Abstract
SnoN (Ski-novel protein) was discovered as a nuclear protooncogene on the basis of its ability to induce transformation of chicken and quail embryonic fibroblasts. As a crucial negative regulator of transforming growth factor-β (TGF-β) signaling and also an activator of p53, it plays an important role in regulating cell proliferation, senescence, apoptosis, and differentiation. Recent studies of its expression patterns and functions in mouse models and mammalian cells have revealed important functions of SnoN in normal epithelial development and tumorigenesis. Evidence suggests that SnoN has both pro-oncogenic and anti-oncogenic functions by modulating multiple signaling pathways. These studies suggest that SnoN may have broad functions in the development and homeostasis of embryonic and postnatal tissues.
Introduction
SnoN is a member of the Ski family of proteins that, in addition to Ski and SnoN, also includes two distantly related proteins Fussel-18 (Functional Smad suppressing element on chromosome 18) [1] and Fussel-15 (or LBXCOR1) [2] with unknown functions. SnoN was identified as a nuclear proto-oncoprotein of 684 amino acid residues that displays extensive sequence homology with Ski in the amino terminal portion of the protein (Figure1). Three alternatively spliced forms, SnoN2, SnoI, and SnoA that share identical first 366 residues [3-6] have been reported. Little is known regarding their expression and function. When overexpressed, both Ski and SnoN induce anchorage-independent growth of quail and chicken embryo fibroblasts and terminal muscle differentiation in quail embryo cells [7-9]. The Ski homology domain appears to be necessary and sufficient for the transformation and differentiation functions of Ski and SnoN [10]. Although Ski and SnoN share extensive sequence homology and a certain degree of functional similarity, they also display important functional and regulatory differences [11,12]. Most of the past studies on these proteins focused on their actions in human cancer cell lines. Little is known about their expression and functions in normal mammalian tissues. In this review, we will summarize recent progresses on SnoN expression and function in normal tissues and cells.
Figure 1.
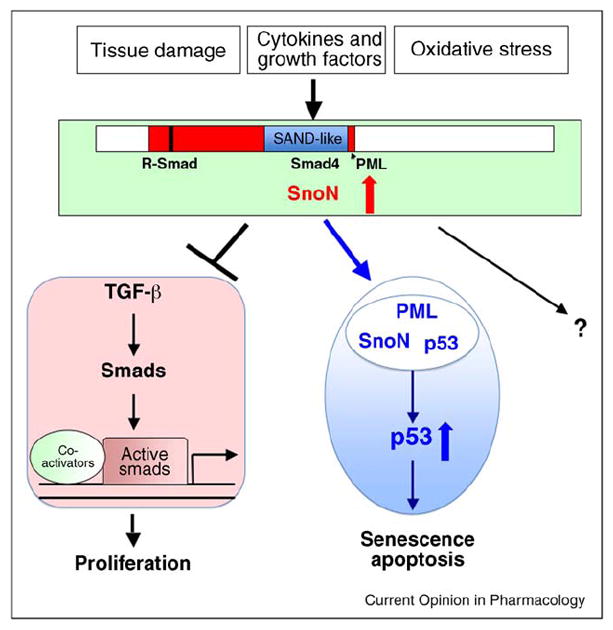
SnoN regulation and function. SnoN expression is induced by cytokines and growth factors such as TGF-β and HGF, tissue damage and oxidative stress. SnoN is a 684 amino acids protein with a conserved ski-homology domain (red) that contains R-Smad binding site, a SAND-like domain (Sp100, AIRE1, NucP41/75, and DEAF1) (blue) that mediates Smad4 binding, and the PML binding region (indicated by the arrowhead). Upon induction, SnoN can bind to the Smad proteins to antagonize TGF-β signaling, leading to cell proliferation. High levels of SnoN also interact with the PML protein and are recruited to PML nuclear bodies where it stabilizes p53 to promote senescence and apoptosis. SnoN may also activate other yet-to-be-identified signaling pathways.
SnoN expression pattern and biological functions
SnoN is ubiquitously expressed in all embryonic and adult tissues but at low levels [3,6,13]. Upregulation of SnoN expression occurs during certain stages of embryonic and postnatal development as well as in some human cancer cell lines and tissues.
SnoN in embryogenesis
In mouse embryos, snoN transcript is induced at 9.5 days post coitum (p.c) in neural tissues and neural crest cells, as well as in the developing limb buds, vasculatures, and mandibular arches. At 11.5 days p.c, snoN expression appears in the developing skeletal muscle and continues to be elevated in tissues with high proliferative potentials [6]. It is also detected around the tip of the growing digit cartilage primordia and in the developing joints and tendons, suggesting a potential role of SnoN in the selection of cell fates during the formation of joints [14].
The function of SnoN in embryonic development has not been well defined. Three lines of snoN knockout mice, two with targeted deletion of exon 1 and a third one with a deletion of the snoN promoter, have been reported [15,16]. While one of the exon 1 deleted line showed embryonic lethality at E3.5 [15], the other two were viable with only a mild defect in T-cell activation [16]. These phenotypes differ significantly from that observed in ski knockout mice with defects in neural tube closure and in skeletal muscle development [17,18]. Thus, Ski and SnoN probably regulate different events during embryonic development. More studies in the future are needed to define the role of SnoN in embryonic development.
SnoN in the mammary gland
The best characterized adult tissue for SnoN expression is the mammary gland. SnoN is detected at a low level in the cytoplasm of luminal epithelial cells of the ducts and in the epithelial cells of the lobuli and lateral ducts in the virgin glands. Its expression is upregulated significantly during late pregnancy, reaching the maximal level at around day 18.5 of gestation and lasting through the beginning of lactation. Interestingly, SnoN expression is dramatically downregulated at 5 days of lactation and by day 3–5 of involution, is returned to the basal level observed in the mature virgin gland [19••]. This suggests that SnoN expression is highly dynamic during mammary gland development and that it may play an important role in the proliferation and structural and functional differentiation of the secretory alveolar epithelial cells.
Indeed, analysis of a transgenic mouse model expressing a fragment of SnoN under the control of the mouse mammary tumor virus (MMTV) promoter and the snoN knockout mice showed that SnoN promotes side-branching and lobular-alveolar proliferation in the virgin glands by increasing epithelial proliferation [19••]. This function is consistent with its activity as a negative regulator of TGF-β signaling as TGF-β has been shown to inhibit lobular-alveolar proliferation and ductal growth [20]. High levels of SnoN also accelerate post-lactational involution by inducing an increase in apoptosis and a decrease in survival cues [19••]. From its peak expression at late pregnancy and early lactation, SnoN is likely to also regulate functional differentiation of mammary epithelial cells. Future analysis of the snoN knockout mammary glands may address this important aspect of SnoN function.
SnoN in the nervous system
In the adult central nervous system, abundant snoN transcripts are found in the cerebrum and cerebellum [6], and SnoN protein is present in the nucleus of the granule neurons of the cerebellum and in both the granule neurons and purkinje cells in the cerebellar cortex [21]. In the rat brain, reducing SnoN in the cerebellar cortex markedly impaired axon growth, suggesting that SnoN is essential for the development of granule neuron parallel fiber [21]. This activity appears to require the actin binding protein Ccd1, which promotes axon growth by activating the JNK kinase at axon termini. SnoN cooperates with p300/CBP to activate Ccd1 transcription in neurons and may serve as a cell intrinsic regulator of axon morphogenesis [22•].
SnoN in other tissues
In the epidermis, SnoN is present in the nucleus and cytoplasm of the keratinocytes in the outer suprabasal layers but not in the basal cell layer. It is also detected in benign seborrheic keratosis and is elevated in the non-metastatic intraepidermal squamous cell carcinoma, especially in the well-differentiated keratinocytes of the superficial epidermal cell layers. By contrast, in invasive squamous cell carcinoma, SnoN positive cells are present largely in the poorly differentiated areas. Thus, SnoN may regulate the differentiation of normal skin and early stages of skin tumors but promote proliferation of undifferentiated squamous cell carcinoma [23•]. The function of SnoN in epidermal differentiation has not been defined.
SnoN has also been reported previously to be expressed in the progenitor and mature cells of the hematopoietic lineages [13] and may also regulate muscle differentiation [9,24,25]. However, these early studies have not been followed up with more in-depth studies in vivo. Clearly much more work is needed to uncover the roles of SnoN in normal tissue development and function.
SnoN in tumorigenesis
SnoN possess both pro-oncogenic and anti-oncogenic activities (Figure 1). Supporting its role as an oncogene, SnoN level is elevated in many cancer cell lines and some tissues. The human snoN gene is located at the chromosome 3q26.2, which is frequently amplified in many cancer types, including esophagus, lung, ovary, cervix, head and neck, and prostate [11]. Two recent studies provided the strongest support for the pro-oncogenic activity of SnoN in human cancer. The first study published in 2005 showed that reduction of SnoN by shRNA in invasive breast cancer and lung cancer cell lines inhibited tumor growth both in vitro and in vivo [26]. More importantly, overexpression of SnoN in the mouse mammary gland accelerates the formation of aggressive multi-focal adenocarcinomas and pulmonary metastasis induced by the Polyoma middle T antigen (PyVmT) [19], providing the first in vivo support for the pro-oncogenic activity of SnoN.
In recent years, more and more evidence have emerged indicating that SnoN can also act as a tumor suppressor. While some human cancer tissues display elevated SnoN levels, many cancer tissues including some colon cancer tissues with microsatellite instability or adenocarcinoma of the esophagus show significant downregulation of SnoN expression [27,28]. More importantly, heterozygous snoN+/− mice are more susceptible to carcinogen-induced tumors than wild-type mice, suggesting that an extra copy of the snoN gene protects against carcinogenesis in vivo [15]. Recently, we have shown that overexpression of SnoN inhibits oncogene-induced transformation of primary mouse embryonic fibroblasts (MEF) by activating a p53-dependent and PML-dependent premature senescence pathway. In an in vivo two-step skin carcinogenesis model, mice expressing high levels of SnoN are resistant to the development of papilloma in vivo, and the small number of papilloma developed in these mice regressed spontaneously, probably owing to increased senescence [29••]. Finally, reducing SnoN by shRNA enhances epithelial to mesenchymal transition (EMT) of lung and breast cancer cells in vitro and tumor metastasis in vivo [26].
Therefore SnoN plays a dual role in tumorigenesis and acts through distinct signaling pathways to elicit the pro-oncogenic or anti-oncogenic responses.
Regulation of SnoN expression
SnoN expression is subjected to stringent regulation in normal mammalian cells at the levels of protein stability, transcriptional activation, and post-translational modification [11,12]. SnoN transcription can be induced by oxidative stress [29] and tissue injury and upon stimulation with certain growth factors and cytokines (Figure 1). The observation that SnoN transcription is upregulated at late pregnancy and early lactation suggests that it might also be regulated by the pregnancy hormones [19]. During liver regeneration following injury, SnoN expression is activated, possibly mediated by the hepatocyte growth factor (HGF) [30,31]. SnoN expression is also altered during obstructive neuropathy [32,33]. The factors and pathways responsible for SnoN induction during these processes have not been determined.
SnoN transcription is potently and rapidly induced by TGF-β through a direct binding of the Smad2/Smad4 complex to the Smad binding elements in the snoN promoter [34]. TGF-β also regulates SnoN stability through an ubiquitin-dependent proteasome degradation pathway. Several E3 ubiquitin ligases that interact with the R-Smads have been shown to be recruited to SnoN for its ubiquitination, including the Smad ubiquitin regulatory factor (Smurf 2), the Anapahase promoting complex (APC/C) and Arkadia [11]. At the post-translational level, SnoN can be modified by Sumoylation and phosphorylation. The SUMO E3 ligase PIAS has been shown to modify lysines 50 and 383 of SnoN in a TGF-β-independent manner [24,35]. SnoN can also be phosphorylated by the TGF-β-activated kinase (TAK1) on serines 115 and 117, and/or threonine 119 [36]. The functional significance of these modifications has not been fully understood.
Intracellular localization constitutes another layer of SnoN regulation. Although originally identified as a nuclear protein in transformed cell lines and fibroblasts [3,13], SnoN is predominantly cytoplasmic in normal mammary gland, epidermis, esophagus, and liver. The predominant cytoplasmic localization is also found in primary mammary epithelial cells, keratinocytes, hepatocytes, and untransformed mammary epithelial cell lines [19••,37]. The mechanism that regulates SnoN localization has not been defined.
Mechanism of SnoN functions
SnoN as a negative regulator of TGF-β signaling
The best understood action of SnoN is its ability to negatively regulate TGF-β signaling [11,12] (Figure 1). TGF-β is a ubiquitously expressed cytokine that regulates cell proliferation, apoptosis, differentiation, and extra cellular matrix production [38]. Upon ligand binding, the activated TGF-β receptor kinases phosphorylate Smad2 and Smad3, leading to their hetero-oligomerization with Smad4 and accumulation in the nucleus. The nuclear Smad complexes interact with co-activators and sequence-specific DNA binding factors to regulate expression of TGF-β-responsive genes [39,40]. SnoN interacts with Smad2, Smad3, and Smad4 in the cytoplasm to prevent their nuclear translocation [19••,37] and in the nucleus to represses their transcription activity by disrupting the active heteromeric Smad complex, blocking the interaction of Smad2/3 with transcriptional co-activators and recruiting a transcriptional co-repressor complex [11,12]. By doing so, SnoN represses the ability of the Smads to activate their TGF-β target genes and mediate the cytostatic response of TGF-β [34,41,42] (Figure 1).
Indeed many biological activities of SnoN are related to its ability to negatively regulate TGF-β signaling. The hyperproliferative phenotype observed in the mammary epithelium of the SnoN transgenic mice as well as the ability of high levels of SnoN to accelerate PyVmT tumor growth and metastasis is similar to the phenotype of mice lacking the type II TGF-β receptor [43,44]. Moreover, primary PyVmT tumors that also express high levels of SnoN are insensitive to TGF-β-induced growth arrest and express TGF-β target genes at a lower level [19••]. In tissue culture, cancer cells with elevated SnoN expression are refractory to TGF-β induced growth arrest, and reducing SnoN expression restores TGF-β responses and reverses oncogenic transformation [26]. More importantly, mutant SnoN unable to bind to the Smad proteins is defective in inducing oncogenic transformation, indicating that the ability of SnoN to antagonize the tumor suppressor pathway of TGF-β/Smad underlies the pro-oncogenic activity of SnoN [29••,41] (Figure 1).
One recent study suggests that under very low TGF-β concentrations, SnoN may also operate as an activator of TGF-β-induced cell cycle arrest in the mink lung epithelial cell line [45]. This activity may be mediated by the interaction of SnoN with ING2 [46]. Whether this is true for other cell lines and the biological significance of these findings remain to be determined.
In addition to promoting tumor cell proliferation, SnoN may also be involved in other Smad-dependent physiological or pathological processes. For example, SnoN can facilitate liver regeneration after injury by antagonizing the inhibitory activity of TGF-β on hepatocyte proliferation [30]. In caveolin-1 deficient mice, a significant increase in SnoN expression can be detected, and this correlates with attenuation of TGF-β signaling and an acceleration of liver regeneration [31]. In proximal tubular cell lines, BMP-7 can inhibit Smad3 signaling by inducing stabilization of SnoN, thereby opposing TGF-β-induced renal fibrosis [47]. Finally, SnoN has been shown to antagonize TGF-β-induced expression of the metalloprotease-disintegrin ADAM12, a gene that is aberrantly induced in many pathological conditions including cancer, cardiovascular disease, and certain inflammatory diseases [48].
SnoN as a regulator of p53 stability
SnoN also regulates pathways beyond TGF-β signaling. Our recent study has revealed a novel function of SnoN in regulation of p53 stability through a PML-dependent mechanism [29••]. In response to oxidative stress, SnoN expression is induced, and this elevated SnoN interacts with the PML protein and is recruited to the PML bodies, where it stabilizes p53, leading to the onset of premature senescence. This ability to stabilize p53 is independent of its ability to antagonize TGF-β signaling since mutant SnoN defective in binding to the Smad proteins still associates with PML and stabilizes p53 to induce senescence [29••] (Figure 1). These results suggest that SnoN may be an important mediator of the stress responses, in particular oxidative stress, in mammalian cells and bridge the oxidative stress signals with p53 activation. The observation that SnoN expression is markedly induced upon tissue damage is also consistent with this model.
Since both PML and p53 are known tumor suppressors, the ability of SnoN to stabilize p53 also provides a molecular mechanism for the anti-tumorigenic activity of SnoN in human cancer (Figure 1). Thus, SnoN possesses both pro-oncogenic and anti-oncogenic activities. When SnoN expression is elevated in tumor cells, those cells with normal p53 are likely to undergo cessation of proliferation through a p53-dependent senescence or apoptosis program. In order to bypass this important block in malignant progression, many tumor cells inactivate p53 or PML, effectively muting the anti-oncogenic function of SnoN. The silencing of the anti-oncogenic activity of SnoN then allows the pro-oncogenic activity of SnoN to be manifested to promote tumor cell proliferation through antagonizing the cytostatic function of TGF-β.
The identification of SnoN as a p53 regulator independent of TGF-β signaling marks the beginning of a new phase of study of SnoN function. It is likely that SnoN may also modulate other intracellular signaling pathways either through its activity as a transcriptional regulator or through direct physical interactions with other signaling molecules.
SnoN as a therapeutic target
SnoN is both an oncogene, through its ability to antagonize the growth inhibitory activity of TGF-β signaling, and a tumor suppressor, through its ability to activate p53. In late stage human cancers that lack a functional p53 or PML protein, the anti-oncogenic activity of SnoN is silenced, and it mainly functions as an oncogene that can be targeted for cancer therapy. Indeed, reducing SnoN levels in human breast and lung cancer cell lines restored the cytostatic responses to TGF-β and significantly reversed malignant transformation in vitro and tumor growth in vivo [26], suggesting that targeting SnoN in human cancer can effectively block tumor progression. shRNA-based drug that reduces SnoN expression or development of small molecules that block the SnoN/Smad interaction may be useful in anti-cancer therapy.
In addition to cancer, SnoN may also play crucial roles in tissue regeneration and aging. For example, high levels of SnoN may promote liver regeneration following injury by dampening the negative effects of TGF-β signaling on hepatocyte proliferation and survival [30,31]. Therapeutic targeting of SnoN may therefore be useful in modulating the regenerative processes.
Future directions
In the past, most studies on SnoN have focused on its role in tumorigenesis as an important regulator of TGF-β signaling. However, it is becoming clear that SnoN also participates in diverse biological processes in embryogenesis as well as development, function, and homeostasis of normal adult tissues and organs. Mechanistically, SnoN can regulate the activity of other signaling pathways in addition to TGF-β signaling. Thus, an important goal in the near future is to determine the roles of SnoN in the embryonic and postnatal development, function and homeostasis of mammalian tissues and organs as well as the molecular mechanisms underlying these events. The complex activities of SnoN in mammalian tumorigenesis also need to be clarified.
Acknowledgments
We apologize to the authors whose work could not be cited in this review owing to space constraints. Research in the authors’ laboratory was supported by NIH RO1 CA101891, Philip Morris External Research Program grant 019016 to K. Luo, and DOD BCRP pre-doctoral fellowship to N. Jahchan.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
of special interest
of outstanding interest
- 1.Arndt S, Poser I, Schubert T, Moser M, Bosserhoff AK. Cloning and functional characterization of a new Ski homolog, Fussel-18, specifically expressed in neuronal tissues. Lab Invest. 2005;85:1330–1341. doi: 10.1038/labinvest.3700344. [DOI] [PubMed] [Google Scholar]
- 2.Arndt S, Poser I, Moser M, Bosserhoff AK. Fussel-15, a novel Ski/Sno homolog protein, antagonizes BMP signaling. Mol Cell Neurosci. 2007;34:603–611. doi: 10.1016/j.mcn.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Nomura N, Sasamoto S, Ishii S, Date T, Matsui M, Ishizaki R. Isolation of human cDNA clones of ski and the ski-related gene, sno. Nucleic Acids Res. 1989;17:5489–5500. doi: 10.1093/nar/17.14.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson-White S. SnoI, a novel alternatively spliced isoform of the ski protooncogene homolog, sno. Nucleic Acids Res. 1993;21:4632–4638. doi: 10.1093/nar/21.19.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearson-White S, Deacon D, Crittenden R, Brady G, Iscove N, Quesenberry PJ. The ski/sno protooncogene family in hematopoietic development. Blood. 1995;86:2146–2155. [PubMed] [Google Scholar]
- 6.Pelzer T, Lyons GE, Kim S, Moreadith RW. Cloning and characterization of the murine homolog of the sno proto-oncogene reveals a novel splice variant. Dev Dyn. 1996;205:114–125. doi: 10.1002/(SICI)1097-0177(199602)205:2<114::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Colmenares C, Stavnezer E. The ski oncogene induces muscle differentiation in quail embryo cells. Cell. 1989;59:293–303. doi: 10.1016/0092-8674(89)90291-2. [DOI] [PubMed] [Google Scholar]
- 8.Colmenares C, Sutrave P, Hughes SH, Stavnezer E. Activation of the c-ski oncogene by overexpression. J Virol. 1991;65:4929–4935. doi: 10.1128/jvi.65.9.4929-4935.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer PL, Colmenares C, Stavnezer E, Hughes SH. Sequence and biological activity of chicken snoN cDNA clones. Oncogene. 1993;8:457–466. [PubMed] [Google Scholar]
- 10.Liu X, Sun Y, Weinberg RA, Lodish HF. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 2001;12:1–8. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 11.Deheuninck J, Luo K. Ski and SnoN, potent negative regulators of TGF-beta signaling. Cell Res. 2009;19:47–57. doi: 10.1038/cr.2008.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo K. Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev. 2004;14:65–70. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Pearson-White S, Crittenden R. Proto-oncogene Sno expression, alternative isoforms and immediate early serum response. Nucleic Acids Res. 1997;25:2930–2937. doi: 10.1093/nar/25.14.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorda-Diez CI, Montero JA, Martinez-Cue C, Garcia-Porrero JA, Hurle JM. Transforming growth factors beta coordinate cartilage and tendon differentiation in the developing limb mesenchyme. J Biol Chem. 2009;284:29988–29996. doi: 10.1074/jbc.M109.014811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shinagawa T, Dong HD, Xu M, Maekawa T, Ishii S. The sno gene, which encodes a component of the histone deacetylase complex, acts as a tumor suppressor in mice. EMBO J. 2000;19:2280–2291. doi: 10.1093/emboj/19.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson-White S, McDuffie M. Defective T-cell activation is associated with augmented transforming growth factor Beta sensitivity in mice with mutations in the Sno gene. Mol Cell Biol. 2003;23:5446–5459. doi: 10.1128/MCB.23.15.5446-5459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berk M, Desai SY, Heyman HC, Colmenares C. Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial, patterning, and skeletal muscle development. Genes Dev. 1997;11:2029–2039. doi: 10.1101/gad.11.16.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colmenares C, Heilstedt HA, Shaffer LG, Schwartz S, Berk M, Murray JC, Stavnezer E. Loss of the SKI proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Ski-/- mice. Nat Genet. 2002;30:106–109. doi: 10.1038/ng770. [DOI] [PubMed] [Google Scholar]
- 19••.Jahchan NS, You YH, Muller WJ, Luo K. Transforming growth factor-beta regulator SnoN modulates mammary gland branching morphogenesis, postlactational involution, and mammary tumorigenesis. Cancer Res. 2010;70:4204–4213. doi: 10.1158/0008-5472.CAN-10-0135.This study investigated the expression pattern and the functions of SnoN in normal mammary gland development using transgenic and knockout mouse models. It also provided the first in vivo evidence of the pro-oncogenic activity of SnoN in breast cancer.
- 20.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 21.Stegmuller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 22•.Ikeuchi Y, Stegmuller J, Netherton S, Huynh MA, Masu M, Frank D, Bonni S, Bonni A. A SnoN-Ccd1 pathway promotes axonal morphogenesis in the mammalian brain. J Neurosci. 2009;29:4312–4321. doi: 10.1523/JNEUROSCI.0126-09.2009.. This study suggested a novel mechanism underlying the function of SnoN in promoting axonal morphogenesis through acting as a transcriptional co-activator.
- 23•.Zhang X, Egawa K, Xie Y, Ihn H. The expression of SnoN in normal human skin and cutaneous keratinous neoplasms. Int J Dermatol. 2009;48:579–583. doi: 10.1111/j.1365-4632.2009.03685.x.. This study documented a systematic analysis of SnoN expression in normal, benign, and metastatic skin tissues.
- 24.Hsu YH, Sarker KP, Pot I, Chan A, Netherton SJ, Bonni S. Sumoylated SnoN represses transcription in a promoter-specific manner. J Biol Chem. 2006;281:33008–33018. doi: 10.1074/jbc.M604380200. [DOI] [PubMed] [Google Scholar]
- 25.Mimura N, Ichikawa K, Asano A, Nagase T, Ishii S. A transient increase of snoN transcript by growth arrest upon serum deprivation and cell-to-cell contact. FEBS Lett. 1996;397:253–259. doi: 10.1016/s0014-5793(96)01165-9. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Q, Krakowski AR, Dunham EE, Wang L, Bandyopadhyay A, Berdeaux R, Martin GS, Sun L, Luo K. Dual role of SnoN in mammalian tumorigenesis. Mol Cell Biol. 2007;27:324–339. doi: 10.1128/MCB.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chia JA, Simms LA, Cozzi SJ, Young J, Jass JR, Walsh MD, Spring KJ, Leggett BA, Whitehall VL. SnoN expression is differently regulated in microsatellite unstable compared with microsatellite stable colorectal cancers. BMC Cancer. 2006;6:252. doi: 10.1186/1471-2407-6-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villanacci V, Bellone G, Battaglia E, Rossi E, Carbone A, Prati A, Verna C, Niola P, Morelli A, Grassini M, et al. Ski/SnoN expression in the sequence metaplasia-dysplasia-adenocarcinoma of Barrett’s esophagus. Hum Pathol. 2008;39:403–409. doi: 10.1016/j.humpath.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 29••.Pan D, Zhu Q, Luo K. SnoN functions as a tumour suppressor by inducing premature senescence. EMBO J. 2009;28:3500–3513. doi: 10.1038/emboj.2009.250.. This study revealed a novel signaling pathway of SnoN in stabilization of p53 in a PML-dependent manner to induce premature senescence and provided the first molecular mechanism for the anti-oncogenic activity of SnoN in mammalian tumorigenesis.
- 30.Macias-Silva M, Li W, Leu JI, Crissey MA, Taub R. Up-regulated transcriptional repressors SnoN and Ski bind Smad proteins to antagonize transforming growth factor-beta signals during liver regeneration. J Biol Chem. 2002;277:28483–28490. doi: 10.1074/jbc.M202403200. [DOI] [PubMed] [Google Scholar]
- 31.Mayoral R, Valverde AM, Llorente Izquierdo C, Gonzalez-Rodriguez A, Bosca L, Martin-Sanz P. Impairment of transforming growth factor beta signaling in caveolin-1-deficient hepatocytes: role in liver regeneration. J Biol Chem. 2010;285:3633–3642. doi: 10.1074/jbc.M109.072900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan R, Zhang J, Tan X, Zhang X, Yang J, Liu Y. Downregulation of SnoN expression in obstructive nephropathy is mediated by an enhanced ubiquitin-dependent degradation. J Am Soc Nephrol. 2006;17:2781–2791. doi: 10.1681/ASN.2005101055. [DOI] [PubMed] [Google Scholar]
- 33.Fukasawa H, Yamamoto T, Togawa A, Ohashi N, Fujigaki Y, Oda T, Uchida C, Kitagawa K, Hattori T, Suzuki S, et al. Ubiquitin-dependent degradation of SnoN and Ski is increased in renal fibrosis induced by obstructive injury. Kidney Int. 2006;69:1733–1740. doi: 10.1038/sj.ki.5000261. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Q, Pearson-White S, Luo K. Requirement for the SnoN oncoprotein in transforming growth factor beta-induced oncogenic transformation of fibroblast cells. Mol Cell Biol. 2005;25:10731–10744. doi: 10.1128/MCB.25.24.10731-10744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wrighton KH, Liang M, Bryan B, Luo K, Liu M, Feng XH, Lin X. Transforming growth factor-beta-independent regulation of myogenesis by SnoN sumoylation. J Biol Chem. 2007;282:6517–6524. doi: 10.1074/jbc.M610206200. [DOI] [PubMed] [Google Scholar]
- 36.Kajino T, Omori E, Ishii S, Matsumoto K, Ninomiya-Tsuji J. TAK1 MAPK kinase kinase mediates transforming growth factor-beta signaling by targeting SnoN oncoprotein for degradation. J Biol Chem. 2007;282:9475–9481. doi: 10.1074/jbc.M700875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krakowski AR, Laboureau J, Mauviel A, Bissell MJ, Luo K. Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-beta signaling by sequestration of the Smad proteins. Proc Natl Acad Sci USA. 2005;102:12437–12442. doi: 10.1073/pnas.0504107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schilling SH, Hjelmeland AB, Rich JN, Wang X-F. TGF-β: a multipotential cytokine. In: Derynck R, Miyazono K, editors. The TGF-beta Family. Cold Spring Harbor Press; 2008. pp. 45–78. [Google Scholar]
- 39.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 40.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 41.He J, Tegen SB, Krawitz AR, Martin GS, Luo K. The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J Biol Chem. 2003;278:30540–30547. doi: 10.1074/jbc.M304016200. [DOI] [PubMed] [Google Scholar]
- 42.Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 43.Gorska AE, Jensen RA, Shyr Y, Aakre ME, Bhowmick NA, Moses HL. Transgenic mice expressing a dominant-negative mutant type II transforming growth factor-beta receptor exhibit impaired mammary development and enhanced mammary tumor formation. Am J Pathol. 2003;163:1539–1549. doi: 10.1016/s0002-9440(10)63510-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forrester E, Chytil A, Bierie B, Aakre M, Gorska AE, Sharif-Afshar AR, Muller WJ, Moses HL. Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res. 2005;65:2296–2302. doi: 10.1158/0008-5472.CAN-04-3272. [DOI] [PubMed] [Google Scholar]
- 45.Sarker KP, Wilson SM, Bonni S. SnoN is a cell type-specific mediator of transforming growth factor-beta responses. J Biol Chem. 2005;280:13037–13046. doi: 10.1074/jbc.M409367200. [DOI] [PubMed] [Google Scholar]
- 46.Sarker KP, Kataoka H, Chan A, Netherton SJ, Pot I, Huynh MA, Feng X, Bonni A, Riabowol K, Bonni S. ING2 as a novel mediator of transforming growth factor-beta-dependent responses in epithelial cells. J Biol Chem. 2008;283:13269–13279. doi: 10.1074/jbc.M708834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo DD, Phillips A, Fraser D. Bone morphogenetic protein-7 inhibits proximal tubular epithelial cell Smad3 signaling via increased SnoN expression. Am J Pathol. 2010;176:1139–1147. doi: 10.2353/ajpath.2010.090459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solomon E, Li H, Duhachek Muggy S, Syta E, Zolkiewska A. The role of SnoN in transforming growth factor {beta}1-induced expression of metalloprotease disintegrin ADAM12. J Biol Chem. 2010;285:21969–21977. doi: 10.1074/jbc.M110.133314. [DOI] [PMC free article] [PubMed] [Google Scholar]


