doi: 10.1016/j.jviromet.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 45.Tucker JB. The Once and Future Threat of Smallpox. New York: Grove/Atlantic Inc.; 2001. [Google Scholar]
- 46.Kjeldsberg E. J Virol Methods. 1986;34:321–333. doi: 10.1016/0166-0934(86)90034-0. [DOI] [PubMed] [Google Scholar]
- 47.Catrysse PB, Fan S. Journal of Nanophotonics. 2008;2 021790. [Google Scholar]
- 48.Shvets G, Trendafilov S, Pendry JB, Sarychev A. Phys. Rev. Lett. 2007;99 doi: 10.1103/PhysRevLett.99.053903. 053903. [DOI] [PubMed] [Google Scholar]
- 49.Ekinci Y, Solak HH, David C. Optics Letters. 2007;32:172–174. doi: 10.1364/ol.32.000172. [DOI] [PubMed] [Google Scholar]
- 50.Ebbesen TW, Lezec HJ, Ghaemi HF, Thio T, Wolff PA. Nature. 1998;391:667. [Google Scholar]
- 51.Barnes WL, Murray WA, Dintinger J, Devaux E, Ebbesen TW. Phys. Rev. Lett. 2004;92:107401. doi: 10.1103/PhysRevLett.92.107401. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, Lalanne P. Nature. 2008;452:728. doi: 10.1038/nature06762. [DOI] [PubMed] [Google Scholar]
- 53.Artar A, Yanik AA, Altug H. Appl. Phys. Lett. 2009;95 051105. [Google Scholar]
- 54.Tetz K, Pang L, Fainman Y. Opt. Lett. 2006;31:1528. doi: 10.1364/ol.31.001528. [DOI] [PubMed] [Google Scholar]
- 55.Adato R, Yanik AA, Amsden JJ, Kaplan DL, Omenetto FG, Hong MK, Erramilli S, Altug H. Proc Natl Acad Sci USA. 2009;106:19227–19232. doi: 10.1073/pnas.0907459106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cubukcu E, Zhang S, Park Y-S, Bartal G, Zhang X. Appl. Phys. Lett. 2009;95 043113. [Google Scholar]
- 57.Cubukcu E, Degirmenci F, Kocabas C, Zimmler MA, Rogers JA, Capasso F. Proc Natl Acad Sci USA. 2009;106:8. doi: 10.1073/pnas.0812217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shumaker-Parry JS, Campbell CT. Anal. Chem. 2004;76:907–917. doi: 10.1021/ac034962a. [DOI] [PubMed] [Google Scholar]
- 59.Hazelton PR, Gelderblom HR. Emerg. Infec. Dis. 2003;9:294. doi: 10.3201/eid0903.020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang M, Yanik AA, Chang T-Y, Altug H. Optics Express. 2009;17:24224–24233. doi: 10.1364/OE.17.024224. [DOI] [PubMed] [Google Scholar]
- 61.Nettikadan SR, Johnson JC, Vengasandra SG, Muys J, Henderson E. Nanotechnology. 2004;15:383. [Google Scholar]
- 62.Eliasson M, Andersson R, Olsson A, Wigzell H, Uhlen M. J. Biol. Chem. 1988;263:4323–4327. [PubMed] [Google Scholar]
- 63.Chackerian B, Briglio L, Albert PS, Lowy DR, Schiller JT. J. Virol. 2004;78:4037–4047. doi: 10.1128/JVI.78.8.4037-4047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dimmock N, Easton A, Leppard K. Introduction to Modern Virology. Wiley-Blackwell; 2007. [Google Scholar]
- 65.Lefrancois L, Lyles DS. J Immunol. 1983;130:394–398. [PubMed] [Google Scholar]
- 66.Lefrancois L, Lyles DS. Virology. 1982;121:168–174. doi: 10.1016/0042-6822(82)90126-x. [DOI] [PubMed] [Google Scholar]
- 67.Golden JW, Hooper JW. Virology. 2008;377:19–29. doi: 10.1016/j.virol.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boltovets PM, Snopok BA, Boyko VR, Shevchenko TP, Dyachenko NS, Shirshov YM. J Virol Methods. 2004;121:101–106. doi: 10.1016/j.jviromet.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 69.Drazen JM. N Engl J Med. 2002;346:1262–1263. doi: 10.1056/NEJM200204253461702. [DOI] [PubMed] [Google Scholar]
- 70.Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Moller P, Wagner R, Volchkov V, Klenk HD, Feldmann H, Stroher U. J Virol. 2004;78:5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moss B. Poxviridae: The Viruses and Their Replication. In: Knipe, P.M DMH, editor. Fields Virology. 5 ed. vol. 2. Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 72.Schoefield DJ, Dimmock NJ. J. Virol. Methods. 1996;62:33–42. doi: 10.1016/0166-0934(96)02086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wittekindt C, Fleckenstein B, Wiesmuller K, Eing BR, Kuhn JE. J. Virol. Methods. 2000;87:133–144. doi: 10.1016/s0166-0934(00)00160-9. [DOI] [PubMed] [Google Scholar]
- 74.Tanaka Y, Shimoike T, Ishii K, Suzuki R, Suzuki T, Ushijima H, Matsuura Y, Miyamura T. Virology. 2000;270:229–236. doi: 10.1006/viro.2000.0252. [DOI] [PubMed] [Google Scholar]