Abstract
Pseudogenes, resulting from duplications of functional genes, contribute to the functional complexity of their parental genes. The glucocerebrosidase gene (GBA), located in a gene-rich region on chromosome 1q 21, is mutated in Gaucher disease. The presence of contiguous, highly homologous pseudogenes for both GBA and metaxin 1 at this locus increases the likelihood of DNA rearrangement. We describe a facile method to identify and analyze recombinant alleles in patients with Gaucher disease. Genomic DNA from 20 patients with recombinant GBA alleles and five controls was evaluated to identify DNA rearrangements or copy number variation using six probes specific for either the GBA gene or pseudogene. Quantitative real-time PCR was performed on genomic DNA, and Southern blot analyses using HincII together with sequencing confirmed the real-time results. Both GBA fusions and duplications could be detected. Different sites of crossover were identified, and alleles resulting from gene conversion could be distinguished from reciprocal recombinant alleles. Quantitative real-time PCR is a sensitive and rapid method to detect fusions and duplications in patients with recombinant GBA alleles. This technique is more sensitive, faster, and cheaper than Southern blot analysis, and can be used in diagnostic laboratories, and to detect other recombinant alleles within the genome.
DNA rearrangement, occurring by chromosomal pairing, breakage, and crossover between two non-sister chromatids during meiosis, is a cellular process essential for genetic diversity. Recombination between homologous sequences may also introduce harmful mutations. DNA rearrangement is influenced by the degree of sequence homology between corresponding segments, and can be prompted by the presence of a pseudogene.
Gaucher disease (GD) (type 1: OMIM #230800, type 2: OMIM #230900, type 3: OMIM #2301000), the most common lipidosis and the most frequently inherited disorder among Ashkenazi Jewish individuals, presents with hepatosplenomegaly, anemia, thrombocytopenia, bone involvement, and neurological symptoms (type 2 and 3). This disorder is caused by the deficiency of the lysosomal enzyme glucocerebrosidase, which degrades glucocerebroside, a membrane glycolipid, to ceramide and glucose. The gene for glucocerebrosidase (GBA) (OMIM #606463) is located on 1q21, a gene-rich region encompassing seven genes and two pseudogenes within 85 kb.1–3 GBA, spanning 7.6 kb, has 11 exons and 10 introns. A highly homologous 5.7-kb pseudogene (GBAP) is located 16 kb downstream, with the same organization of exons and introns.3 Another gene, metaxin (MTX), encoding for a mitochondrial membrane protein, is convergently transcribed and located immediately downstream of GBAP, with a pseudogene (pMTX) located between GBA and GBAP (Figure 1A).4,5
Figure 1.
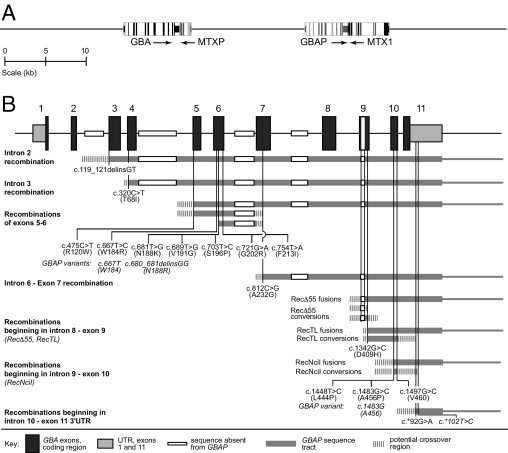
A: The location of glucocerebrosidase gene (GBA) and its pseudogene, metaxin and metaxin pseudogene on chromosome 1. B: Exonic and intronic structure of GBA, pseudogene-derived mismatches, and amino acid changes associated with the allele are indicated. Typical nonreciprocal (gene conversion) and reciprocal (gene fusion or duplication) recombinants are shown. The sequences absent in pseudogene are also shown.
GBA and GBAP share more than 96% exonic sequence homology, enhancing the likelihood of homologous recombination.6 Such recombinations can result in deletions, duplications, inversions, and fusions leading to genetic disorders.7 Recombination events between GBAP and GBA or MTX and MTXP are responsible for several different identified mutant alleles.8,9 Crossover has been reported at sites ranging from intron 2 in GBA to MTX (Figure 1B), causing both reciprocal and nonreciprocal recombination.8
To accurately describe a recombinant allele, a combination of direct sequencing and Southern blot analyses, often performed using several restriction enzymes, has been required.8 Since both of these techniques are time consuming and expensive, they are not commonly used in diagnostic laboratories, and the detection of recombinant alleles in diagnostic settings has been challenging. We report the optimization of a facile method to identify and analyze recombinant alleles in patients with GD using quantitative real-time PCR (real-time qPCR).
Materials and Methods
Sample Preparation
DNA samples collected under National Institutes of Health institutional review board–approved protocols from 20 patients with recombinant GBA alleles and from five controls were extracted following standard protocols.
Real-Time Analysis
Six sets of primers and probes were designed targeting sequence present in the GBA gene, but absent from GBAP, synthesized by Applied Biosystems (Table 1) to detect any variation in the gene structure caused by a partial or complete deletion or duplication (Applied Biosystems, Foster City, CA). Six additional primer sets and probes were designed targeting GBAP. Each GBA/GBAP probe was labeled with FAM. A labeled β-globin probe was co-amplified as an internal control.
Table 1.
GBA and GBAP primer and probe sequences
| GBA | GBAP |
|---|---|
| Intron 2 | Intron 2 |
| Probe: 5′-CCCAGCTAATTTTTGTATTT-3′ | Probe: 5′-CAGCTGTGTCCGTTCT-3′ |
| Forward primer: 5′-ACAGGCGGCCACCACTAC-3′ | Forward primer: 5′-TGAGGATTTTTGAAACCGTCTTC-3′ |
| Reverse primer: 5′-CATGGTGAAACCCCGTCTCTA-3′ | Reverse primer: 5′-GGTGAGGTCTGACAAGGATGTG-3′ |
| Intron 4 | Intron 4 |
| Probe: 5′-ATGATATACCTGCCTTGGC-3′ | Probe: 5′-TCTTTGCCAATTCTTT-3′ |
| Forward primer: 5′-CATGTTGACCAGGCTAGTCTTAAACTC-3′ | Forward primer: 5′-GGATGACATAGAGCCACTGACACT-3′ |
| Reverse primer: 5′-CGGTGGCTCACACTTGTAATTC-3′ | Reverse primer: 5′-GGTGAGGTCTGACAAGGATGTG-3′ |
| Intron 6 | Intron 6 |
| Probe: 5′-CTGCCTCCTGAGTTCA-3′ | Probe: 5′-TTCTCTTGCCAGGTTC-3′ |
| Forward primer: 5′-CGCCATCTTCACTCACTGTAACC-3′ | Forward primer: 5′-TCGGTCTGGCCCACTTTCT-3′ |
| Reverse primer: 5′-AAGGCTGAGGCAGGAGAATTG-3′ | Reverse primer: 5′-GTGCTCAGCATAGGCATCCA-3′ |
| Intron 7 | Intron 7 |
| Probe: 5′-CACTGCACTCCAGCC-3′ | Probe: 5′-TTCTGGAGGCTAAGGC-3′ |
| Forward primer: 5′-TTGCGGTGAGCCGAGATC-3′ | Forward primer: 5′-GGCACGCCTATAGTCCCAAGT-3′ |
| Reverse primer: 5′-CCCCCAGCCTAGATAGTTTTGTT-3′ | Reverse primer: 5′-CCTGAAATTCTAGGCTCAACTAAACCT-3′ |
| Exon 9 | Exon 9 |
| Probe: 5′-ACAGTCCCATCATTG-3′ | Probe: 5′-AGACATCACCAAGCAC-3′ |
| Forward primer: 5′-CAATTGGGTGCGTAACTTTGTC-3′ | Forward primer: 5′-CCGACTGGAACCCATCATTG-3′ |
| Reverse primer: 5′-AAAACGTGTCCTTGGTGATGTCT-3′ | Reverse primer: 5′-GAACATGGGCTGTTTGTAAAACG-3′ |
| 3′ UTR (pMTX) | 3′ UTR (MTX) |
| Probe: 5′-CCCATTGAGAAGAAGG-3′ | Probe: 5′-AAGGAAGGCTCCGCC-3′ |
| Forward primer: 5′-CCCGCTGGCCGTCAA-3′ | Forward primer: 5′-GTCCAGCGCCCAATGC-3′ |
| Reverse primer: 5′-TTGCGGCCCAACCTTTC-3′ | Reverse primer: 5′-TTCTGGGAGCTGCTCCTAGTTT-3′ |
| β-Globin | |
| Probe: 5′-CTCATGGCAAGAAAGTGCTCGGTGC-3′ | |
| Forward primer: 5′-TGGGCAACCCTAAGGTGAAG-3′ | |
| Reverse primer: 5′-GTGAGCCAGGCCATCACTAAA-3′ | |
Quantitative real-time PCR reactions were optimized and performed using an Applied Biosystems StepOnePlus real-time PCR System. Reactions in a 20-μL volume included 10 μL of TaqMan Universal PCR Master mix (Applied Biosystems), 0.5 μL of each probe (10 μmol/L), 0.5 μL of β-globin probe (10 μmol/L), 1.8 μL of primer mix for each target (20 μmol/L), 1.8 μL of β-globin primer mix (20 μmol/L), and 10 ng of genomic DNA. Real-time PCR was performed with one cycle at 50°C for 2 minutes, one cycle at 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. Three replicates were used for each sample, and each reaction was performed twice.
The normal range for relative quantitation (RQ) was determined based on 2-ΔΔCt for each allele, using 10 samples with fusion alleles, and 10 samples with duplications compared to five normal controls. On the basis of these preliminary results, it was determined that the normal copy number ranged from 0.8 to 1.25 RQ, whereas fusion alleles were less than 0.5 RQ, and duplications were greater than 1.75 RQ (Figure 2A).
Figure 2.
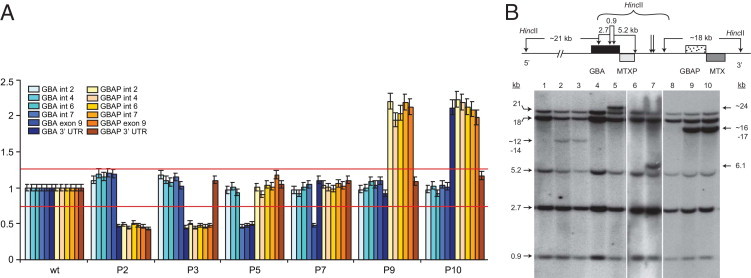
A: Results of quantitative real-time PCR of GBA and GBAP performed in samples from the patients listed in Table 2. The probes are in order, beginning from upstream of GBA to the 3′ end of GBAP. WT: Control sample with no known recombination, used to normalize the data; P2and P3: patients with fusion alleles; P5: patient with a large deletion; P7: patient with recΔ55 [c.1263-1317del (55-bp deletion)]; P9 and P10: patients with GBA duplications. The y axis reflects relative quantitation. The red lines indicate the range in copy number found among controls. B: Southern blot using HincII-digested DNA samples, corresponding to the cases described in Table 2 (except P4, where insufficient DNA was available for real-time qPCR). Normal individuals have five bands as shown, whereas patients with recombinant alleles demonstrate extra bands.
Southern Blot Analyses
HincII-digested genomic DNA (10 to 15 μg) was electrophoresed on a 0.6% I.D.NA agarose gel (FMC Bioproducts, Rockland, ME) and hybridized with 32P-labeled GBA cDNA.8
Sequencing of GBA
Exons and flanking intronic regions of GBA were sequenced using a 3130 Genetic Analyzer (Applied Biosystems),10 and data analyzed using Sequencing Analysis v.5.1.1 and SeqScape v.2.1.1 software programs (Applied Biosystems). Intronic regions of GBA with polymorphic sites were sequenced to confirm amplification of both alleles.
Results
Quantitative comparative (ΔΔCt) real-time qPCR identified both GBA fusion alleles, where the 5′ portion of the allele is derived from the gene and the 3′ from the pseudogene, and duplicated alleles, with an extra copy of the pseudogene using a normal sample as a calibrator. Moreover, unlike Southern blot analysis, this technique could discern mutant alleles resulting from gene conversion from those due to deletions, and detect the specific crossover site. All GBA fusions and duplications identified by Southern blot analysis were confirmed with real-time qPCR. However, real-time qPCR was more sensitive, enabling the identification and definition of several complex recombinant alleles as summarized in Table 2 and described below:
Table 2.
The Genotypes of the Cases with Recombinations Studied
| Patient | Phenotype | Genotype⁎ | Site of cross over initiation | Southern blot | Real-time qPCR |
|---|---|---|---|---|---|
| 1, 6, 8 | Wild type | Normal controls (wt/wt) | |||
| 2 | Type 2 | p.Leu483Pro (L444P) + p.Glu365Lys (E326K)/ p.Leu483Pro (L444P)+ Rec F | MTX | Fusion in MTX | Fusion in MTX |
| 3 | Type 1 | p.Arg502Cys (R463C)/Rec F | Intron 9 | Fusion | Fusion of GBA and GBAP gene |
| 4 | Type 1 | p.Asn409Ser (N370S)/Rec F | Exon 10 | Normal | NA |
| 5 | Type 2 | c.1505 + 2T>A (IVS10 + 2)/Rec D | Exon 7 | Fusion | Partial GBA deletion |
| 7 | Type 2 | p.Arg296Gln (R257Q)/ RecΔ55 (c.1263-1317del) | Intron 8-exon 9 | Missing exon 9 site on Southern blot | 55-bp deletion (gene conversion) |
| 9 | Type 1 | p. Arg209Cys (R170C)/ p.Leu483Pro (L444P)+Rec G | 3′ UTR (PMTX) | Duplication in pseudogene | Duplication in pseudogene |
| 10 | Type 1 | c.222-224delTAC/Rec G | Intron 10-exon 11 | Duplication in pseudogene | Duplication in pseudogene |
GBA cDNA nucleotides (c.) are numbered designating the adenine of the first ATG translation initiation codon as the first nucleotide (GenBank reference sequence NM_000157). Amino acid designated with (p.) are based on the primary GBA translation product, including the 39-residue signal peptide. Recombinant alleles mentioned here are described in more detail in Tayebi et al and Hruska et al.8,9
NA, not available; wt, wild type.
Rec D: recombination at the end of exon 7. Rec F: recombination at the end of intron 9 or beginning of exon 10, Rec G: recombination in intron10-exon 11 or the 3′ untranslated region.
1) Large deletions: In Patient 5, the Southern blot analysis indicated a recombinant fusion allele (Figure 2, A and B), where the crossover site was approximated to be between the end of intron 6 and the beginning of exon 7. Real-time qPCR demonstrated that this patient actually had a partial deletion allele extending from intron 6 to the 3′ UTR (Figure 2A).
2) Allelic conversion: Real-time qPCR could detect nonreciprocal sequence exchanges, including gene conversions, which occur frequently with homologous sequences. Probes from different regions of the GBA locus were used to detect gene conversion events. For example, a common Gaucher mutation, rec Δ55 in exon 9, resulting from gene conversion, is easily identified by real-time qPCR, since the exon 9 probe detects only one copy (Figure 2, A and B, patient 7).
3) Recombinations in the 3′ UTR region: Because the number of DNA restriction sites is limited, different recombinant alleles with crossover sites that are close by look the same on Southern blot analysis. Figure 2B, lane 9, shows a duplication, with the site of initiation of the crossover in the 3′ UTR of GBA. A second patient (lane 10) appeared to have the same banding pattern. Using a probe in this region, real-time qPCR narrowed down the site of crossover initiation, demonstrating that these were two different mutant alleles. In lane 9, the 3′ UTR of GBA is normal, whereas in lane 10, the probe indicates that this region is duplicated (Figure 2A). Since an enhancer is found in the 3′ UTR, duplication of this region could impact the phenotype.11
Similarly, the cases in lanes 2 and 3 (Figure 2, A and B) both carry fusion alleles, resulting from recombination occurring in the 3′ UTR and intron 9. Although by Southern blot analysis both patients have the same bands, real-time qPCR confirms the presence of the GBAP 3′ UTR (MTX) probe in lane 3, although not in lane 2. The phenotypes of these two cases were extremely different, reflecting the different length of the deleted segment.
Discussion
We demonstrate that real-time qPCR is a more sensitive alternative method to detect recombinant alleles previously only identified with Southern blots and extensive sequencing. Reciprocal crossover between homologous regions can result in fusions with RQ values less than 0.5, and duplications with values of over 1.75. When different copy numbers are detected with two nearby probes, it can be concluded that the crossover lies in the region between the probes.
The presence of a highly homologous pseudogene sequence greatly complicates genotypic analysis in many disorders. In the case of Gaucher disease, the high degree of sequence homology between the exons of GBA and GBAP result in many of the mutant alleles encountered in patients, complicating the accurate identification and description of mutant GBA alleles. The development of real-time qPCR as a means to detect copy number variation provides a new tool for evaluating genomic duplications, deletions, and other rearrangements, facilitating the identification and definition of complex GBA alleles. Other advantages of this method over Southern blot analysis is that it requires far less DNA and can detect all known GBA recombinant alleles in a single reaction, whereas previously, two or more restriction enzyme digests were necessary.8 Southern blot analyses are also limited by available restriction sites, leading to false-negative results, whereas with real-time qPCR, our ability to design more specific probes increases the sensitivity.
In Gaucher disease, the development of diagnostic kits has proven to be quite challenging due to the homologous nearby GBA pseudogene and the occurrence of recombinant alleles. The few kits commercially available screen for a limited number of mutations, and can generate false-positive and false-negative results, leading to diagnostic errors.12,13
Because of limitations in genotype–phenotype association in Gaucher disease, it has been postulated that other genes within the GBA locus, such as MTX, transcription factor binding sites, or enhancers may modify patient phenotype. Real-time qPCR may detect sequence rearrangements that impact gene function.
In the case of GBA, the design of primers and probes posed a major challenge, due to the high homology between GBA and GBAP. Multiple sets were optimized and tested, and the primers presented here were found to be the most useful for differentiating the various GBA recombinations. Our ability to compare the real-time results to traditional Southern blot analyses confirmed the value of this technique. However, as with Southern blot analysis, this approach may still miss some recombinations, especially in the upstream region of the gene, and may not discern whether recombinations result from double or reciprocal crossover.
We demonstrate that quantitative real-time PCR is a sensitive and rapid method to detect fusions and duplications in patients with recombinant GBA alleles. Since this technique is faster and less costly than Southern blot analysis, it may have wide applicability in diagnostic laboratories. Moreover, this technique may prove valuable in evaluating other genes in which mutations commonly result from recombination events.
Acknowledgments
We thank Jae H. Choi for his help in preparing the figures.
Footnotes
Supported by the Division of Intramural Research of the National Human Genome Research Institute, and the National Institutes of Health.
References
- 1.Barneveld R.A., Keijzer W., Tegelaers F.P., Ginns E.I., Geurts van Kessel A., Brady R.O., Barranger J.A., Tager J.M., Galjaard H., Westerveld A., Reuser A.J. Assignment of the gene coding for human beta-glucocerebrosidase to the region q21-q31 of chromosome 1 using monoclonal antibodies. Hum Genet. 1993;64:227–231. doi: 10.1007/BF00279398. [DOI] [PubMed] [Google Scholar]
- 2.Winfield S.L., Tayebi N., Martin B.M., Ginns E.I., Sidransky E. Identification of three additional genes contiguous to the glucocerebrosidase locus on chromosome 1q21: implications for Gaucher disease. Genome Res. 1997;7:1020–1026. doi: 10.1101/gr.7.10.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horowitz M., Wilder S., Horowitz Z., Reiner O., Gelbart T., Beutler E. The human glucocerebrosidase gene and pseudogene: structure and evolution. Genomics. 1989;4:87–96. doi: 10.1016/0888-7543(89)90319-4. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong L.C., Komiya T., Bergman B.E., Mihara K., Bornstein P. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J Biol Chem. 1997;272:6510–6518. doi: 10.1074/jbc.272.10.6510. [DOI] [PubMed] [Google Scholar]
- 5.Long G.L., Winfield S., Adolph K.W., Ginns E.I., Bornstein P. Structure and organization of the human metaxin gene (MTX) and pseudogene. Genomics. 1996;33:177–184. doi: 10.1006/geno.1996.0181. [DOI] [PubMed] [Google Scholar]
- 6.Chen K.S., Manian P., Koeuth T., Potocki L., Zhao Q., Chinault A.C., Lee C.C., Lupski J.R. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- 7.Mazzarella R., Schlessinger D. Pathological consequences of sequence duplications in the human genome. Genome Res. 1998;8:1007–1021. doi: 10.1101/gr.8.10.1007. [DOI] [PubMed] [Google Scholar]
- 8.Tayebi N, Stubblefield BK, Park JK, Orvisky E, Walker JM, LaMarca ME, Sidransky E: Reciprocal and nonreciprocal recombination at the glucocerebrosidase gene region: implications for complexity in Gaucher disease. Am J Hum Genet 72, 519–534 [DOI] [PMC free article] [PubMed]
- 9.Hruska K.S., LaMarca M.E., Scott C.R., Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29:567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 10.Stone D.L., Tayebi N., Orvisky E., Stubblefield B., Madike V., Sidraksy E. Glucocerebrosidase gene mutations in patients with type 2 Gaucher disease. Hum Mutat. 2000;15:181–188. doi: 10.1002/(SICI)1098-1004(200002)15:2<181::AID-HUMU7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 11.Blech-Hermoni Y.N., Ziegler S.G., Hruska K.S., Stubblefield B.K., Lamarca M.E., Portnoy M.E., NISC Comparative Sequencing Program. Green E.D., Sidransky E. In silico and functional studies of the regulation of the glucocerebrosidase gene. Mol Genet Metab. 2010;99:275–282. doi: 10.1016/j.ymgme.2009.10.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi J.H., Velayati A., Stubblefield B.K., Orr-Urtreger A., Gan-Or Z., Tayebi N., Sidransky E. False-positive results using a Gaucher diagnostic kit—RecTL and N370S. Mol Genet Metab. 2010;100:100–102. doi: 10.1016/j.ymgme.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torralba M.A., Alfonso P., Pérez-Calvo J.I., Cenarro A., Pastores G.M., Giraldo P., Civeira F., Pocoví M. High prevalence of the 55-bp deletion (c. 1263del55) in exon 9 of the glucocerebrosidase gene causing misdiagnosis (for homozygous N370S (c. 1226A > G) mutation) in Spanish Gaucher disease patients. Blood Cells Mol Dis. 2002;29:35–40. doi: 10.1006/bcmd.2002.0535. [DOI] [PubMed] [Google Scholar]


