Abstract
DNA from archived FFPE can be used for papillomavirus genotyping, but potential problems include paraffin as a physical barrier, DNA cross-linking, and PCR inhibitors. To address these complications, we combined a commercially available DNA isolation kit (Qiagen DNeasy) with a heat treatment and evaluated the resulting DNA with regards to HPV typing. DNA was extracted from 10-μm sections from 150 FFPE cancer samples. One protocol followed the manufacturer's recommendation, including paraffin removal by xylene and tissue lysis at 56°C. A second section was directly incubated at 120°C and subsequently lysed at 65°C. After spin-column purification, both extracts were tested with a linear array HPV genotyping assay. Additionally, cellular DNA yield, HPV16 DNA copies, and PCR inhibitors were assessed by real-time qPCR assays. Inadequate linear array HPV genotyping assay results were significantly more frequent (P = 0.0003) in xylene-treated (29/150, 19.3%) than in heat-treated extracts (8/150, 5.3%). HPV detection also differed, with 94/150 (62.7%) and 110/150 (73.3%) positive results, respectively (P = 0.0026). The heat method also yielded more PCR-amplifiable cellular DNA (8.2-fold; P < 0.001) and HPV16 copies (6.5-fold; P = 0.009), although PCR inhibitors also had a greater effect (P = 0.035). Aggressive heat treatment demonstrated an advantage over traditional xylene purification protocols, resulting in higher DNA yields and increased sensitivity for HPV testing.
Molecular analysis such as HPV detection and genotyping of formalin-fixed, paraffin-embedded (FFPE) tissues has become of great interest in recent years. As new or refined and more sensitive analytic methods become available, FFPE tissue collections provide vast sample repositories for research projects and retrospective epidemiology studies. Although the DNA in archived tissues is generally preserved over long periods of time, fragmentation and DNA-protein cross-linking by formaldehyde exposure, as well as the presence of paraffin, can have significant negative effects on DNA yield and amplification efficiency in subsequent PCR applications.
A number of methods and protocols to eliminate or reverse these factors have been proposed and evaluated, with varying success.1 DNA cross-links can be reversed to some extent by heat, and various temperature and incubation times have been proposed.2–4 The majority of protocols for nucleic acid extraction from FFPE tissues traditionally incorporate a pretreatment with xylene and ethanol to physically remove the paraffin wax.5 The disadvantages of this procedure include labor intensity, chemical toxicity, and the risk of accidentally removing small tissue fragments during the process.
For HPV genotyping assays, relatively small amounts of DNA suitable for conventional endpoint PCR are needed. Population-based studies, however, require the ability to process many samples simply, and with standardized reliable results. We found that the current published protocols are either too laborious or do not reliably produce DNA in sufficient quantity for this objective. We therefore combined a 120°C heat treatment, as suggested by Shi et al,3 with a commercially available DNA isolation kit. The resulting extracts were compared with DNA isolated in accordance with the manufacturer's protocol and were evaluated with regards to HPV typing. Additionally, we also assessed the quantity of PCR-amplifiable cellular genomic DNA (gDNA) and DNA quality in extracts from both methods.
Materials and Methods
Sample Collection
To minimize the potential for PCR contamination, the histotechnologists were instructed to wear gloves when handling blocks and to clean the microtome and the area around the cutting station before beginning work, and to use a new disposable microtome blade for each block. Six serial 5-μm sections were cut from each block. The first and last sections were stained with H&E. The intervening sections were transferred directly from the microtome blade, using a new disposable wooden applicator for each block, as two 5-μm sections each into a 2-mL conical screw-cap tube with tether cap (Simport, St-Michel-de-Bellechasse, QC, Canada), yielding two tubes per block.
The H&E slides were reviewed to confirm the presence of sufficient diagnostic tissue. A total of 2517 specimens received from three ongoing studies of HPV type-specific prevalence in HPV-associated invasive and preinvasive disease met this criterion. The samples came from seven different states in the United States and had been archived for 2 to 4 years. A set of 150 samples was randomly selected using a pseudo-random number generator; of these, 50 were cervical biopsies of intraepithelial neoplasia (grade 2/3) and 100 were squamous cell carcinomas (SCC) of the anus, cervix, vagina, penis, or head and neck (tonsil, tongue, larynx).
DNA Extraction
Total DNA was extracted from the replicate tubes by two different methods, based on the DNeasy blood and tissue kit (Qiagen, Valencia, CA). One tube was processed according to the manufacturer's protocol, using xylene treatment to remove paraffin as outlined in the DNeasy user manual (“Purification of Total DNA from Animal Tissues” as found in DNeasy Blood & Tissue Handbook, Qiagen, Valencia, CA; July 2006). Briefly, the paraffin was removed by vortexing and 10 minutes incubation with 1.2 mL xylene, followed by two washes with pure (200 proof) ethanol. The air-dried pellet was then incubated with 20 μL proteinase K and 180 μL ATL lysis buffer (from the DNeasy kit) for 16 hours in a heat block at 56°C. The lysed emulsion was further purified with the DNeasy spin-column kit. DNA was finally recovered in a single elution step with 100 μL AE solution from the kit.
The other tube was processed using a high-heat treatment. Instead of xylene extraction, 180 μL ATL lysis buffer from the DNeasy kit was added directly to the paraffin sections, followed by incubation of the tightly closed tube at 120°C for 20 minutes. Within 5 minutes after the paraffin had melted, the emulsion was mixed by finger-flicking, to ensure that all tissue fractions were submerged in the buffer. After this initial incubation, tubes were briefly centrifuged to remove condensate. Proteinase K (20 μL) was added to the liquid, and the closed tubes were incubated at 65°C for 16 hours in another heat block. Subsequent steps with DNeasy and DNA elution were identical to the first protocol described above.
HPV Genotyping
HPV detection and typing used the linear array (LA) HPV genotyping test (Roche Molecular Systems, Branchburg, NJ). The assay is based on L1 consensus PCR with PGMY primers yielding a 450-bp amplicon, with type-specific hybridization to detect 37 individual types (6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72 73, 81, 82, 83, 84, 89, IS39). In addition, the assay includes β-globin primers (150-bp amplicon) detected on the HPV typing strip as positive control for amplifiable sample DNA. The manufacturer's protocol was followed, except that a 10-μL aliquot of each DNA extract was used in the 100-μL PCR, and an automated BeeBlot instrument (Bee Robotics, Caernarfon, UK) was used for hybridization and detection. Each assay included controls for PCR contamination (water blanks taken through extraction and reagent water) and low-positive HPV control. Two technologists independently interpreted the HPV strips after a 1-hour drying period. Extracts prepared by the two methods were interspersed into the regular workflow of the laboratory and were tested independently.
Quantification by Real-Time PCR
Quantitative real-time PCR (qPCR) assays with 5′nuclease TaqMan technology were developed to quantify yield of human DNA (gDNA) and HPV16 copy number, as well as to detect the presence of inhibitors in the extracts. Primers and probes were designed with Primer Express 3.0 software (Applied Biosystems, Foster City, CA) (Table 1).
Table 1.
Primers and Probes Used in the Real-Time PCR Assays
| Oligo | Sequence | Amplicon |
|---|---|---|
| Target: β-globin (Homo sapiens) | ||
| Fwd primer | 5′-GGTGAGCCAGGCCATCACTA-3′ | 71 bp |
| Rev primer | 5′-TATGGGCAACCCTAAGGTGAA-3′ | |
| Probe | HEX-5′-CACCGAGCACTTTCTTGCCATGAGC-3′-BHQ1 | |
| Target: L1 HPV16 | ||
| Fwd primer | 5′-TTGCAGATCATCRAGAACACGTAGA-3′ | 175 bp |
| Rev primer | 5′-CTTGTCCAGCTGGACCATCTATTT-3′ | |
| Probe | FAM-5′-AATCATGCATGGAGATACACCTACATTGCATGA-3′-BHQ1 | |
| Target: rubisco activase (Glycine max) | ||
| Fwd primer | 5′-TTCAAGAGGCTAAGACCGCATAC-3′ | 72 bp |
| Rev primer | 5′-GCACTTGGCCGAACGTTGTC-3′ | |
| Probe | QUAS705-5′-CCAACGGCTTCATCCGTATCATCGG-3′-BHQ2 |
TaqMan probes were labeled with fluorescent tags FAM, HEX, or QUAS705 in combination with black-hole quencher 1 or 2 (BHQ1, BHQ2).
For quantification of gDNA, a 71-bp fragment of the human β-globin gene (HBB) was amplified. A standard curve was prepared from human genomic DNA (Roche Diagnostics, Indianapolis, IN) in fivefold dilutions ranging from 20 ng to 32 pg per reaction. The assay to quantify HPV16 targeted a 175-bp fragment within viral L1 region. The standard curve was prepared from a previously quantified HPV16-positive sample and ranged from 1 × 107 to 1 × 101 copies per PCR reaction in 10-fold dilutions. Presence of PCR inhibitors in the extracts was assessed though an exogenous control, a 502-bp fragment of the Glycine max ribulose-1,5-bisphosphate carboxylase oxygenase gene (rubisco activase) in pGEM-T Easy vector (Promega, Madison, WI) (courtesy of Dr. Harold Trick, Kansas State University) detected with PCR targeting a 72-bp region. The plasmid was diluted so that the Ct value was 26.0 in the presence of 20 ng highly purified human gDNA (Roche Diagnostics). This amount of rubisco DNA was spiked into each PCR reaction. Differences in Ct value increase >26 were used directly as measurement for comparison.
All qPCR assays were performed in a C1000 thermal cycler with a CFX96 real-time system (Bio-Rad, Hercules, CA). Reactions for the β-globin and rubisco amplification were prepared in a 50-μL volume containing 1× PCR TaqMan buffer A, 3 mmol/L MgCl2 (both Applied Biosystems), 0.3 mmol/L dNTP mix, 1.5 U FastStart Taq polymerase (both Roche Diagnostics), 0.8 μmol/L of each forward and reverse primer, 0.25 μmol/L probe, and 10 μL DNA template. After heat activation at 95°C for 4 minutes, a two-step thermal profile was applied for 40 cycles of 15 seconds at 95°C and 30 seconds at 61°C. Reactions targeting HPV16-L1 contained 0.17 μmol/L of the forward primer, 0.15 μmol/L of the reverse primer, and 0.1 μmol/L probe. The thermal profile was 95°C for 9 minutes of heat activation and then 40 cycles of 15 seconds at 95°C and 1 minute at 62°C. A no-template control was included on each plate for each assay. All reactions were performed in duplicate, and quantitative results were accepted only if the coefficient of variation was <2.5%.
Statistical Analysis
Success of extraction was evaluated based on the number of samples yielding adequate DNA in LA (ie, β-globin and/or HPV detected on typing strip), number with HPV detected and number of HPV types detected per sample in LA, the yield of human DNA (qPCR), and number of samples with inhibitors (qPCR). Statistical analysis for independence of discrepant LA HPV results was done with McNemar's test. Student's t-test for paired samples was applied to evaluate differences in gDNA yield, PCR inhibitors, and HPV16 copies as measured by real-time PCR. All P values were two-sided and were considered to be statistically significant at P < 0.05. All calculations were performed with SPSS software, version 15.0 for Windows (SPSS, Chicago, IL).
Results
A direct comparison of the overall HPV results obtained from xylene-treated versus heat-treated extracts is given in Table 2. The xylene method was more susceptible to inadequate results than the heat-treated method: 29/150 vs. 8/150 samples (P = 0.0003, McNemar's test). HPV was also detected in more of the heat-extracted DNA samples (P = 0.0026). Among the 116 samples with adequate LA results from both methods, HPV was detected in 90 of the xylene-treated extracts, compared with 101 of the heated extracts. This difference was also seen in a type-specific comparison between the extracts. The 121 adequate xylene-treated extracts averaged 1.11 types per positive sample. By contrast, the adequate heat-treated extracts averaged 1.35 types per positive sample.
Table 2.
Linear Array HPV Genotyping Test Results from DNA Extracted Using Heat versus Xylene Treatment
| Xylene treated | Heat treated |
|||
|---|---|---|---|---|
| Pos | Neg | Inad | Total | |
| Pos | 90 | 0 | 4 | 94 |
| Neg | 11 | 15 | 1 | 27 |
| Inad | 9 | 17 | 3 | 29 |
| Total | 110 | 32 | 8 | 150 |
Inad, inadequate (neither HPV nor β-globin detected); Neg, negative (no HPV detected, but β-globin positive); Pos, positive (HPV detected).
Overall, concordance of individual HPV types between the two sets of extracts was 99.3%. Type discordance occurred 37 times in 26 samples; the majority (32/37, 86.5%) resulting from additional types being detected in the heat-treated extracts.
gDNA yields were quantified from 100 pairs of extracts and valid results were obtained from 96 of them (the remaining 4 had DNA below the level of the qPCR detection). The median gDNA yield was 23 pg/μL (range, 0 to 780) in xylene-treated extracts and 157 pg/μL (range, 3 to 4651) in heat-treated extracts. In 95 samples, gDNA yield was higher in the heat-treated extracts (P < 0.001), with a median difference of 8.2-fold between the pairs.
HPV16 copies were quantified in all 62 pairs of extracts with concordant positive results for HPV16 by LA. Four samples were excluded because of failed amplification or irreproducible (CV >2) qPCR results. In the remaining 58 pairs that passed quality standards, xylene-treated extracts had a median copy number of 405, compared with 1639 per μL in heat-treated extracts (Figure 1). By pairwise analysis, heat-treated extracts had a higher viral DNA yield (P = 0.009); the median increase was 6.5-fold.
Figure 1.
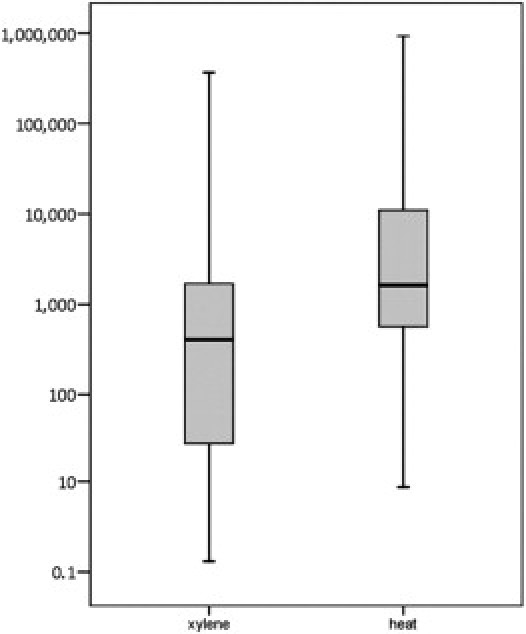
Boxplot of distribution of measured HPV16 DNA copies in 58 xylene- and heat-treated extracts from FFPE tissues. Data are expressed as copy number per 10 μL in log10 scale.
PCR inhibitors were assessed in 32 pairs: 22 pairs with inadequate LA results from both methods and 10 samples positive for HPV in both extracts. The Ct values for the exogenous template in all 64 extracts tested were higher than the reference containing highly purified DNA. However, xylene-treated extracts were significantly less affected than heat-treated extracts (P = 0.035). The median Ct increase over the reference reaction was 0.41 and 1.14 for the xylene and heat extraction methods, respectively.
Discussion
The present findings demonstrate the advantage of incorporating high-heat treatment into the extraction protocol for FFPE tissue. Tissues extracted with the heat method were less likely to yield inadequate results in the LA assay, compared with the xylene method (P = 0.0003, McNemar's test). The yield of gDNA and HPV16 DNA was also higher (8.2- and 6.5-fold, respectively), HPV was more likely to be detected, and more types per sample were detected with the heat method. In most instances, gDNA yield correlated with adequacy of LA results, although there were exceptions. The overall improvement in performance with heat treatment, combined with reduced sample handling, commends this approach to laboratories embarking on population-based studies of HPV in FFPE.
Amplification of an external template in all extracts indicated a slightly greater reduction in PCR efficiency in the majority of the heat-treated extracts. Because applying heat treatment leaves melted paraffin and other possible components in the lysis emulsion, some residues might remain with the eluted DNA and interfere with the subsequent PCR reaction; however, this disadvantage (also recognized by Stanta and Schneider6) is clearly compensated by the increase in amplifiable DNA yield.
The improvement might be due to reversal of formaldehyde-induced cross-linking of nucleic acids through the 120°C incubation. Some improvements may also be due to the increased heat during the proteinase K treatment, which we had previously found more effective for tissue lysis. However, the amount of available PCR template might still be limited by fractionation of gDNA occurring in fixed tissues. Length of DNA fragments might be an important factor, and other investigators have reported difficulties with amplicons >300 bp.7 The relatively large target of 450 bp in the HPV L1 region spanned by the LA PGMY primer system8 might be at the upper limit for amplification of DNA from FFPE tissue samples. Other HPV genotyping tests that rely on smaller targets, such as SPF109 (65 bp) or GP5+/6+10 (150 bp), might avert this problem and be better suited for FFPE tissues.11,12 Use of the LA assay as a basis for comparison of the extraction methods was specifically chosen because the larger amplicon would emphasize small differences in fragment size.
Eliminating the traditional xylene wash steps from the protocol has considerable advantages, beyond the improved yields. Manual pipetting time and effort are greatly reduced, because the tedious separation of liquids from tissue fragments is eliminated. Moreover, the danger of accidentally removing small tissue fragments by pipetting is also reduced. We noticed that tissue sections may occasionally stick to the upper walls of the tube during the 120°C incubation step, so that temperature and buffer contact are less than optimal. We therefore recommend to shake or finger-flick the tubes gently and visually verify tissue fractions floating in the lysis buffer, shortly after paraffin has melted.
Henning et al13 also described a xylene-free method, but their method separates the lysate physically from preheated paraffin in an extra step. Such a method applies a less efficient lower heat treatment3 and relies on the availability of an automated VERSANT kPCR system (Siemens, Erlangen, Germany) with silica-coated iron oxide beads. Our protocol can likely be adapted to most automated methods for DNA extraction. For example, we have extracted samples in a Chemagic MSM1 system (Chemagen, Baesweiler, Germany) after lysis by both methods. The HPV results were identical by LA, and real-time quantification of gDNA yields showed at least the same increase in samples that were prepared with the heat method (data not shown).
Although the present study directly compared a modest number of samples (150 blocks), these were contributed from a variety of different pathology laboratories from seven different states, indicating that differences in tissue processing are included in the analysis. Furthermore, we have been using the heat-treatment extraction method in ongoing studies of HPV in FFPE with continued good results in nearly 5000 FFPE samples to date. Most samples were <5 years old, and the effect of longer storage times has not been evaluated. In addition we had only limited opportunities to evaluate the heat method in tissues processed with fixatives other than formalin; however, results from six glyoxaline-fixed tissues showed a trend concurrent with the present findings (data not shown).
We conclude that adding an aggressive heat treatment to the commercially available DNeasy kit resulted in an efficient and standardized method for DNA extraction and HPV typing from FFPE tissues. Greater amplifiable DNA yield though the reversal of cross-linking might be the most significant factor, but this method is also applicable to limited starting material, and it eliminates both the laborious tissue washing process and the associated risk of accidentally removing tissue fragments.
Acknowledgments
We thank Mariela Zamarron for assistance with DNA extraction and HPV genotyping and thank Dr. Harold Trick (Kansas State University, Manhattan, KS) for supplying the plasmid containing the rubisco activase DNA.
Footnotes
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
CME Disclosure: None of the authors disclosed any relevant financial relationships.
References
- 1.Gilbert M.P., Haselkorn T., Bunce M., Sanchez J.J., Lucas S.B., Jewell L.D., Van Marck E., Worobey M. The isolation of nucleic acids from fixed, paraffin embedded tissues–which methods are useful when? PLoS One. 2007;6:e537. doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Man Y.G., Moinfar F., Bratthauer G.L., Kuhls E.A., Tavassoli F.A. An improved method for DNA extraction from paraffin sections. Pathol Res Pract. 2001;197:635–642. doi: 10.1078/0344-0338-00138. [DOI] [PubMed] [Google Scholar]
- 3.Shi S.R., Cote R.J., Wu L., Liu C., Dater R., Shi Y., Liu D., Lim H., Taylor C.R. DNA Extraction from archival formalin-fixed paraffin-embedded tissue sections based on the antigen retrieval principle: heating under the influence of pH. J Histochem Cytochem. 2002;50:1005–1011. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- 4.Dedhia P., Tarale S., Dhongde G., Khadapkar R., Das B. Evaluation of DNA extraction methods and real time PCR optimization on formalin -fixed paraffin-imbedded tissues. Asian Pac J Cancer Prev. 2007;8:55–59. [PubMed] [Google Scholar]
- 5.Goelz S.E., Hamilton S.R., Vogelstein B. Purification of DNA from formaldehyde fixed and paraffin embedded human tissues. Biochem Biophys Res Commun. 1985;130:118–126. doi: 10.1016/0006-291x(85)90390-0. [DOI] [PubMed] [Google Scholar]
- 6.Stanta G., Schneider C. RNA extracted from paraffin-embedded human tissues is amenable to analysis by PCR amplification. Biotechniques. 1991;11:304. 306, 308. [PubMed] [Google Scholar]
- 7.Bonin S., Petrera F., Niccolini B., Stanta G. PCR analysis in archival postmortem tissues. J Clin Pathol Mol Pathol. 2003;56:184–186. doi: 10.1136/mp.56.3.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravitt P.E., Peyton C.L., Alessi T.Q., Wheeler C.M., Coutlee F., Hildesheim A., Schiffman M.H., Scott D.R., Apple R.J. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000;38:357–361. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleter B., van Doorn L.J., ter Schegget J., Schrauwen L., van Krimpen K., Burger M., ter Harmsel B., Quint W. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol. 1998;153:1731–1739. doi: 10.1016/S0002-9440(10)65688-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Brule A.J., Pol R., Fransen-Daalmeijer N., Schouls L.M., Meijer C.J., Snijders P.J. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of Human papillomavirus genotypes. J Clin Microbiol. 2002;40:779–787. doi: 10.1128/JCM.40.3.779-787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dal Bello B., Spinillo A., Alberizzi P., Cesari S., Cardella B., Silini E.M. Validation of the SPF10 LiPA human papillomavirus typing assay using formalin-fixed paraffin-embedded cervical biopsy samples. J Clin Microbiol. 2009;47:2175–2180. doi: 10.1128/JCM.00286-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan S.E., Garland S.M., Rumbold A.R., Tabrizi S.N. Human papillomavirus genotyping using archival vulva dysplastic or neoplastic biopsy tissues: comparison between the INNO-LiPA and linear array assays. J Clin Microbiol. 2010;48:1458–1460. doi: 10.1128/JCM.02311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henning G., Gehrmann M., Stropp U., Brauch H., Fritz P., Eichbaum M., Schwab M., Schroth W. Automated extraction of DNA and RNA from a single formalin-fixed paraffin-embedded tissue section for analysis of both single-nucleotide polymorphisms and mRNA expression. Clin Chem. 2010;56:1845–1853. doi: 10.1373/clinchem.2010.151233. [DOI] [PubMed] [Google Scholar]


