Abstract
Feline breeding colonies face genetic constraints involving founder effects. A Siamese-founded colony used to study primary congenital glaucoma displayed coat colors additional to the Siamese coat. Genes affecting pigment can exhibit pleiotropy on ocular development and function. To remove potentially confounding phenotypes from our colony, we documented the source and frequency of the Siamese allele at the gene for tyrosinase (TYR), the dilution allele at melanophilin (MLPH), and the brown allele at tyrosinase-related protein 1 (TYRP1). We used PCR–RFLP diagnostics to genotype cats in our colony for the published alleles. A commercially acquired phenotypically normal tom was the source of the dilute allele. A founding Siamese queen was the source of the brown allele. Founders also were blood-typed and screened for disease-associated alleles segregating in Siamese cats at 3 loci (ASB, GLB1, and CEP290). Siamese founders were normal at all loci except ASB, at which both animals carried the hypomorphic allele. Current stock is being managed to limit production of glaucomatous cats with brown, dilute, or Siamese phenotypes or homozygosity for the ASB hypomorphic allele. Genotyping will aid in the elimination of these alleles. The clinical effect of these phenotypes and alleles on the glaucoma phenotype is uncertain, but their elimination will remove potentially confounding effects. In conclusion, when founding a colony, stock should be selected or screened to limit potentially confounding phenotypes. When studying the immune, nervous, and visual systems, screening stock for alleles known to be associated with coat color may be warranted.
Abbreviations: TYR, tyrosinase; MLPH, melanophilin; TYRP1, tyrosinase-related protein 1; PCG, primary congenital glaucoma; ASB, arylsulfatase B; MPS, mucopolysaccharidosis
Nonrodent large-animal genetic disease models are both useful and clinically relevant models of the corresponding human disease, particularly feline models of ophthalmologic disease. Cats are well suited as an eye disease model due to their history as a well-characterized and heavily used visual systems model4,37,44 and because they are a long-lived, relatively outbred, and large-eyed species. Several spontaneous feline models of inherited eye disease are now characterized at the genetic level,34,35 including a model of primary congenital glaucoma (PCG).12,13,28-31,43 This disease appears very similar in description and presentation to a condition reported in Siamese cats in both the United States and Europe and in fact may represent the same condition.1,36 The feline PCG line is maintained in a research breeding colony at Iowa State University. This colony is used as a glaucoma research resource and segregates PCG due to a mutation in latent TGFβ binding protein 2,12 a feline homolog of the mutant protein that causes PCG in humans.2
The foundation of nonrodent large-animal breeding colonies for studying spontaneous genetic diseases frequently includes the cosegregation of additional genetic diseases within the same pedigree.7 The occurrence of additional genetic diseases or traits in such colonies may complicate both the study of the phenotypes of primary interest and animal production. The genes for tyrosinase (TYR), melanophilin (MLPH), and tyrosinase-related protein 1 (TYRP1) all encode proteins of pigmentation (reviewed in reference 42). Two of these loci (TYR and TYRP1) have been associated with conditions affecting normal ocular function: oculocutaneous albinism type IA (OMIM 203100)18 and oculocutaneous albinism type III (OMIM 203290),6 respectively. To date MLPH mutations are associated with a discrete defect affecting only hair color.32 Of specific concern was the potential of genes influencing pigment to affect the phenotype of PCG, as has been described for the TYR and TYRP1 loci of mice.3,5,23 The PCG identified in Siamese cats has been shown to segregate independently of the Siamese coat color trait, and glaucomatous Siamese- and nonSiamese-colored cats appear to have a similar course of disease.13
Our PCG colony was founded in 2004 by using PCG-affected Siamese cats and phenotypically normal cats from a commercial source. Since that time, we have noted brown and gray hair coats segregating within our colony, ostensibly due to mutations in the feline TYRP1 and MLPH genes that were introduced by founders of the colony. The presence of 3 coat-color variants (Figure 1 B through D) in our PCG colony was a concern, because exhaustive studies to determine definitively the influence of coat and eye color on PCG phenotype were beyond the scope of our ongoing program of glaucoma research. For practical reasons, we therefore elected to characterize this colony at the molecular level for the feline mutations known to cause these traits22,25,26,41 and subsequently use these genotypes to limit the future production of PCG animals to those with ‘normal’ eye and coat color. Herein we report our investigation of the Siamese (cs) allele at TYR,45 the brown (b) allele at TYRP1,39 and the dilution allele (d) at MLPH11 within our PCG colony. Animals were genotyped by using novel (MLPH) or modified (TYR and TYRP1) PCR–RFLP-based assays. Using these diagnostics, we documented the introduction of these 3 alleles into the colony and have developed a simple screening test for these alleles that can be used to limit the associated phenotypes in our colony and to aid in characterizing the coat color genetics in other feline colonies. In addition, founding animals were screened and genotyped for blood type and at loci with disease-associated alleles known to segregate in Siamese cats.
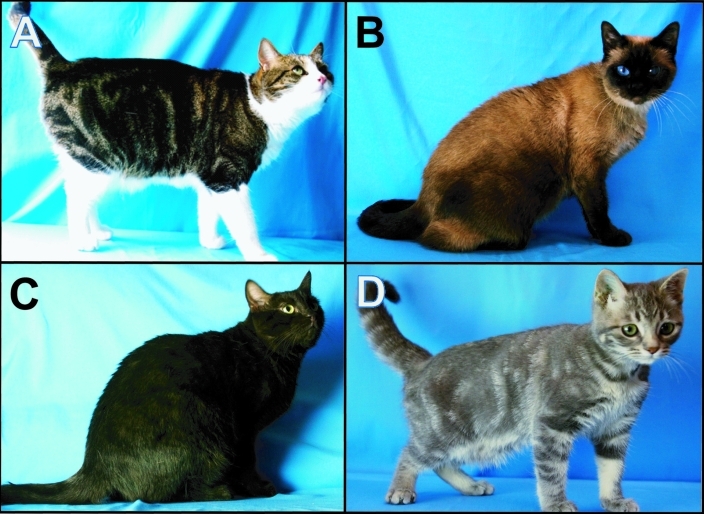
Figure 1.
Photos of the (A) phenotypically normal founding tom and (B–D) 3 coat-color variants in the PCG colony. (B) Siamese coat coloration. (C) The brown coat coloration in a nonagouti animal. (D) The gray, color-dilute coat coloration on a blotched tabby animal.
Materials and Methods
Cats used in this study were produced and housed at Iowa State University (Ames, Iowa). Animals and animal samples were generated by using approved IACUC protocols. DNA samples from blood and tissue were extracted by using a standard phenol–chloroform protocol.40
The founding glaucomatous Siamese cats were acquired from a private owner, and normal (nonglaucomatous) founder cats were obtained from a commercial laboratory animal supplier (Harlan Laboratories, Indianapolis, IN). All normal founding stock was blood-typed (Clinical Pathology Laboratory, Iowa State University), and all founders tested negative on consecutive ELISA tests for feline leukemia virus and feline immunodeficiency virus (Clinical Pathology Laboratory, Iowa State University). In addition, all founders were genotyped for the CMAH b allele that is associated with the feline B blood type. CMAH exon 2 was PCR-amplified by using the published forward primer6 and a new reverse primer (5′ CCC CTG AGA GAG AGG AAA CC 3′) that was designed by using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). The resultant amplicon includes the G139A site that is consistent with the A and b CMAH alleles. Each PCR reaction (20 µL) used 1× buffer (Denville Scientific, Metuchen, NJ), 0.2 mM each dNTP, 1 µM each primers, and 0.1 U Taq polymerase (Denville). PCR conditions were initial denaturation at 95 °C for 5 min; followed by 35 cycles of 95 °C for 45 s, 63 °C for 30 s, and 72 °C for 45 s; and a final extension step of 72 °C for 10 min. PCR product purification, sequencing, and alignment was performed as described.17 Siamese founders were genotyped (Section of Medical Genetics, University of Pennsylvania School of Veterinary Medicine, Philadelphia, PA) for the severe feline mucopolysaccharidosis type VI (MPS VI) allele (L476P) of the feline arylsulfatase B (ASB) gene.10 The Siamese founders also were genotyped for the D520N hypomorphic allele of the ASB gene, which only causes disease when found in combination with the L476P allele,10 for the feline GM1 gangliosidosis allele at the feline GLB1 locus,27 and for the retinal degeneration in Abyssinian and Somali cats (rdAc) allele of the feline CEP290 gene,34 according to the published protocols. PCR amplicons generated from affected animal samples were sequenced (Core Sequencing Facility, Iowa State University) to confirm that the mutations of target genes that were segregating in our colony were identical to those previously published.
The TYR diagnostic (Figure 2) was generated based on the published PCR assay and primers for exon 2,26 with the exception that the primer used was corrected for a slight error relative to the published primer's sequence. Comparing the forward primer with the cited sequence of the Felis catus tyrosinase gene (GenBank accession nos. AY012029 and U40716) revealed a typographic error: the published forward primer lacked a guanine residue at primer position 16 from the 5′ end, when compared with referenced accession sequence. We added the missing guanine residue to our forward primer; the reverse primer was not altered. These primers amplify a region of exon 2 containing a point mutation (nucleic acid, 940G>A; protein, G302R); this mutation obliterates an HpaII restriction site, which when present results in amplicon digestion into 62 and 117 bp fragments. PCR amplification was performed in a 20-μL reaction volume with final concentrations of 2.0 mM MgCl2, 0.2 mM each dNTP, 0.63 μM each primer, 1 × of supplied buffer, 0.8 U GoTaq Flexi DNA polymerase (Promega, Madison, WI), and approximately 5 to 50 ng genomic template DNA in a Techne Thermal Cycler (model no. TC312, MidSci, St Louis, MO) by using the published profile.26 Resultant amplicons were digested with 5 U HpaII in supplied 1× buffer 1 (New England Biolabs, Ipswich, MA) in a 25-μL volume for 3 h at 37 °C. Reaction products were separated electrophoretically and assessed on 3% agarose gels (Figure 2).
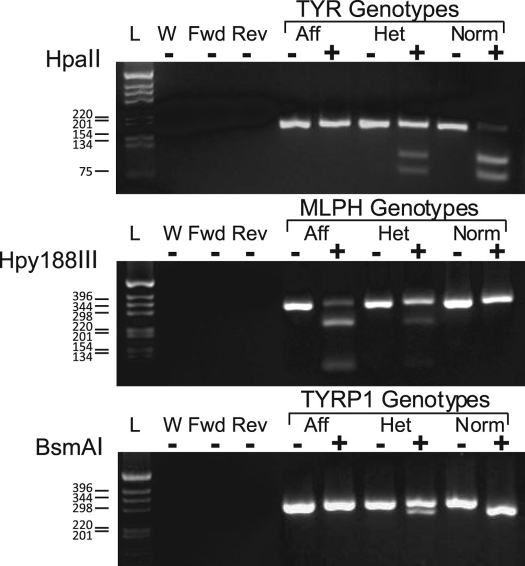
Figure 2.
Scheme of the TYR, MLPH, and TYRP1 PCR–RFLP diagnostics. The images are of the electrophoresis gels of the PCR amplicons and resultant fragments. Control PCR reactions included: complete reaction mixtures minus DNA template (W), complete reactions with only the forward (Fwd) or reverse (Rev) primer, and control DNA samples from known affected (Aff), carrier (Het), and normal (Norm) cats. For each representative genotyped, there are 2 lanes: 1 containing the uncut PCR product (–) and the other the PCR product cut with the enzyme indicated at the left of the illustration (+). On the left is the DNA ladder with ladder marker sizes indicated.
We developed the MLPH diagnostic by using a published22 single-nucleotide deletion in exon 2 (nucleic acid, 83delT; protein, L28RfsX11) that generates a frameshift, introduces a premature stop codon and an Hpy188III restriction site, and leads to truncation of the predicted protein. Primers (forward, 5′ TGA CAG GCA GAG ATG GGG AAA AA 3′; reverse, 5′ GGA ATG CAG GCT GGG GAG TCG 3′) were designed by using Lasergene (version 7.2, DNASTAR, Madison, WI). The reaction was performed as described for TYR, with substitution of the MLPH primers and a reaction profile of initial denaturation at 95 °C for 10 min; 40 cycles of 98 °C for 10 s, 65 °C for 30 s, and 72 °C for 1 min; final extension at 72 °C for 15 min; and cooling to 4 °C for 5 min. This reaction yielded a 323-bp product, which was digested with 5 U Hpy188III in 1× digestion buffer 4 (New England Biolabs) in 30 μL at 37 °C for 4 h. This digestion resulted in predicted fragment lengths of 227 and 96 bp for the dilute allele (Figure 2).
The TYRP1 diagnostic also was based on the published assay and primers.25 The mutation occurs in intron 6 at the exon 6 downstream splice–donor recognition signal (c. 1262+5G>A), leading to large and variable inframe insertions (c.1262ins51/54) and introduction of a BsmAI restriction site.25,41 The PCR reaction used the same components and concentrations as listed for MLPH, with substitution of the TYRP1 primers and a reaction profile of denaturation at 95 °C for 10 min; 30 cycles of 95 °C for 30 s, 60 °C for 30 s decreasing by 0.5 °C every cycle, and 72 °C for 30 s; 20 cycles of 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s; final extension at 72 °C for 15 min; and cooling to 4 °C for 10 min. This procedure yielded a predicted 318-bp amplicon, which was digested with 5 U BsmAI (New England Biolabs) in 25 μL for 3 h at 55 °C, resulting in predicted fragments of 268 and 50 bp for normal (nonbrown) animals (Figure 2).
Results
The normal founding cats for our PCG colony were shown to have blood type A, and all founders were found to be free of the b allele at CMAH, except for the normal tom, which was heterozygotic at this locus. Further genotyping of all current breeders identified 2 queens that were carriers for the b allele at CMAH (Figure 3 C). Genotyping of the Siamese founders revealed them to be neither affected by nor carriers for MPS VI, feline GM1 gangliosidosis, or retinal degeneration (the rdAc mutation). The Siamese founders were both heterozygous for the D520N allele at ASB, which is an allele not documented to cause disease on its own.10 Sequencing of the PCR amplicons from samples of affected coat-color variants within our colony confirmed the presence of the published mutations in TYR, TYRP1, and MLPH within our colony.22,25,41 On the basis of pedigree analysis, the glaucomatous Siamese queen and the ostensibly normal (nonglaucomatous) tabby tom were implicated as potential sources of the brown and dilute alleles within our PCG feline colony (Figure 3 A and B). Genotyping confirmed these assumptions: the Siamese queen was the source of the brown allele, and the normal tom was the source of the dilute allele. The introduction of the Siamese allele by Siamese founder stock was self-evident.
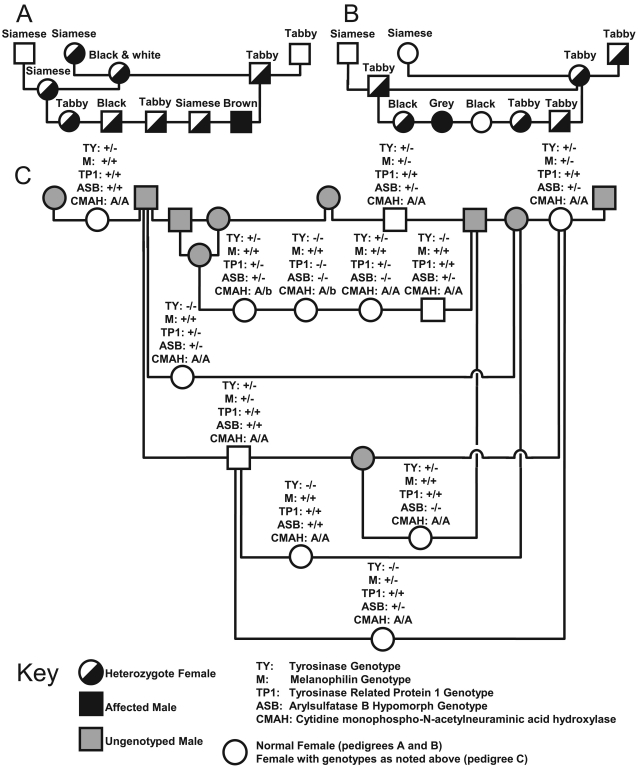
Figure 3.
Pedigrees detailing the history and segregation of the brown and dilute alleles within the feline PCG colony and the pedigree of the current breeding animals within the PCG colony. The key contains information on the sex and genotype of individual presented. Each animal within the (A) brown and (B) dilute pedigrees is listed by their coat color phenotype with their genotype indicated symbolically as indicated by the key; genotyping for pedigree A assessed TYRP1, and that for pedigree B used MLPH. Pedigree A clearly implicates the founding Siamese queen as the source of the brown allele, and pedigree B clearly implicates the commercial tom as the source of the dilute allele within the colony. Animals in pedigree C are current breeding stock. In pedigree C, the shaded symbols indicate animals that are no longer used in the breeding program, which therefore were not genotyped, whereas open symbols represent current breeding stock with their genotypes indicated in text above the symbol. The frequencies of the Siamese, brown, dilute, and D520N alleles within the colony are 77%, 23%, 18%, and 9%, respectively.
With confirmation of the path of introduction of the alleles in question, current breeding animals in the feline colony were genotyped to screen for the presence of the Siamese, brown, dilute, and D520N alleles. Of the 14 cats used in the breeding program, 12, 4, 5, and 9 cats were found to segregating the Siamese, brown, dilute, and D520N alleles, respectively. Overall, allele frequencies within the current colony stock were 61% (TYR), 18% (TYRP1), 18% (MLPH), and 43% (D520N; Figure 3 C).
Discussion
The creation of a research breeding colony for a single disease of interest can often be complicated by the presence of confounding phenotypes. Indeed, nonrodent large-animal genetic disease research colonies often segregate multiple genetic diseases, as recently reported and reviewed.7 This phenomenon is not uncommon, as demonstrated by the feline model of severe MPS VI (2 allelic disorders), 10 canine model of MPS I (2 disorders),7 Brittany spaniel model of spinal muscular atrophy (3 disorders),8,24,38 and giant schnauzer model of canine Imerslund–Gräsbeck syndrome (3 disorders).14-16,21 We screened our feline PCG colony for known disease-causing alleles identified in, or known to segregate in, Siamese cats, namely those alleles causing or contributing to feline MPS VI (both the severe and attenuated forms), feline GM1 gangliosidosis, and rdAc.9,10,33,34 Of particular concern were the D520N allele, which has an allelic frequency of greater than 11%, and the rdAc allele, which occurs in Siamese at an allelic frequency exceeding 26%.9,33 Of these alleles of concern, only the D520N allele of ASB was present in our colony. No clinical phenotype is associated exclusively with the D520N allele, for which the colony founders were heterozygous. This allele causes attenuated MPS VI only in cats also carrying the severe allele at ASB. Homozygosity for the D520N allele has thus far only been associated with variable WBC granulation and mild, occasional inclusions seen histologically in chondrocytes.10 This allele has been speculated, but not substantiated, to lead to late-age onset of arthritides.10 Nevertheless, we are eliminating the D520N allele from the colony to avoid potential confounding effects on ocular phenotype. Founders of the colony were blood-typed to avoid the complication of neonatal isoerythrolysis, which can result from breeding animals discordant for A and B blood types.20 Subsequent molecular testing predicted the blood type of all founders to be A, although the normal tom was a carrier for the b allele, which persists at a low and manageable frequency in the current breeding stock. These findings are consistent with the observation that B blood type has not been documented in Siamese cats.19
Our PCG feline colony is an example of the introduction of potentially confounding phenotypes, by a founder effect, into a colony created to study a single disease of interest. In the case of the tyrosinase allele, we were aware from the onset of the need to limit this genotype, given the locus involved and its known effect on eye phenotype. In contrast, we were unaware of the brown and dilute alleles in our colony, which were introduced by carriers. In retrospect, had we anticipated the need, we could have tested the founders and F1 crosses (if needed) to fully limit the penetration of these alleles within the pedigree. Through this study we were able to confirm by sequencing and restriction enzyme digests that the mutations leading to the Siamese, brown, and dilute coat coloration observed within our colony were consistent with the published mutations.22,25,26,41 We first confirmed this finding by sequencing, because colony pedigree data were unclear regarding whether the brown and dilute alleles originated with the female Siamese or male commercially acquired cat. Conceivably one of the alleles might have been novel, especially if the brown allele had been associated with the normal cat, because the published brown allele has not been documented in nonpedigreed or randombred cats. A novel allele was less likely for the dilute allele, which is well recognized in the nonpedigreed–randombred cat population.22,25,41 Using published and newly developed protocols, we were able to create reliable PCR–RFLP diagnostics based on unique restriction sites that identify carrier animals and trace the introduction of the brown allele to the founding Siamese queen and the dilute allele to the phenotypically normal, commercially acquired tom.
Identification of carrier cats will allow us eventually to eliminate the Siamese, brown, dilute, and D520N alleles from the breeding colony. We have already documented that the inheritance and phenotype of feline PCG is not influenced by the Siamese coat phenotype. We would not predict prima facie that the brown or dilute phenotypes would influence the feline PCG phenotype, but studies in mice indicate a need for caution in making such an assumption.3,5,23 Furthermore, we cannot reliably model or predict the effect of the 3 coat-color phenotypes in combination on the feline PCG phenotype. The testing presented here will allow selective breeding of current stock of known genotype and the selection of additional fully homozygous, normally pigmented animals as future breeding stock. These practices will ensure a more thoroughly genetically defined and phenotypically homogeneous nonrodent large-animal model of glaucoma. In most cases, we pursued molecular diagnostics using inhouse testing due to efficiency concerns and because of the need for ongoing testing for the coat colors we knew were segregating within the colony. Other research institutions could very easily access commercial providers of these molecular tests for one-time screening purposes.
In conclusion, founder animals should be screened appropriately when establishing research colonies. Blood-type screening should be considered for any cats except those breeds in which the B blood type is not recognized. Breed risk should influence the testing of disease-causing alleles; in the current study, we focused on disease alleles known to occur at high frequency in Siamese cats. In light of the pleiotropic effects of various genes involved in pigment production on the development and function of the visual, nervous, and immune systems, screening founding cats for coat color alleles is prudent. Although the 3 phenotypes of concern have not been implicated individually as confounding factors, one cannot discount that their combination may lead to unanticipated effects. In addition, the alleles in question are not uncommon, as evidenced by our current findings. The PCR–RFLP diagnostics that we used or developed can facilitate such screening. Finally, these diagnostic resources can be accessed by laboratory animal resource facilities and are available to community cat breeders who may wish to select for desirable coat-color traits.
Acknowledgments
All laboratory analysis was conducted at the Department of Animal Science (Iowa State University) or Department of Population Health and Reproduction (University of California–Davis)
We acknowledge foundation and grant support from The Center for Integrated Animal Genomics (NME and GJM), the Merck–Merial Summer Scholars Fellowship Program (MMR-M, SAC), The Glaucoma Research Foundation (MHK and GJM), The Glaucoma Foundation (MHK), and the NIH (5K08EY018609 to GJM and P30EY016665 to CR Brandt). We thank Leslie A Lyons (University of California–Davis) for assistance in CMAH genotyping, photographer Michaela Auffart, and a cadre of devoted, talented, and caring ISU undergraduate students, whose work and dedication makes possible the continued maintenance of this breeding colony.
References
- 1.Aguirre GD, Bistner SI. 1973. Microphakia with lenticular luxation and subluxation in cats. Vet Med Small Anim Clin 68:498–500 [PubMed] [Google Scholar]
- 2.Ali M, McKibbin M, Booth A, Parry DA, Jain P, Riazuddin SA, Hejtmancik JF, Khan SN, Firasat S, Shires M, Gilmour DF, Towns K, Murphy AL, Azmanov D, Tournev I, Cherninkova S, Jafri H, Raashid Y, Toomes C, Craig J, Mackey DA, Kalaydjieva L, Riazuddin S, Inglehearn CF. 2009. Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet 84:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson MG, Smith RS, Hawes NL, Zabaleta A, Chang B, Wiggs JL, John SW. 2002. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet 30:81–85 [DOI] [PubMed] [Google Scholar]
- 4.Bahn CF, Meyer RF, MacCallum DK, Lillie JH, Lovett EJ, Sugar A, Martonyi CL. 1982. Penetrating keratoplasty in the cat. A clinically applicable model. Ophthalmology 89:687–699 [DOI] [PubMed] [Google Scholar]
- 5.Bidinost C, Hernandez N, Edward DP, Al-Rajhi A, Lewis RA, Lupski JR, Stockton DW, Bejjani BA. 2006. Of mice and men: tyrosinase modification of congenital glaucoma in mice but not in humans. Invest Ophthalmol Vis Sci 47:1486–1490 [DOI] [PubMed] [Google Scholar]
- 6.Boissy RE, Zhao H, Oetting WS, Austin LM, Wildenberg SC, Boissy YL, Zhao Y, Sturm RA, Hearing VJ, King RA, Nordlund JJ. 1996. Mutation in and lack of expression of tyrosinase-related protein 1 (TRP1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as ‘OCA3.’ Am J Hum Genet 58:1145–1156 [PMC free article] [PubMed] [Google Scholar]
- 7.Carlstrom LP, Jens JK, Dobyns M, Passage M, Dickson PI, Ellinwood NM. 2009. Inadvertent Propagation of factor VII deficiency in a canine mucopolysaccharidosis type I research breeding colony. Comp Med 59:378–382 [PMC free article] [PubMed] [Google Scholar]
- 8.Cork LC, Griffin JW, Munnell JF, Lorenz MD, Adams RJ. 1979. Hereditary canine spinal muscular atrophy. J Neuropathol Exp Neurol 38:209–221 [DOI] [PubMed] [Google Scholar]
- 9.Crawley AC, Muntz FH, Haskins ME, Jones BR, Hopwood JJ. 2003. Prevalence of mucopolysaccharidosis type VI mutations in Siamese cats. J Vet Intern Med 17:495–498 [DOI] [PubMed] [Google Scholar]
- 10.Crawley AC, Yogalingam G, Muller VJ, Hopwood JJ. 1998. Two mutations within a feline mucopolysaccharidosis type VI colony cause 3 different clinical phenotypes. J Clin Invest 101:109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doncaster L. 1904. On the inheritance of tortoiseshell and related colours in cats. Proc Camb Philol Soc 13:35–38 [Google Scholar]
- 12.Ellinwood NM, Deckman KH, Zhao Z, Rutz-Mendicino MM, Jens JK, David VA, Kuehn MH, O'Brien SJ, Menotti-Raymond MA, McLellan GJ. [Internet] 2010. Candidate gene analysis of a feline model of primary congenital glaucoma implicates LTBP2 as the causative locus. [Cited 12 May 2011]. Available at: http://www.iovs.org/gca?allch=&submit=Go&gca=iovsmtg%3B51%2F5%2F6390
- 13.Ellinwood NM, Jens JK, Rutz MM, Faylon MP, Snella EM, Kuehn MH, McLellan GJ. [Internet] 2009. Preliminary characterization of the genetics and inheritance of primary congenital glaucoma in a spontaneous feline model. [Cited 12 May 2011]. Available at: http://www.iovs.org/gca?allch=&submit=Go&gca=iovsmtg%3B50%2F5%2F883
- 14.Fyfe JC, Al-Temaimi RA, Castellani R, Rosenstein D, Goldowitz D, Henthorn PS. 2010. Inherited neuroaxonal dystrophy in dogs causing lethal, fetal-onset motor system dysfunction and cerebellar hypoplasia. J Comp Neurol 518:3771–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fyfe JC, Giger U, Hall CA, Jezyk PF, Klumpp SA, Levine JS, Patterson DF. 1991. Inherited selective intestinal cobalamin malabsorption and cobalamin deficiency in dogs. Pediatr Res 29:24–31 [DOI] [PubMed] [Google Scholar]
- 16.Fyfe JC, Lassaline ME, Graham PA. Canine congenital hypothyrotropinism, p 553–555 Proceedings of the 19th Annual Veterinary Medical Forum of the American College of Veterinary Internal Medicine; Denver, CO [Google Scholar]
- 17.Gandolfi B, Outerbridge CA, Beresford LG, Myers JA, Pimentel M, Alhaddad H, Grahn JC, Grahn RA, Lyons LA. 2010. The naked truth: sphynx and devon Rex cat breed mutations in KRT71. Mamm Genome 21:509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giebel LB, Strunk KM, King RA, Hanifin JM, Spritz RA. 1990. A frequent tyrosinase gene mutation in classic, tyrosinase-negative (type IA) oculocutaneous albinism. Proc Natl Acad Sci USA 87:3255–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giger U, Bucheler J, Patterson DF. 1991. Frequency and inheritance of A and B blood types in feline breeds of the United States. J Hered 82:15–20 [DOI] [PubMed] [Google Scholar]
- 20.Giger U, Casal ML. 1997. Feline colostrum—friend or foe: maternal antibodies in queens and kittens. J Reprod Fertil Suppl 51:313–316 [PubMed] [Google Scholar]
- 21.Greco DS, Feldman EC, Peterson ME, Turner JL, Hodges CM, Shipman LW. 1991. Congenital hypothyroid dwarfism in a family of giant schnauzers. J Vet Intern Med 5:57–65 [DOI] [PubMed] [Google Scholar]
- 22.Ishida Y, David VA, Eizirik E, Schaffer AA, Neelam BA, Roelke ME, Hannah SS, O'Brien SJ, Menotti-Raymond M. 2006. A homozygous single-base deletion in MLPH causes the dilute coat color phenotype in the domestic cat. Genomics 88:698–705 [DOI] [PubMed] [Google Scholar]
- 23.Libby RT, Smith RS, Savinova OV, Zabaleta A, Martin JE, Gonzalez FJ, John SW. 2003. Modification of ocular defects in mouse developmental glaucoma models by tyrosinase. Science 299:1578–1581 [DOI] [PubMed] [Google Scholar]
- 24.Lorenz MD, Cork LC, Griffin JW, Adams RJ, Price DL. 1979. Hereditary spinal muscular atrophy in Brittany spaniels: clinical manifestations. J Am Vet Med Assoc 175:833–839 [PubMed] [Google Scholar]
- 25.Lyons LA, Foe IT, Rah HC, Grahn RA. 2005. Chocolate-coated cats: TYRP1 mutations for brown color in domestic cats. Mamm Genome 16:356–366 [DOI] [PubMed] [Google Scholar]
- 26.Lyons LA, Imes DL, Rah HC, Grahn RA. 2005. Tyrosinase mutations associated with Siamese and Burmese patterns in the domestic cat (Felis catus). Anim Genet 36:119–126 [DOI] [PubMed] [Google Scholar]
- 27.Martin DR, Rigat BA, Foureman P, Varadarajan GS, Hwang M, Krum BK, Smith BF, Callahan JW, Mahuran DJ, Baker HJ. 2008. Molecular consequences of the pathogenic mutation in feline GM1 gangliosidosis. Mol Genet Metab 94:212–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLellan GJ, Betts DM, Sigle K, Grozdanic S. [Internet] 2005. Congenital glaucoma in the Siamese cat—a novel spontaneous animal model for glaucoma research. [Cited 12 May 2011]. Available at: http://www.iovs.org/gca?allch=&submit=Go&gca=iovsmtg%3B46%2F5%2F134
- 29.McLellan GJ, Kim CBY, Seo K, Heyne GW, Ver Hoeve JN. [Internet] 2010. Pattern ERG deficits in a feline model of primary congenital glaucoma. [Cited 12 May 2011]. Available at: http://www.iovs.org/gca?allch=&submit=Go&gca=iovsmtg%3B51%2F5%2F5482
- 30.McLellan GJ, Kuehn MH, Ellinwood NM, Kim CY, Jens J, Sigle K, Petersen C. [Internet] 2006. A feline model of primary congenital glaucoma—histopathological and genetic characterization. [Cited 12 May 2011]. Available at: http://www.iovs.org/gca?allch=&submit=Go&gca=iovsmtg%3B47%2F5%2F175
- 31.McLellan GJ, Lin TL, Hildreth S, Petersen C, Leon A, Jens JK, Ellinwood NM. [Internet] 2009. Diurnal intraocular pressure and response to topically administered 1% brinzolamide in a spontaneous feline model of primary congenital glaucoma. [Cited 12 May 2011]. Available at: http://www.iovs.org/gca?allch=&submit=Go&gca=iovsmtg%3B50%2F5%2F4059
- 32.Menasche G, Ho CH, Sanal O, Feldmann J, Tezcan I, Ersoy F, Houdusse A, Fischer A, de Saint Basile G. 2003. Griscelli syndrome restricted to hypopigmentation results from a melanophilin defect (GS3) or a MYO5AF–exon deletion (GS1). J Clin Invest 112:450–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menotti-Raymond M, David VA, Pflueger S, Roelke ME, Kehler J, O'Brien SJ, Narfstrom K. 2010. Widespread retinal degenerative disease mutation (rdAc) discovered among a large number of popular cat breeds. Vet J 186:32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menotti-Raymond M, David VA, Schaffer AA, Stephens R, Wells D, Kumar-Singh R, O'Brien SJ, Narfstrom K. 2007. Mutation in CEP290 discovered for cat model of human retinal degeneration. J Hered 98:211–220 [DOI] [PubMed] [Google Scholar]
- 35.Menotti-Raymond M, Deckman KH, David V, Myrkalo J, O'Brien SJ, Narfstrom K. 2010. Mutation discovered in a feline model of human congenital retinal blinding disease. Invest Ophthalmol Vis Sci 51:2852–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molleda JM, Martin E, Ginel PJ, Novales M, Moreno P, Lopez R. 1995. Microphakia associated with lens luxation in the cat. J Am Anim Hosp Assoc 31:209–212 [DOI] [PubMed] [Google Scholar]
- 37.Narfstrom K, Nilsson SE. 1987. Hereditary rod–cone degeneration in a strain of Abyssinian cats. Prog Clin Biol Res 247:349–368 [PubMed] [Google Scholar]
- 38.Richtsmeier JT, Sack GH, Jr, Grausz HM, Cork LC. 1994. Cleft palate with autosomal recessive transmission in Brittany spaniels. Cleft Palate Craniofac J 31:364–371 [DOI] [PubMed] [Google Scholar]
- 39.Robinson R. 1977. Genetics for cat breeders, 2nd ed, p 107–108New York (NY): Pergamon Press [Google Scholar]
- 40.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. [Google Scholar]
- 41.Schmidt-Kuntzel A, Eizirik E, O'Brien SJ, Menotti-Raymond M. 2005. Tyrosinase and tyrosinase-related protein 1 alleles specify domestic cat coat-color phenotypes of the albino and brown loci. J Hered 96:289–301 [DOI] [PubMed] [Google Scholar]
- 42.Schmutz SM, Berryere TG. 2007. Genes affecting coat colour and pattern in domestic dogs: a review. Anim Genet 38:539–549 [DOI] [PubMed] [Google Scholar]
- 43.Seo K, Rasmussen CA, Finch AK, Xiong K, Kaufman PL, McLellan GJ. [Internet] 2010. SD-OCT imaging of the retina and optic nerve in normal and glaucomatous cats. Association for Research in Vision and Ophthalmology 2010 Annual Meeting. [Cited 12 May 2011]. Available at: http://www.iovs.org/gca?allch=&submit=Go&gca=iovsmtg%3B51%2F5%2F2138
- 44.Van Sluyters RC. 1978. Recovery from monocular stimulus deprivation amblyopia in the kitten. Ophthalmology 85:478–488 [DOI] [PubMed] [Google Scholar]
- 45.Wright S. 1918. Color inheritance in mammals, X. The cat. J Hered 9:139–144 [Google Scholar]