Abstract
Murine norovirus (MNV) is endemic in mouse research facilities in the United States and Europe, with a prevalence as high as 58% to 64%. Because of MNV's orofecal route of infection, clinically silent persistent infections in some mouse strains, and proclivity for macrophage and dendritic cells, its presence in mouse colonies has potential to alter phenotypes in experimental mouse models, particularly those involving inflammation and immunologic responses. Although MNV is subclinical, not causing overt disease in immunocompetent mice, we found that MNV infection can accelerate bacteria-induced inflammatory bowel disease (IBD) progression in Mdr1a−/− mice. The studies presented here examined whether MNV infection also affects the phenotype of a bacterially driven mouse model of inflammation-associated colon cancer in genetically susceptible Smad3−/− mice. In vitro culture of bone-marrow–derived macrophages (BMDM) was used to determine whether MNV4 influenced macrophage cytokine production. For in vivo studies, Smad3−/− mice were infected with MNV4 one week prior to infection with Helicobacter. Mice were monitored for 17 to 32 wk for development of IBD and colon cancer, and tissues were analyzed histopathologically. Although in vitro infection of BMDM with MNV4 led to increased inflammatory cytokine production, infection with MNV4 in vivo did not result in any statistically significant differences in survival, IBD scores, tumor incidence, or tumor phenotype in Smad3−/− mice. In addition, MNV infection alone did not result in IBD or colon cancer. Therefore MNV infection alone or in conjunction with Helicobacter does not alter the development or progression of IBD or colon cancer in Smad3−/− mice.
Abbreviations: BMDM, bone-marrow–; derived macrophages; IBD, inflammatory bowel disease; MNV, murine norovirus
The variability within an in vivo animal model depends on multiple factors, including pathogen status. Many bacterial and viral agents that can infect mice have been shown to be intercurrent variables in murine models of human disease. Inapparent infections may cause undesirable variability, unexplainable data variation, or misinterpretation of data.1,18 Since the discovery and recognition of murine norovirus (MNV) as prevalent in mouse colonies9,11 and the identification of numerous strains of MNV that are biologically and genetically distinct,11,14,24 questions have been raised about how infection with this virus might affect mouse models of human disease.
MNV1 initially was reported in Stat1−/− mice.13 Although Stat1−/− mice, which have abnormal signaling through interferon pathways, develop lethal disease when infected with MNV, MNV does not appear to cause clinical disease in wildtype mice or in several other strains of immunodeficient mice, such as recombination-activating gene-deficient (Rag−/−) or inducible nitric oxide synthase-deficient mice.13,29 Like human noroviruses, which cause inflammatory changes in the gastrointestinal tract,16 MNV appears to be transmitted easily by the orofecal route13 and has been associated with histologic changes in the intestine of immunocompetent mice.25 MNV has a tropism for dendritic cells and macrophages,30 which are important players in immunologic and inflammatory responses. Studies in Rag−/− mice demonstrate that adaptive immune responses play an important role in clearance of virus and that CD4+ and CD8+ T cells and B cells are important in this process as well.13,31 Only a few studies have examined the potential effects of MNV infection on mouse models. A recent study6 found that MNV has the subtle but definite effect of depressing CD8+ T cell responses in a model of murine cytomegalovirus infection. In contrast, another study10 found that MNV infection had no significant effect on adaptive immune responses to vaccinia virus or influenza A virus. Our laboratory demonstrated that MNV4 infection accelerated the progression of Helicobacter-induced inflammatory bowel disease (IBD) in the FVB.129P2-ABC1atm1BorN7 (Mdr1a−/−) mouse and that this induction was mediated in part through alteration of dendritic and T-cell cytokine responses.17
Our laboratory studies bacteria-induced IBD3 and inflammation-associated colon cancer20,22 by using SMAD3-deficient mice. Smad3−/− mice are deficient in TGFβ signaling due to the absence of SMAD3, a molecule involved in signal transduction from the membrane-bound TGFβ receptor to the nucleus. TGFβ dysregulation results in the development of transient bacteria-induced colitis when mice are infected with Helicobacter; this transient colitis then progresses to colon cancer.22 The model has a strong inflammatory component and is characterized by dysregulation of both adaptive22 and innate21 immune responses.
Because MNV can reside in both macrophages and dendritic cells, which are pivotal in inflammatory responses, we reasoned that this model could potentially be altered by concurrent MNV infection. In contrast to the previous report on Mdr1a−/− mice,17 we report here that MNV4 infection does not affect the progression or phenotype of tumors in Helicobacter-infected Smad3−/− mice. However, in vitro studies using bone-marrow–derived macrophages (BMDM) from Smad3−/− and wildtype mice did show differences in cytokine profiles after infection with MNV4 and H. bilis.
Materials and Methods
Animals, infections, and experimental design.
129-Smad3tm/Par/J (Smad3−/−) mice initially were obtained from The Jackson Laboratory (Bar Harbor, ME). Animals were housed at the University of Washington in a specific pathogen-free facility in static micro-isolator (Alternative Design, Siloam Springs, AR) or individually ventilated cages (Allentown, Allentown, NJ) containing corncob bedding (Andersons, Maumee, OH) and cotton nesting pads. Mice were fed irradiated rodent chow (Animal Specialties, Portland, OR) and autoclaved, acidified water. All supplies entering animal rooms were autoclaved or gas-sterilized, and rooms were maintained at 20 to 23 °C, with a 12:12-h light:dark cycle. To prevent cross-contamination of Helicobacter-uninfected and -infected mice, cages were changed in dedicated changing stations. Sentinel mice (Crl:CD1[ICR]; Charles River, Wilmington, MA) were tested quarterly for endo- and ectoparasites, MNV, mouse hepatitis virus, mouse parvovirus, and rotavirus and annually for Mycoplasma pulmonis, pneumonia virus of mice, reovirus 3, Sendai virus, and Theiler murine encephalomyelitis virus. In addition, yearly fecal colon samples were screened for Citrobacter rodentium, nonlactose-fermenting Escherichia coli, Salmonella spp., Klebsiella spp., and Clostridium spp. (Phoenix Laboratories, Everett, WA). During the study period, all sentinels were negative for the listed pathogens, except for sentinels on the infected racks, which seroconverted to MNV and were PCR-positive for Helicobacter spp. All animal procedures were approved by the University of Washington Institutional Animal Care and Use Committee.
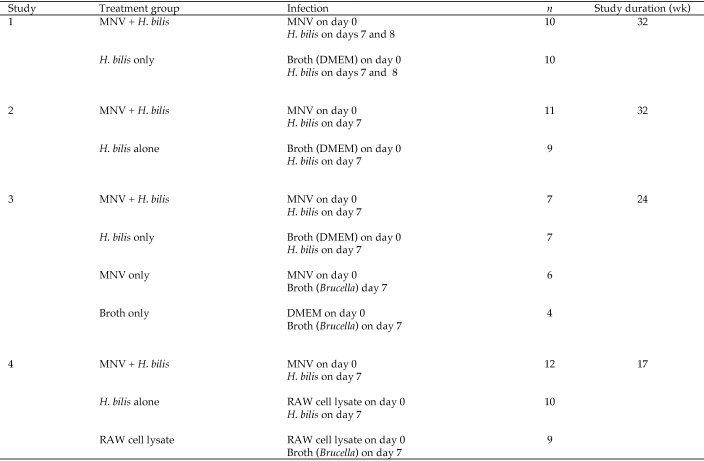
Age-matched Smad3−/− mice (male and female, 2 to 5 mo old) were used in 4 different infection studies. Study treatment groups, animal numbers and duration are outlined in Figure 1. MNV4 (a kind gift from L Riley, University of Missouri, Columbia, MO) stocks were propagated in RAW264.7 cells, a transformed murine macrophage cell line. RAW264.7 cells were PCR-negative for murine leukemia virus.8 Subconfluent cultured cells were infected with MNV at a multiplicity of infection of 0.05 and incubated for 48 to 72 h, as described.12 MNV was recovered by successive freeze–thaw cycles, concentrated by centrifugation at 90,000 × g for 3 h, and titered by plaque assay with RAW264.7 cells.
Figure 1.
Design of studies.
Mice were inoculated with either 1 × 106 or 5 × 106 plaque-forming-units MNV4 in 0.2-mL RAW264.7 (RAW) cell lysate clarified by centrifugation at 3000 x g for 10 min or sham-inoculated with 0.2 mL DMEM or RAW cell lysate on day 0. H. bilis was a natural isolate kindly provided by L Riley (University of Missouri). Organisms were prepared for infection as previously described,17 and mice were inoculated 1 wk after MNV infection with 0.2 mL H. bilis in Brucella broth for a dose of 2 × 107 or 1 × 105 cfu per mouse. Note that in study 1, H. bilis was given in 2 doses, on day 7 and 8 after infection with MNV or sham-inoculation.
Mice were euthanized by CO2 when they developed severe diarrhea, 20% body weight loss, significant anemia, or signs of illness during the study (Figure 1); remaining mice were euthanized at the end of the study. At necropsy, blood samples were obtained through cardiac puncture, and tissue samples for histopathology were taken from the cecum and colon.
Serology and fecal PCR.
Serum was separated from cardiac blood collected at necropsy, frozen, and stored at −80 °C until processing. Seroconversion to MNV was confirmed in studies 1 and 2. Antibodies to MNV were detected by microsphere-based serologic multiplexed fluorescent immunoassay with secondary testing by indirect fluorescent antibody analysis (Research Animal Diagnostic Laboratory, Columbia, MO, or Department of Comparative Medicine, University of Washington, Seattle, WA).
For confirmation of MNV in feces, RT–PCR analysis was done on RNA extracted from feces4 collected from animals in studies 1 (8 wk after infection), 2 (1 wk after infection), and 3 (2 to 3wk after infection). RT–PCR for MNV was performed as previously described with slight modification.12 Briefly, the RNA was amplified by using a commercial kit (OneStep RT-PCR Kit, Qiagen, Valencia, CA). The RT-PCR mixture was heated at 50 °C for 30 min and at 95 °C for 15 min, followed by 40 cycles consisting of 94 °C for 1 min, 59 °C for 30 s, and 72 °C for1 min, with a final primer extension at 72 °C for 10 min.
For confirmation of H. bilis infection in studies 1 through 3, fecal samples were collected at least 7 d after infection. DNA was extracted by using a commercial kit (QIAamp DNA mini-kit, Qiagen). Helicobacter spp. PCR was performed as described.3
Pathology.
Necropsy, tissue sampling, processing, and histologic examination (with some modifications) were done as described previously.3 Cecum, colon, and rectum were fixed in 10% buffered formalin. The colon was prepared in a ‘Swiss roll’ technique to evaluate the entirety of the proximal, middle, and distal colon on the same section. Tissues were processed routinely in neutral-buffered formalin, embedded in paraffin, sectioned at 4 to 5 μm, and stained with hematoxylin and eosin. Tissues were scored for inflammation and dysplasia by a pathologist, and each mouse was scored for severity of mucosal epithelial changes, degree of inflammation, and extent of pathology. The total inflammation and dysplasia score for each mouse (IBD score)21 was derived by summing the scores from the individual segments and, subsequently, the mean was calculated for each treatment group. Tumors were counted and classified as invasive (mucinous adenocarcinomas penetrating the tunica muscularis) or noninvasive (adenomas with high-grade dysplasia and intraepithelial carcinomas).2,22
In vitro infection of bone marrow-derived macrophages and cytokine analyses.
Bone marrow was isolated from Smad3−/− and Smad3+/+ (wildtype) mice. RBC were lysed by incubating cells in Gey solution (ammonium chloride RBC lysis buffer) for 5 min at room temperature. After RBC lysis, remaining cells were differentiated to macrophages by culture for 10 to 12 d in the presence of conditioned medium from L929 cells expressing macrophage colony-stimulating factor.7 Aliquots of 1 × 106 macrophages were cultured overnight in RPMI containing 5% FCS and gentamycin at 37 °C. MNV was added at a multiplicity of infection of 2.5 and incubated for 6 h. H. bilis sonicate with a total protein concentration of 16 mg/mL (determined by absorbance at 280 nm) was added to cultures at a final concentration of 0.2 ng/mL (low Hb) and 4 ng/mL (high Hb), and cultures were incubated for another 24 h. RNA was extracted (RNeasy Mini Kit, Qiagen, and Qiashredder columns, Qiagen), converted into cDNA (SuperScript SSIII kit, Invitrogen, Carlsbad, CA) by using oligodT primers, and assayed for TNFα, IL6, and IL1β expression by using real-time PCR (PowerSybrGreen PCR master mix; Life Technologies–ABI, Carlsbad, CA) on a Mx3005P apparatus (Stratagene, La Jolla, CA). Cytokine levels in treated macrophages are expressed relative to those of the control (uninfected macrophages of the same genotype). Relative expression of cytokine genes compared with control samples was calculated by using MxPro software (Agilent, Santa Clara, CA). Primer sequences were: TNFα forward, 5′AGC CGA TGG GTT GTA CCT TGT CTA; TNFα reverse, 5′ TGA GAT AGC AAA TCG GCT GAC GGT; IL1β forward, 5′ CTT CAG GCA GGC AGT ATC ACT CAT; IL1β reverse, 5′ AAG AAG GTG CTC ATG TCC TCA TCC; IL6 forward, 5′ ATC CAG TTG CCT TCT TGG GAC TGA; IL6 reverse, 5′ TAA GCC TCC GAC TTG TGA AGT GGT.
Statistics.
For tissue culture experiments (Figure 2), the Student t test was used to show significant differences compared with control samples. One-way ANOVA followed by the Tukey multiple comparison test was used to determine significant increases in H. bilis-treated tissue culture samples compared with H. bilis- and MNV-cotreated samples. Kaplan–Meier survival curves were generated, and significance determined with the log-rank (Mantel–Cox) test statistic by using GraphPad Prism 5 (GraphPad Software, La Jolla, CA). Animals that died due to reasons unrelated to IBD or cancer or that lived to the endpoint of the study were censored in the analysis. Differences between group IBD scores and tumor scores were analyzed with Kruskall–Wallis ANOVA with Dunn posttest (studies 3 and 4) or by the Mann–Whitney test (studies 1 and 2). Fisher exact contingency statistics were used to compare tumor incidence between groups. Animals that died before 10 wk after infection with H. bilis that did not have tumors were excluded from tumor and IBD analysis due to uncertainty of whether tumors would have developed with more time. Animals that died of complications not related to IBD or cancer also were excluded from the analysis. A value of P ≤ 0.05 was considered significant for all tests.
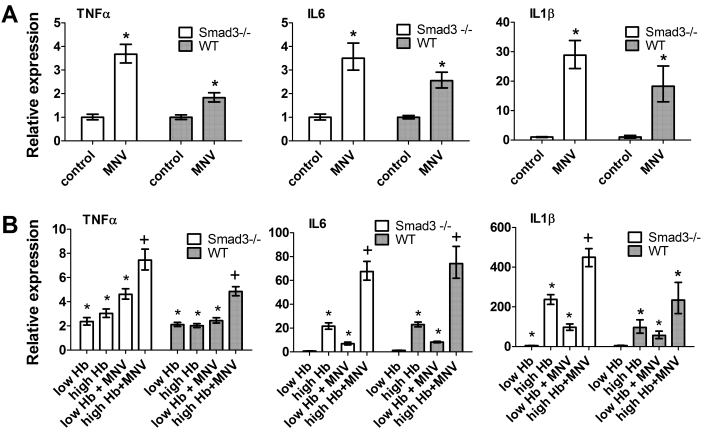
Figure 2.
Inflammatory cytokine expression in BMDM is altered with MNV infection in vitro. (A) Relative expression of the indicated cytokine genes in BMDM incubated with MNV compared with control cultures. (B) Relative gene expression of indicated cytokines in BMDM incubated with H. bilis sonicate (Hb) alone or with MNV and 1 of 2 doses (low, 0.2 ng/mL; high, 4 ng/mL) of Hb. Cytokine expression was determined by quantitative RT–PCR. Cytokine levels in treated macrophages are expressed relative to those of controls (uninfected macrophages). Error bars indicate 1 SEM (n = 4) in a single experiment; data for 1 of 2 independent experiments are shown. *, Value significantly (P < 0.05, unpaired t test) different from that of control; +, value significantly (P < 0.05, ANOVA with Tukey post test) different from value after Hb treatment alone.
Results
Cytokine profiles of macrophages from wildtype and Smad3−/− mice infected with MNV4 in vitro.
Because MNV can persistently infect macrophage cell lines,5 we cultured BMDM from Smad3−/− and Smad3+/+ (wildtype) mice with MNV4 at a multiplicity of infection of 2.5. Because our in vivo IBD-induced cancer model in Smad3−/− mice involves infection with H. bilis, we tried to mimic double infection in vitro; therefore some treatment groups also were cultured in the presence of H. bilis sonicates. After 30 h of incubation, RNA was extracted and cytokine expression was determined by quantitative real-time PCR. Incubation of BMDM with MNV alone caused increases in expression of the inflammatory cytokines TNFα, IL6, and IL1β (Figure 2 A), ranging from a 1.8- (wildtype TNFα) to 28- (Smad3−/− IL1β) fold increase in cytokine gene expression compared with untreated controls. In addition, MNV enhanced the cytokine expression of cells treated with H. bilis sonicates (Figure 2 B), and the magnitude of the effect differed with a high or low concentration of H. bilis. In both wildtype and Smad3−/− macrophages, the largest increases with MNV infection were in levels of IL1β at the low dose of H. bilis compared with H. bilis alone (21-fold increase in wildtype and 12-fold increase in Smad3−/−) and IL6 (7-fold increase in wildtype and 6.4-fold increase in Smad3−/−). Because macrophage-derived inflammatory cytokines are important in the Smad3−/− mouse model of IBD-associated colon cancer, these data suggested that concurrent MNV infection might alter the phenotype. We, therefore, evaluated whether concurrent MNV infection altered the in vivo model in Smad3−/− mice.
Survival of Smad3−/− mice infected with Helicobacter and MNV4.
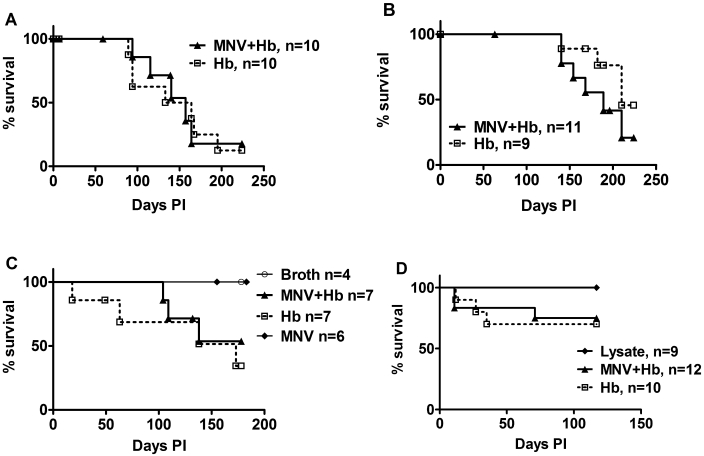
Four independent studies were performed to examine the effect of MNV infection on IBD and colon cancer development in Helicobacter-infected Smad3−/− mice. As an overall measure of the progression of disease, we examined the effect of MNV4 infection on survival until the experimental endpoint. In study 1, Smad3−/− mice first were inoculated with MNV or broth on day 0 followed by H. bilis infection on days 7 and 8, and mice were euthanized and necropsied at 32 wk after infection. Median survival did not differ between groups (median survival: coinfected mice, 157.0 d; singly infected mice, 148.5 d; P = 0.1643; Figure 3 A). In studies 2, 3, and 4, Smad3−/− mice were infected with MNV4 and then infected with H. bilis on day 7 only. Again, median survival was not different between groups (Figure 3 B through D). In study 4, an additional experimental group received RAW264.7 cell lysate only. Neither RAW cell lysate nor MNV4 infection alone affected survival in this model (Figure 3 C and D).
Figure 3.
Survival of Smad3−/− mice coinfected with MNV and H. bilis (MNV+Hb) or infected with H. bilis only (Hb). Survival curves from studies 1 to 4 are shown (A through D, respectively). There was no significant difference in survival between coinfected and singly infected animals in any of the 4 studies. In C and D, survival of either infected group was significantly (P ≤ 0.05) different from that of control animals (broth, MNV, or lysate).
Effects of MNV4 infection on the severity of endpoint IBD, tumor burden, and tumor phenotype in Smad3−/− mice.
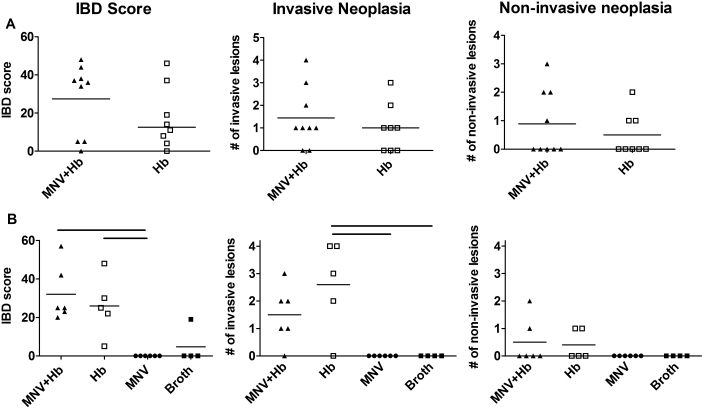
Smad3−/− mice develop IBD after infection with Helicobacter, characterized by colonic epithelial cell hyperplasia and inflammation which may persist until late in the disease process.22 Because we previously found that the severity of Helicobacter-induced IBD was affected by MNV4 infection in Mdr1a−/− mice,17 we compared the IBD scores between MNV4-infected and -uninfected Smad3−/− mice. Histologic scoring of the cecum and colon found no statistically significant differences between coinfected (MNV and H. bilis) and singly infected (H. bilis only) groups in any of the 4 studies (Figure 4, Table 1). IBD scores in study 4 were somewhat lower than those in other studies, possibly due to the shortened duration of this particular study; nonetheless IBD scores did not differ significantly between the 2 groups. Because Smad3−/− mice can develop a variety of tumors throughout the colon and rectum, our histopathologic analysis differentiated between invasive and noninvasive neoplasia. In all studies, the overall average tumor burden (number of distinct invasive or noninvasive tumors per mouse) was not statistically different. In addition, the number of mice developing either noninvasive or invasive tumors was not significantly different between groups (Figure 4, Table 1). No tumors developed in Smad3−/− mice given broth or MNV alone; however, 1 of 9 mice given RAW cell lysate in study 4 developed an invasive adenocarcinoma, an unusual occurrence in this model.
Figure 4.
IBD and neoplasia in Smad3−/− mice infected with H. bilis alone or coinfected with MNV and H. bilis. IBD scores (left), number of invasive lesions (middle), and number of noninvasive lesions (right) are shown for (A) study 2 and (B) study 3. In study 3, 2 additional control groups of mice were infected with MNV alone (n = 6) or broth only (n = 4) are shown. Black bars above graphs in (B) indicate statistically significant differences between groups as tested by nonparametric one-way ANOVA and a Dunn post test. There were no statistically significant differences between mice coinfected with both MNV and H. bilis or with H. bilis only (P > 0.05 for Mann–Whitney tests of IBD and tumor scores; P > 0.05 for Fisher exact test for tumor incidence).
Table 1.
IBD scores and incidence of adenoma and adenocarcinoma in Smad3−/−mice infected with H. bilisand/or MNV4
| Mean IBD scorea | No. of mice with lesions/total no. in group (tumor incidence)b |
Time after infection (wk) | |||
| Study | Treatment | Adenoma (noninvasive) | Adenocarcinoma (invasive) | ||
| 1 | H. bilis | 23.6 | 5/8 (62%) | 5/8 (62%) | 32 |
| MNV + H. bilisb | 24.8 | 6/6 (100%) | 3/6 (50%) | ||
| P = 0.21 | P = 0.66 | ||||
| 2 | H. bilis | 17.4 | 3/8 (38%) | 5/8 (62%) | 32 |
| MNV+ H. bilisb | 27.4 | 4/9 (44%) | 7/9 (78%) | ||
| P = 0.41 | P = 0.62 | ||||
| 3 | H. bilis | 26 | 2/6 (33%) | 5/6 (83%) | 24 |
| MNV + H. bilisb | 32 | 2/5 (40%) | 4/5 (80%) | ||
| P = 1.0 | P = 1.0 | ||||
| MNV alonec | 0 | 0/6 (0%) | 0/6 (0%) | ||
| P = 0.015 | P = 0.015 | ||||
| Brothc | 4.7 | 0/4 (0%) | 0/4 (0%) | ||
| P = 0.048 | P = 0.048 | ||||
| 4 | H. bilis | 5.5 | 2/8 (25%) | 3/8 (38%) | 17 |
| MNV + H. bilisb | 9 | 0/10 (0%) | 3/10 (30%) | ||
| P = 0.18 | P = 1.0 | ||||
| RAW cell lysate | 2.2 | 0/9 (0%) | 1/9 (11%) | ||
IBD score comparisons between H. bilis and MNV + H. bilis were all nonsignificant by Mann–Whitney t test
P values shown compare tumor incidence of H. bilis with that of MNV+ H. bilis by using the Fisher exact test and are all nonsignificant.
Mice given broth and MNV alone did not develop tumors (study 3) and were significantly (P < 0.05) different from H. bilis and MNV + H. bilis groups.
Infection status of MNV4-inoculated Smad3−/− mice.
Fecal samples were collected at 2 time points in 2 different studies, and the presence of MNV4 was demonstrated by using fecal RT-PCR. In study 1, all 11 MNV4-infected mice were positive for MNV4 in feces at 8 wk after infection, and all 10 mice infected with H. bilis only remained negative for MNV4 in feces. MNV4 was detected in feces as early as 1 wk after infection. In study 3, 11 of 12 MNV4-infected mice were positive for MNV4 in feces, whereas none of the control or mice infected with H. bilis alone was positive for MNV4. Endpoint serology was used in studies 1 and 2 to determine seroconversion to MNV4. The combined results from studies 1 and 2 showed that all 16 mice that were infected with H. bilis only were seronegative for MNV at 1 to 32 wk after infection, and all 21 mice infected with MNV seroconverted to MNV as early as 1 to 32 wk after infection.
Discussion
MNV is endemic in many animal facilities in the United States, Canada,9,27 Europe,24 and Asia,15,32 with prevalence as high as 58% to 64%. Whereas the first isolate, MNV1, caused fatal disease in immunocompromised mice, such as Stat1−/− and interferon-signaling–deficient (IFNαβγR−/−) mice,13 MNV does not cause any overt disease in immunocompetent mice. However, we found that MNV induces subtle changes in lymphoid tissue of immunocompetent wildtype mice26 and that MNV infection can exacerbate IBD in a commonly used mouse model, Mdr1a−/− mice.17 Effects of MNV on IBD in the Mdr1a−/− model were due in part to changes in antigen-presenting cells, such as dendritic cells and macrophages, for which MNV has a demonstrated tropism.30 Because macrophages are important players in inflammatory responses (particularly inflammation-associated cancer)28 and because heightened macrophage responses and enhanced cytokine elaboration play a role in bacteria-induced inflammation in the Smad3−/−22 and Smad3 −/− Rag2 −/− models21 of colon cancer, we wondered whether MNV infection would alter the phenotype of the Smad3−/− model. Because MNV can cause persistent infection and subtle, subclinical immunologic changes—even in immunocompetent mice—and replicates in both epithelium and antigen-presenting cells in the gut, there is considerable concern that this virus could alter mouse models of disease, especially those involving intestinal inflammation.
In vitro experiments in both wildtype and Smad3−/− BMDM indicated that MNV alone could alter inflammatory cytokine production by macrophages. Increased cytokine production has recently been demonstrated in MNV1-infected bone-marrow–derived dendritic cells of wildtype animals and was shown to be dependent on viral interaction with pattern recognition receptors Mda5 and possibly TLR3.23 Our data demonstrate that infection of BMDM with MNV4 can enhance production of cytokines in response to bacterial (H. bilis) antigens. BMDM treated with a low dose of H. bilis extract responded weakly in their production of IL6 and IL1β RNA. However, infection of BMDM with MNV4 prior to treatment with low-dose H. bilis extract increased cytokine production 8.6- (IL6) and 21- (IL1β) fold.
Despite the results from our in vitro experiments, MNV4 infection prior to infection with H. bilis in vivo did not significantly alter endpoint IBD and subsequent cancer in Smad3−/− mice. Although IBD scores were not increased at the 17- to 32-wk endpoints in the coinfected Smad3−/− mice, IBD in this model is primarily tumor-associated, and inflammation is being driven by tumor development instead of or in addition to the ongoing Helicobacter infection. Perhaps MNV4 infection was associated with increased inflammation earlier in the disease process that was being driven by Helicobacter, similar to what we observed in the MDR1a−/− model.17 However, if this increase did occur, it was not robust enough to cause significant changes in the endpoint experimental outcomes in this model, as reflected in overall survival, tumor burden, or phenotype between Smad3−/− mice coinfected with MNV and H. bilis or with H. bilis alone.
Our studies were limited with regard to several aspects of infection (infectious dose, virus isolate, and timing of infection) that could affect the degree of the response of the Smad3−/− mouse model to a concurrent MNV infection. We tested 2 infectious doses of MNV (1 × 106 and 5 × 106 pfu) and used a single naturally occurring isolate of MNV, MNV4.11 The MNV infection was initiated prior to the main trigger of disease in our model (infection with H. bilis) but not during or late in the disease process. We cannot exclude the possibility that a different dose, strain, or timing of infection would have altered the phenotype of the model. In addition, multiple strains of MNV are likely present in mouse colonies in the United States, and the biology of MNV can vary substantially with each virus isolate.14 Previous infection with MNV does not necessarily confer protection from subsequent infections even with the same strain, as shown in C56BL/6 and 129Sv/Ev mice.19 In an infected colony, some mice likely would become infected or reinfected at different time points during an experimental course, especially if they are being brought in from a MNV-free commercial vendor into an endemically infected colony.
Studies of the pathophysiologic effects and immunologic responses of this ubiquitous virus are still in their infancy. Although we did not observe an effect of MNV infection on the Smad3−/− mouse model, these results cannot necessarily be extrapolated to other mouse models of IBD or cancer progression. As we are gradually acquiring more information on the biology and effect of MNV infection in specific mouse models, researchers should be cautious when dealing with MNV-positive mouse colonies and consider intercurrent viral infection as a potential cause of experimental variability. Studies that find no apparent effect of MNV infection on the model being studied are as important to report as those that detect positive effects.
Acknowledgments
KLC was supported by training grant T-32 RR007019 from the NIH. We thank Lela Riley (University of Missouri) for providing the MNV4, and we are grateful to Aimee McMillan for animal colony and technical support.
References
- 1.Baker DG. 1998. Natural pathogens of laboratory mice, rats, and rabbits and their effects on research. Clin Microbiol Rev 11:231–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin GP, Washington K, Yang K, Ward JM, Pretlow TP, Russell R, Besselsen DG, Godfrey VL, Doetschman T, Dove WF, Pitot HC, Halberg RB, Itzkowitz SH, Groden J, Coffey RJ. 2003. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology 124:762–777 [DOI] [PubMed] [Google Scholar]
- 3.Burich A, Hershberg R, Waggie K, Zeng W, Brabb T, Westrich G, Viney JL, Maggio-Price L. 2001. Helicobacter-induced inflammatory bowel disease in IL10- and T-cell–deficient mice. Am J Physiol Gastrointest Liver Physiol 281:G764–G778 [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed] [Google Scholar]
- 5.Daughenbaugh KF, Wobus CE, Hardy ME. 2006. VPg of murine norovirus binds translation initiation factors in infected cells. Virol J 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doom CM, Turula HM, Hill AB. 2009. Investigation of the impact of the common animal facility contaminant murine norovirus on experimental murine cytomegalovirus infection. Virology 392:153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Englen MD, Valdez YE, Lehnert NM, Lehnert BE. 1995. Granulocyte–macrophage colony-stimulating factor is expressed and secreted in cultures of murine L929 cells. J Immunol Methods 184:281–283 [DOI] [PubMed] [Google Scholar]
- 8.Hartley JW, Evans LH, Green KY, Naghashfar Z, Macias AR, Zerfas PM, Ward JM. 2008. Expression of infectious murine leukemia viruses by RAW264.7 cells, a potential complication for studies with a widely used mouse macrophage cell line. Retrovirology 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson KS. 2008. Murine norovirus, a recently discovered and highly prevalent viral agent of mice. Lab Anim (NY) 37:314–320 [DOI] [PubMed] [Google Scholar]
- 10.Hensley SE, Pinto AK, Hickman HD, Kastenmayer RJ, Bennink JR, Virgin HW, Yewdell JW. 2009. Murine norovirus infection has no significant effect on adaptive immunity to vaccinia virus or influenza A virus. J Virol 83:7357–7360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu CC, Riley LK, Wills HM, Livingston RS. 2006. Persistent infection with and serologic cross-reactivity of 3 novel murine noroviruses. Comp Med 56:247–251 [PubMed] [Google Scholar]
- 12.Hsu CC, Wobus CE, Steffen EK, Riley LK, Livingston RS. 2005. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse-transcriptase–PCR assay to detect murine norovirus 1 infection in mice. Clin Diagn Lab Immunol 12:1145–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karst SM, Wobus CE, Lay M, Davidson J. Virgin HW 4th 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299: 1575–1578 [DOI] [PubMed] [Google Scholar]
- 14.Kelmenson JA, Pomerleau DP, Griffey S, Zhang W, Karolak MJ, Fahey JR. 2009. Kinetics of transmission, infectivity, and genome stability of 2 novel mouse norovirus isolates in breeding mice. Comp Med 59:27–36 [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M, Lee H, Chang KO, Ko G. 2010. Molecular characterization of murine norovirus isolates from South Korea. Virus Res 147:1–6 [DOI] [PubMed] [Google Scholar]
- 16.Ko G, Jiang ZD, Okhuysen PC, DuPont HL. 2006. Fecal cytokines and markers of intestinal inflammation in international travelers with diarrhea due to noroviruses. J Med Virol 78:825–828 [DOI] [PubMed] [Google Scholar]
- 17.Lencioni KC, Seamons A, Treuting PM, Maggio-Price L, Brabb T. 2008. Murine norovirus: an intercurrent variable in a mouse model of bacteria-induced inflammatory bowel disease. Comp Med 58:522–533 [PMC free article] [PubMed] [Google Scholar]
- 18.Lindsey JR, Boorman GA, Collins MJ, Hsu C, Van Hoosier GL, Wagner JE. 1991. Infectious diseases of mice and rats. Washington (DC): National Academies Press. [Google Scholar]
- 19.Liu G, Kahan SM, Jia Y, Karst SM. 2009. Primary high-dose murine norovirus 1 infection fails to protect from secondary challenge with homologous virus. J Virol 83:6963–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maggio-Price L, Shows D, Waggie K, Burich A, Zeng W, Escobar S, Morrissey P, Viney JL. 2002. Helicobacter bilis infection accelerates and H. hepaticus infection delays the development of colitis in multiple drug resistance-deficient (mdr1a−/−) mice. Am J Pathol 160:739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maggio-Price L, Treuting P, Bielefeldt-Ohmann H, Seamons A, Drivdahl R, Zeng W, Lai L, Huycke M, Phelps S, Brabb T, Iritani BM. 2009. Bacterial infection of Smad3–Rag2 double-null mice with transforming growth factor-β dysregulation as a model for studying inflammation-associated colon cancer. Am J Pathol 174:317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maggio-Price L, Treuting P, Zeng W, Tsang M, Bielefeldt-Ohmann H, Iritani BM. 2006. Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res 66:828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. 2008. MDA5 recognition of a murine norovirus. PLoS Pathog 4:e1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller B, Klemm U, Mas Marques A, Schreier E. 2007. Genetic diversity and recombination of murine noroviruses in immunocompromised mice. Arch Virol 152:1709–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mumphrey SM, Changotra H, Moore TN, Heimann-Nichols ER, Wobus CE, Reilly MJ, Moghadamfalahi M, Shukla D, Karst SM. 2007. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J Virol 81:3251–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paik J, Fierce Y, Drivdahl R, Treuting PM, Seamons A, Brabb T, Maggio-Price L. 2010. Effects of murine norovirus infection on a mouse model of diet-induced obesity and insulin resistance. Comp Med 60:189–195 [PMC free article] [PubMed] [Google Scholar]
- 27.Perdue KA, Green KY, Copeland M, Barron E, Mandel M, Faucette LJ, Williams EM, Sosnovtsev SV, Elkins WR, Ward JM. 2007. Naturally occurring murine norovirus infection in a large research institution. J Am Assoc Lab Anim Sci 46:39–45 [PubMed] [Google Scholar]
- 28.Solinas G, Germano G, Mantovani A, Allavena P. 2009. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol 86:1065–1073 [DOI] [PubMed] [Google Scholar]
- 29.Ward JM, Wobus CE, Thackray LB, Erexson CR, Faucette LJ, Belliot G, Barron EL, Sosnovtsev SV, Green KY. 2006. Pathology of immunodeficient mice with naturally occurring murine norovirus infection. Toxicol Pathol 34:708–715 [DOI] [PubMed] [Google Scholar]
- 30.Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2:e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wobus CE, Thackray LB. Virgin HW 4th. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol 80:5104–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeom SC, Yu SA, Choi EY, Lee BC, Lee WJ. 2009. Prevalence of Helicobacter hepaticus, murine norovirus, and Pneumocystis carinii and eradication efficacy of cross-fostering in genetically engineered mice. Exp Anim 58:497–504 [DOI] [PubMed] [Google Scholar]