Abstract
Microarray-based transcriptome analysis of peripheral blood as surrogate tissue has become an important approach in clinical implementations. However, application of gene expression profiling in routine clinical settings requires careful consideration of the influence of sample handling and RNA isolation methods on gene expression profile outcome. We evaluated the effect of different sample preservation strategies (eg, cryopreservation of peripheral blood mononuclear cells or freezing of PAXgene-stabilized whole blood samples) on gene expression profiles. Expression profiles obtained from cryopreserved peripheral blood mononuclear cells differed substantially from those of their nonfrozen counterpart samples. Furthermore, expression profiles in cryopreserved peripheral blood mononuclear cell samples were found to undergo significant alterations with increasing storage period, whereas long-term freezing of PAXgene RNA stabilized whole blood samples did not significantly affect stability of gene expression profiles. This report describes important technical aspects contributing toward the establishment of robust and reliable guidance for gene expression studies using peripheral blood and provides a promising strategy for reliable implementation in routine handling for diagnostic purposes.
In recent years, the concept of integrating genomic and molecular information into the standard of medical care has gained more and more acceptance.1 It is expected that future decisions in medicine will be guided substantially by the patient's individual genetic constitution, as well as by measurement of specific biomarkers. The goal of current medical approaches is thus to define the appropriate disease-associated genes and pathways for targeted therapy and to discover associated markers for prediction of particular treatment outcomes.2 Genomic and genetic molecular diagnostics of highest quality may thus form the basis of what has been termed personalized or individualized medicine.3
Affected tissues can often be obtained only by invasive intervention. It would be desirable, therefore, to use more easily accessible surrogate tissues, such as peripheral blood, sputum, or urine (among others) for biomarker development.4 Moreover, it would be most attractive to develop and apply a biomarker detection platform that could be used for a variety of very different clinical questions. Blood-based gene expression profiling (GEP) represents one of these promising approaches.5 It can be applied to detect diagnostic, prognostic, predictive, and pharmacodynamic biomarkers by integrating the quantitative and qualitative information on transcription levels of thousands of genes simultaneously. GEP of blood-based leukemia samples revealed reproducible detection of robust leukemia-associated signatures almost irrespective of preanalytical conditions6 and of involvement of different laboratories.7 However, when blood is used as surrogate tissue, the blood-based GEP result is significantly influenced by sample handling and storage.8,9 This might be of particular importance when the surrogate blood-based GEP reflects only subtle changes with respect to the disease under investigation.
For viable peripheral blood mononuclear cells (PBMCs) in particular, cryopreservation in liquid nitrogen is a harsh procedure. It induces significant changes in the immunophenotype, performance, and cell viability of stored cells, compared with freshly isolated cells.10–14 Dimethyl sulfoxide (DMSO), which is one of the critical components of the cryopreservation medium, is also known to induce neutrophil differentiation.15,16 Not surprisingly, cryopreservation of PBMCs significantly interferes with the stability of the transcriptome, as was demonstrated, for example, by the loss of the transcript of Charcot-Leyden crystal protein in cryopreserved PBMC samples. A similar effect can be induced by applying only a short pulse with DMSO, which indicates that the freezing medium was in part responsible for this phenomenon.17 Although cryopreservation of PBMCs is commonly used in many clinical trials,18 this approach apparently does not fully assure stabilization of the RNA composition ex vivo. Moreover, it is a time-consuming procedure that cannot easily be integrated into routine clinical workflows. Alternative technologies have therefore been established, such as PAXgene RNA collection tubes (PreAnalytiX; Qiagen, Hilden, Germany). These tubes contain reagents for immediate stabilization of the blood-based RNA already during the blood drawing procedure.19–21 Although this approach seems to be a promising alternative for blood preservation prior to transcriptome analysis, the stability of gene expression profiles of long-term frozen PAXgene samples has not yet been sufficiently investigated.
To obtain greater insight into the effect of different blood-based preservation methods in altering gene expression profiles, we determined the effect of cryopreservation of PBMCs as well as of PAXgene-based stabilized blood samples on GEP. First, we elucidated short-term cryopreservation-induced transcriptional changes of PBMC samples in comparison with their noncryopreserved counterparts. Second, we assessed the effect of long-term cryopreservation periods (up to 60 months) on the stability of gene expression signatures within a clinical PBMC data set. Finally, we analyzed the stability of gene expression profiles of long-term frozen PAXgene samples using whole-genome microarray analysis. We provide evidence that cryopreservation of PBMCs, in contrast to PAXgene-based storage of peripheral whole blood samples, causes significant changes in transcriptional profiles, compared with noncryopreserved PBMC samples. Furthermore, we found significant alterations in GEP of PBMC samples with prolonged cryopreservation time periods. Our results provide the basis for a standard operating procedure for sample handling in GEP studies that rely on prolonged sample storage of peripheral blood as surrogate tissue.
Materials and Methods
Sample Collection and RNA Preparation
Healthy Subjects
Blood samples from apparently healthy female or male blood donors were collected in Vacutainer cell preparation tubes (CPT tubes; Becton Dickinson, Heidelberg, Germany) with sodium citrate or in PAXgene RNA collection tubes (Qiagen) after written informed consent had been obtained and after approval by the institutional review board. In the case of PAXgene, four PAXgene RNA tubes were collected from each donor and stored for 24 hours at room temperature to allow complete lysis of the cells for each of six blood donors. The incubation time was chosen according to the PreAnalytiX product circular (PAXgene Blood RNA Kit Handbook, Version 2) and results obtained by Rainen et al.22The RNA of one of the PAXgene tubes from each blood donor was subsequently isolated, and the other three tubes were stored at −20°C for 6 weeks, 6 months, and 12 months until further processing.
PBMCs of 12 healthy blood donors were isolated by using CPT tubes according to the manufacturer's recommendations and then were subsequently split. In brief, after blood withdrawal, CPT tubes were inverted 10 times and centrifuged within 10 minutes at 1650 × g for 20 minutes at 20°C. After centrifugation, CPT tubes were gently inverted 10 times and supernatant was transferred to a fresh centrifugation tube containing 30 mL PBS (room temperature) and gently mixed. Tubes were centrifuged at 500 × g for 10 minutes at 20°C to pellet cells. Supernatant was removed and cells were resuspended in 10 mL PBS; cells were counted with a Trypan Blue method. At this step, the cell suspension was split into two portions, cells were centrifuged (500 × g for 5 minutes at 20°C), and supernatant was removed. To evaluate the influence of freezing cells in fetal calf serum with 10% (v/v) DMSO and storage in liquid nitrogen on gene expression profiles, one portion of the freshly isolated PBMCs was lysed in TRIzol reagent (1 mL/1 × 107 PBMC) (Invitrogen, Karlsruhe, Germany; Carlsbad, CA) and stored at −80°C until further processing. The other portion of isolated PBMCs was resuspended in fetal calf serum with 10% (v/v) DMSO (1 mL/1 × 107 PBMC) and slowly frozen at a cooling rate of −1°C/minute to −80°C using devices designed for optimal cryopreservation. The next day, frozen PBMCs were transferred to the vapor phase of liquid nitrogen and were cryopreserved for 31 to 41 days (38.08 ± 3.99 days, mean ± SD) until further processing.
Melanoma Patients
PBMCs of 53 melanoma patients were isolated from leukapheresis products as described previously23 and were stored in liquid nitrogen, after written informed consent had been obtained and after approval by the institutional review board. In brief, isolated PBMCs were resuspended in 20% human serum albumin (1 × 108 PBMC in 1.5 mL) and were transferred into freezing vials (2.25 mL cell suspension per one 4.5-mL cryovial) resting on wet ice. Cooled freezing medium (consisting of human serum albumin plus 20% DMSO and 10% glucose) was added to cooled cell suspension (2.25 mL freezing medium per one 4.5 mL cryovial). Final concentration in the vial was 1.5 × 108 PBMC in 4.5 mL human serum albumin + 10% DMSO + 5% glucose. Cells were slowly frozen at a cooling rate of −1°C/minute to −80°C by using a cryofreezing container (Nalgene Cryo 1°C freezing container; Labor Center, Nuremberg, Germany). Finally, cells were transferred to the vapor phase of liquid nitrogen until use.
All patients presented with metastatic stage III or IV melanoma with either resected or measurable tumor, displaying HLA type A1 and/or A2 (0201). According to their stage of disease, patients had received various therapies (eg, surgery, radiation therapy, immune modulating therapy with interferon-α, or chemotherapy) before they were enrolled in a dendritic cell vaccination trial.
Patient recruitment within the dendritic cell vaccination trial occurred over a prolonged time period, which was partitioned into four different time frames. Within the blood genomic study, patients were divided into groups according to their initial blood draw within these four time frames. Thus, PBMC samples were stored in liquid nitrogen for 20 to 60 months before RNA extraction and GEP analysis. Patient characteristics and sample-related cryopreservation time periods are given in Supplemental Table S1 (available at http://jmd.amjpathol.org).
RNA Isolation
For RNA isolation from cryopreserved PBMCs, cells were removed from liquid nitrogen and transferred to a 37°C water bath until thawing. Samples were always thawed in <5 minutes. The thawed cell suspension was quickly transferred to 40 mL chilled RPMI 1640 medium and centrifuged at room temperature at 400 × g for 10 minutes. Supernatant was removed and the PBMCs were washed once with 50 mL of RPMI 1640 at room temperature and then were centrifuged at 400 × g for 10 minutes, also at room temperature. Supernatant was completely removed and cells were subsequently lysed in TRIzol reagent. The procedure described here was performed according to our own standard operating procedures developed for the optimal recovery of viable cells and therefore also providing high integrity of cellular RNA for subsequent analysis. RNA isolation was then performed according to the manufacturer's protocol, followed by a purification step with the RNeasy MiniElute cleanup kit (Qiagen).
Cellular RNA from whole-blood samples (PAXgene RNA tubes) was manually prepared with the PAXgene blood RNA kit (Qiagen) according to the manufacturer's instruction (PAXgene Blood RNA Kit Handbook, version April 2001), including an optional DNase digestion step. PAXgene RNA tubes enable the isolation of intracellular RNA of circulating leukocytes, including B cells, T cells, neutrophils, monocytes, and other less abundant cell types. Furthermore, a large proportion of reticulocyte-derived globin mRNA is prepared from PAXgene blood RNA tubes, as has been demonstrated previously.8,24
Analysis of RNA Samples
Total RNA was quantified by UV spectroscopy at 260 nm. The quality of the isolated RNA samples was determined by measuring the A260/A280 ratio, and the integrity of the ribosomal 28S and 18S bands was determined by agarose-gel electrophoresis.
Microarray Procedure
cDNA and biotin-labeled cRNA synthesis was generated from 100 ng total RNA using the Illumina TotalPrep RNA amplification kit (Applied Biosystems, Darmstadt, Germany; Foster City, CA). cRNA (1.5 μg) was hybridized to Human-6 Expression BeadChips V2 (Illumina, San Diego, CA) and was scanned on an Illumina BeadStation 500X system. To minimize technical induced batch effects, the same reagents and array lots were used within each group of samples for target preparation and microarray hybridization. Furthermore, experiments were performed by one or two very well trained technicians, based on highly standardized operation procedures.
Data Analysis
Raw data collection for Illumina BeadChips was performed using Illumina BeadStudio software version 3.1.3.0. All data analysis was performed using R statistical language and packages from the Bioconductor project25 and IlluminaGUI.26 Data sets were normalized using the quantile normalization method. Microarray data are available at http://www.ncbi.nlm.nih.gov/geo (record number GSE24758).
Unsupervised hierarchical cluster analysis was performed using correlation distance measurement and centroid linkage method using open source software dChip 2008 (Department of Biostatistics, Harvard School of Public Health, Cambridge, MA). Principal components analysis (PCA), determination of differentially expressed genes, and calculation of variable genes [coefficient of variation (ie, SD/mean) between 0.4 and 10] was performed with IlluminaGUI software. Determination of present calls was based on the detection P value assessed by BeadStudio software; an mRNA transcript was called present if the detection P value was <0.05; otherwise, the mRNA transcript was called absent. Differentially expressed genes were assessed by applying t-test with P < 0.05, different fold change (FC) filters (FC ≥ 1.5 or FC ≥ 2), and difference of means >100 and passing 10% false discovery rate. Paired and unpaired t-tests were calculated using SigmaPlot 10.0 (Systat Software, San Jose, CA) and analysis of variance combined with fold change filters was calculated using R statistical language.25 Correlation coefficients were calculated using Excel 2007 spreadsheet software and testing significance of Pearson correlation with Bonferroni correction was performed using R statistical language. Gene ontology analyses were performed with GeneTrail.27
Results
Short-Term Cryopreservation in Liquid Nitrogen Affects the GEP of PBMCs
One preferred and widely used method to conserve PBMCs for functional as well as analytical analysis is to isolate these cells from the blood and to freeze these viable cells in a DMSO-containing solution in liquid nitrogen. As a first step, we therefore investigated to what extent the freezing procedure itself influences the cellular transcriptome. PBMCs of 12 healthy controls were isolated and subsequently split into portions. One portion was lysed in TRIzol reagent immediately after isolation, to allow RNA stabilization, and was subsequently stored at −80°C before GEP; the other portion was frozen and stored for 31 to 41 days in fetal calf serum supplemented with 10% DMSO in liquid nitrogen until GEP. Because no high-quality RNA was obtained from PBMCs frozen without addition of DMSO, or after addition of DMSO without freezing, the particular effect of DMSO in this scenario was not further addressed.
Unsupervised analysis of gene expression profiles of these samples using the most variable genes across the dataset (SD/mean, 0.4 to 10) revealed a clear distinction between cryopreserved cells and freshly lysed cells, resulting in clusters showing a distinct separation of these two groups, with one exceptional case (Figure 1A). Similar results were obtained when performing PCA (Figure 1B). None of the related samples derived from the same individual grouped together, indicating a strong effect of the cell cryopreservation procedure on gene expression profiles. A supervised comparison analysis between the two sample groups (P < 0.05 t-test, difference of means >100, false discovery rate 0.1) resulted in 54 differentially expressed genes (FC ≥ 2). These comprised 35 transcripts with decreased signal intensities in cryopreserved samples and 19 transcripts with increased signal intensities in cryopreserved samples; a heat map of the 54 differentially expressed genes is presented in Figure 1C. Whereas transcripts with increased signal intensities exhibited a maximal fold change of 3.2, transcripts with decreased signal intensities exhibited fold changes up to −27.7. Moreover, among transcripts with decreased signal intensities, seven transcripts were identified in which mean signal intensities dropped to background levels: the Charcot-Leyden crystal protein transcript, the hemoglobin transcripts HBG1, HBG2, and HBD, and three additional and as yet not annotated transcripts (see Supplemental Table S2 at http://jmd.amjpathol.org). Decreasing the fold-change filter to 1.2 increased the number of differentially expressed genes to 1088 (see Supplemental Table S2 at http://jmd.amjpathol.org), further indicating remarkable changes in expression patterns with cryopreservation of PBMCs. Additionally, we also observed a significant overall increase of present call rates in the frozen PBMC samples, compared with the present call rates obtained from nonfrozen PBMC samples (30.65 ± 0.72 vs. 27.63 ± 1.03, P = 1.834 × 10−7 paired t-test). This increase of present call rates predominantly results from a shift from absent state to present state for many transcripts at background level in nonfrozen PBMC samples, without significant quantitative changes in signal intensity (data not shown).
Figure 1.
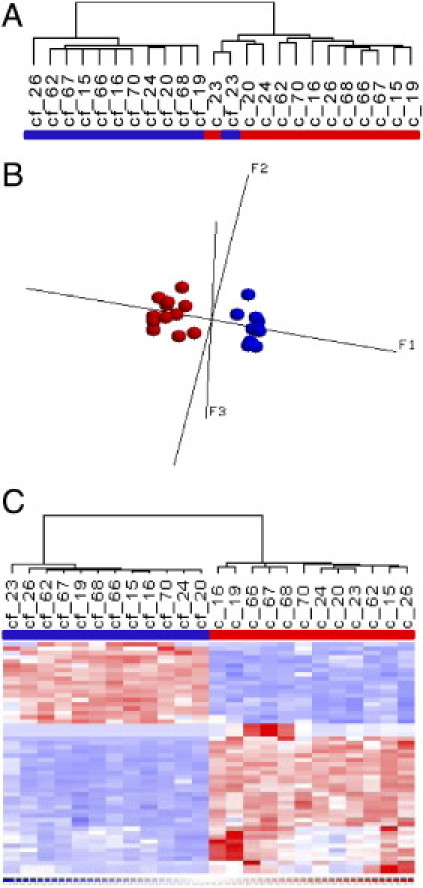
A: Unsupervised hierarchical cluster analysis using the most variable genes across the data set (SD/mean, 0.4 to 10; n = 193 transcripts). c, nonfrozen PBMCs; cf, frozen PBMCs (31 to 41 days). B: PCA using the most variable genes across the data set (SD/mean, 0.4 to 10; n = 193 transcripts) for nonfrozen PBMCs (red) and frozen PBMCs (blue). C: Hierarchical cluster analysis with heat map of PBMCs using differentially expressed genes between nonfrozen and cryopreserved samples (P < 0.05 t-test, difference of means >100, false discovery rate 0.1). c, nonfrozen PBMCs; cf, frozen PBMCs.
To elucidate whether the cryopreservation procedure has an effect on specific gene categories or signal transduction pathways, a GeneTrail analysis of genes differentially expressed between cryopreserved and nonfrozen PBMC samples (FC > 2) was performed. Thereby, the KEGG (http://www.genome.jp/kegg/pathway.html) annotated Toll-like receptor signaling pathway was identified based on the decline of the IRF3 transcripts and increase of CCL5 transcripts. This indicated that either the freezing procedure or the subsequent thawing of cells might interfere with pattern recognition receptor signaling processes, or that these changes in transcript abundance are related to variations in cell populations expressing these transcripts at constant level (see Supplemental Table S3 at http://jmd.amjpathol.org for the complete results of the GeneTrail analysis). Moreover, identified gene ontology categories included genes associated with oxygen transport (HBA1, HBG2, HBB, HBG1), genes of the platelet α granule membrane (ITGA2B, GP1BB, GP9), and other membrane-associated genes (DGKA, TNK2, IGSF6). Oxygen transport and platelet-associated genes were all decreased in cryopreserved PBMCs, indicating a loss of residual erythrocytes and platelets with the cryopreservation procedure. Indeed, we have observed that PBMC preparations with CPT-tubes can lead to a higher contamination of PBMCs with red blood cells, which do not properly penetrate the gel barrier during centrifugation. Furthermore, platelets are also isolated in conjunction with the PBMC fraction as it is elucidated in the BD-CPT product insert (BD Vacutainer CPT Cell Preparation Tube with Sodium Citrate). Apparently, red blood cells and platelets are more prone to damage during the freezing-thawing procedure, explaining our observation of loss of signals for these transcripts. In addition 20 different cluster of differentiation (CD) markers that are typically expressed on PBMCs showed an overall trend of decreased expression in cryopreserved PBMCs at FC ≥ 1.2 (see Supplemental Table S2 at http://jmd.amjpathol.org), which was not related to a particular cell subtype. Thus we cannot establish a significantly different effect on any given cell type.
Prolonged Cryopreservation of PBMCs Provokes Substantial Changes in GEP
The cryopreservation of PBMCs in clinical trials is a widely used approach to performing retrospective analysis. This kind of analysis is based on the assumption that under cryopreservation conditions PBMCs do not undergo significant changes and that, on thawing, these cells are similar in physiology and behavior to their nonfrozen initial state. Although the influence of cryopreservation on the function and phenotype of PBMCs has been addressed in several studies, only limited data on the effect of cryopreservation of PBMCs on whole genome transcriptome analysis are available,17 and the effect of long-term cryopreservation on the stability of GEP in PBMCs had hitherto not been investigated.
To address this issue, we performed GEP of PBMC samples that were collected from melanoma patients in the context of a vaccine study and were subsequently cryopreserved over a period of 20 to 60 months (Figure 2A; see also Supplemental Table S1 at http://jmd.amjpathol.org). According to the different specimen collection periods, PBMC samples were categorized into four groups, by length of cryopreservation time: group 1, storage time 21 to 30 months (mean, 26.38); group 2, 31 to 40 months (mean, 35.56); group 3, 41 to 50 months (mean, 46.22); and group 4, 51 to 60 months (mean, 55.28). An overview of sample distributions according to their cryopreservation time period is presented in Figure 2A, and detailed documentation for each sample is given in Supplemental Table S1 (available at http://jmd.amjpathol.org).
Figure 2.
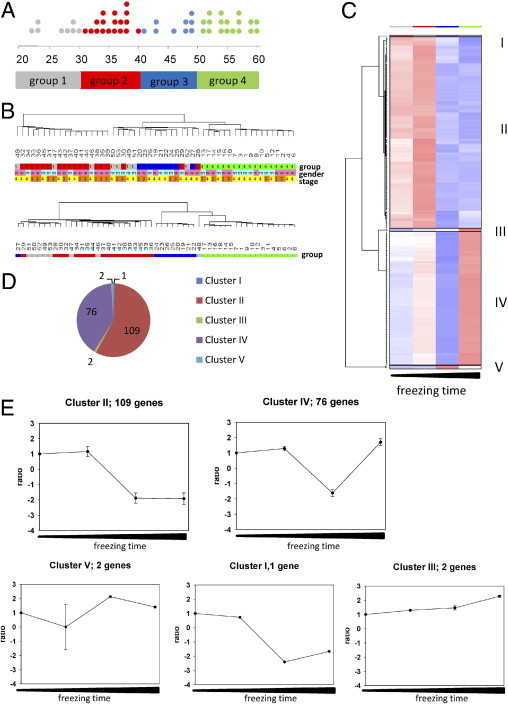
A: Schematic overview of cryopreservation time distributions. Each dot represents one PBMC sample indicated by its individual cryopreservation time (months). Dot colors match four groups ranked according to increasing cryopreservation time: group 1, 21 to 30 months (gray); group 2, 31 to 40 months (red); group 3, 41 to 50 months (blue); group 4, 51 to 60 months (green). B: Hierarchical cluster trees of PBMC samples using either the most variable genes across the data set (SD/mean, 0.4 to 10, n = 440 transcripts) (upper panel) or an analysis of variance gene list assessed across the four groups defined by cryopreservation time (analysis of variance, P < 0.05, 491 genes FC ≥1.5) (lower panel). Color key: Cryopreservation groups 1 to 4, gray, red, blue, and green (as in A). Sex: rose (w), female; blue (m), male. Disease stage: yellow, stage 4; orange, stage 3. C: Analysis of variance was performed across groups defined by their cryopreservation time (groups 1 to 4; analysis of variance, P < 0.05, 190 genes FC ≥ 2). For each group, the mean expression values of these 190 genes were calculated and subjected to hierarchical cluster analysis. The resulting gene clusters with similar changes in gene expression patterns are delineated by black box outline and are indicated by roman numerals. Color coding of cryopreservation groups (indicated above heat map columns) is as in A. D: Pie chart of gene distribution within identified clusters I to V of the analysis in C. E: For each gene cluster identified as described in C (clusters I to V), a pairwise comparison of cryopreservation time for groups 2 to 4 against group 1 was performed. The mean fold changes per comparison ± SD were calculated and plotted, whereas group 1 was defined as baseline and set to 1.
An unsupervised gene selection approach with the most variable genes across the dataset, combined with hierarchical cluster analysis, indicated a cryopreservation time period-dependent clustering pattern of the PBMC samples, because subclusters mostly represent the different groups, as described above (Figure 2B, upper panel). In contrast, there was no clustering according to other factors, such as disease stage or sex distribution (Figure 2B). We next assessed differentially expressed genes across the four groups defined by the cryopreservation period, performing an analysis of variance analysis with Benjamini-Hochberg correction (P < 0.05). To exclude genes with very small differences in mean expression values, a fold change filter was integrated into the analysis of variance. Using these criteria, we identified 491 (FC ≥ 1.5) and 190 (FC ≥ 2) variant genes. The use of these gene lists for hierarchical clustering resulted again in a distinct separation of the PBMC samples according to their cryopreservation time period (Figure 2B, lower panel; FC ≥ 1.5). Samples stored for more than 40 months (group 3 and 4) were grouped in the right cluster tree and samples stored up to 40 months (group 1 and 2) were grouped in the left cluster tree, with only two exceptions (samples 27 and 48). These findings indicate that the cryopreservation time period is the main factor leading to distinction of the different PBMC groups, whereas there were no significant differences between age and sex distribution across the different groups as assessed by Fisher's exact test (data not shown).
To identify characteristic groups of genes with similar profiles of gene expression changes across the dataset, the mean expression values of the 190 genes assessed by analysis of variance (FC ≥ 2) were used for hierarchical cluster analysis. By this approach, two main gene clusters and three minor gene clusters were identified (Figure 2, C and D) that exhibited different kinetics (Figure 2, C and E). Genes of cluster II showed a relative constant expression in samples stored for <40 months, with a subsequent decline of signal intensity with increasing cryopreservation time. We correlated the signal intensities of these genes against the exact cryopreservation time (see Supplemental Table S1 at http://jmd.amjpathol.org) and found significant negative correlation of signal intensity with increasing cryopreservation time for 105 out of 109 genes (mean correlation, −0.69; range, −0.52 to −0.82).
The second main profile in gene expression changes also exhibited a relative constant expression in samples stored for <40 months, followed by a bump in signal intensities (often below background levels) in group 3. Expression levels in group 4 were characterized by an increase to maximum (genes of cluster IV; Figure 3, C and E). These findings indicate a certain degree of variance for this data set. Because of the variation in signal intensities among the four groups, there was no significant correlation with increasing cryopreservation time for these cluster IV genes. However, the signal intensities of cluster IV genes were all in a low to moderate range (50.8 to 498.7; see Supplemental Figure S1A at http://jmd.amjpathol.org); thus, the observed absolute quantitative changes in signal intensities were rather moderate, in comparison with cluster II genes (see Supplemental Figure S1B at http://jmd.amjpathol.org).
Figure 3.
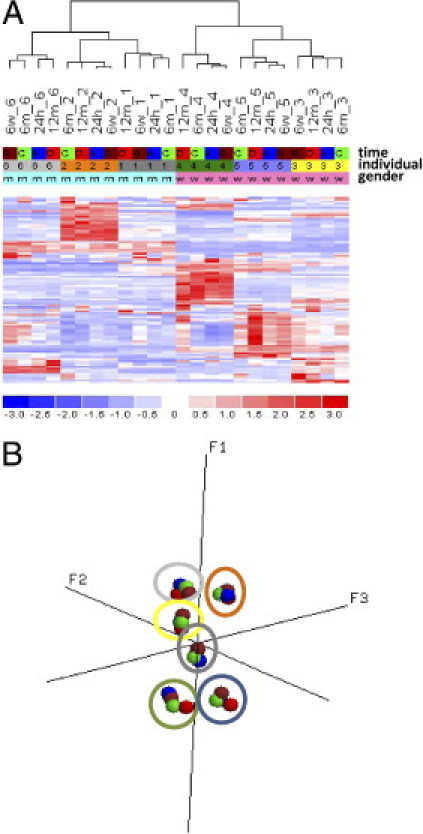
A: Hierarchical cluster analysis of PAX samples using the most variable genes across the data set (SD/mean, 0.5 to 10; n = 97 transcripts). Color key: Storage time and temperature: blue, 24 hours at room temperature; brown, 6 weeks at −20°C; green, 6 months at −20°C; red, 12 months at −20°C. Individuals 1 to 6: dark gray, 1; orange, 2; yellow, 3; green, 4; lilac, 5, light gray, 6. Sex: rose (w), female; blue (m), male. B: PCA using the most variable genes across the data set (SD/mean, 0.5 to 10; n = 97 genes). Color key: Storage time and temperature (solid circles) and individuals (outline circles) as in A.
Taken together, these data indicate that long-term cryopreservation of viable PBMCs in liquid nitrogen did not maintain stability of gene expression profiles over time. Rather, substantial changes in GEP were identified, characterized by pronounced loss of signal over time for many transcripts. This finding is in line with a pronounced decline in present calls in PBMC samples cryopreserved for more than 40 months in liquid nitrogen (data not shown).
Stability of GEP of Long-Term Stored PAXgene Blood Samples
GEP of isolated PBMCs is hampered by the ex vivo effects of sample handling, but the use of PAXgene blood collection tubes enabling direct RNA stabilization during the blood withdrawal process might provide a way out of this dilemma. GEP of freshly isolated PAXgene-stabilized blood reveals highly reproducible results.21,24,28 To further analyze the influence of long-term storage of PAXgene-stabilized blood samples on GEP, PAXgene-stabilized peripheral blood was collected from six healthy volunteers. All PAXgene samples derived from each of the six healthy donors were stored at room temperature for 24 hours before processing. Afterward, RNA was either directly isolated or PAXgene tubes were frozen for different time periods up to 12 months at −20°C prior to further processing for GEP experiments.
Gene expression profiles of these PAXgene samples were compared using unsupervised hierarchical cluster analysis, resulting in a sex- and individual-dependent clustering of the samples in which all four PAXgene samples of each volunteer clustered together (Figure 3A). This result clearly indicated that the variability in this data set depends predominantly on interindividual differences; prolonged freezing periods of the PAXgene blood samples had no major effect on gene expression profiles, and the results were highly similar to those of samples incubated at room temperature for 24 hours. Similar results were obtained performing PCA (Figure 3B). Again, all samples of one individual closely grouped together, indicating that interindividual differences remain consistent with the use of PAXgene tubes even after prolonged storage at −20°C. To assess differentially expressed genes among the four PAXgene groups according to their cryopreservation time period, an analysis of variance with Benjamini-Hochberg correction (P < 0.05) was applied to this data set. With this approach, no genes were identified matching these criteria, again strongly supporting the notion that storage of stabilized blood samples does not affect GEP. Moreover, in analysis of the present call rates of PAXgene samples stored for different time periods, no significant differences were observed in comparison of different cryopreservation time periods using a paired t-test (data not shown). These findings further indicate that prolonged freezing of PAXgene samples has no significant effect on GEP over time.
Discussion
Recently, we and others established blood-based GEP as a promising platform for identifying pharmacodynamic as well as predictive29–31 biomarkers related to the application of biologically active compounds. Moreover, it has been suggested that blood-based GEP can be used as a powerful tool for early diagnosis of certain solid cancer entities,32–36 as well as infectious,37,38 autoimmune,39,40 and neurodegenerative41 disease. Nevertheless, prospective clinical studies are still needed to validate these promising but still preliminary results in larger cohorts of patients or in individuals at risk for the respective diseases. Within clinical settings, blood samples are often collected at multiple sites but are analyzed at a single reference laboratory. Thus, processing and analysis of a large sample series at the same time point is nearly impossible, and so cryopreservation of such samples during a collection period is widely used, and samples are often stored for long time periods prior to further analysis. In this context, sample handling is a major issue that needs to be highly standardized prior to any genome-wide analysis, such as GEP.8,42–44 However, the general effect of cryopreservation of PBMCs as a routine method of sample storage and the effect of long-term cryopreservation on the stability of blood-based GEP had not hitherto been addressed, although much is known about cryopreservation-induced functional changes in PBMCs and subsets thereof.10–14
In the present study, we demonstrated that even short-term cryopreservation of PBMCs in DMSO-based freezing medium results in a distinct separation between all nonfrozen and cryopreserved counterpart samples. By applying unsupervised gene selection approaches combined with hierarchical cluster algorithms and PCA, a substantial introduction of technical-based variation in microarray data was shown. Cryopreservation of PBMCs resulted in differential expression of several genes, in part with dramatic fold changes (up to −27.7). For instance, several transcripts with high signal intensities in nonfrozen PBMCs (eg, LOC728358 and the Charcot-Leyden crystal protein) exhibited diminished signal intensities at background levels in the cryopreserved counterpart samples. Our results provide evidence that cryopreservation of PBMCs dramatically affects the detectability of individual transcripts, in line with a recent study investigating particular Charcot-Leyden crystal protein expression in cryopreserved PBMCs.17
In addition to the dramatic decrease in signal intensity leading to undetectability of several transcripts, we found an overall increase of the mean present call rate. This was related to a switch of absent calls to present calls slightly above background level, indicating increased noise in the microarray data of cryopreserved samples. With regard to long-term cryopreservation of PBMCs, we demonstrate an alteration of the gene expression profile over time, which was characterized by significant negative correlation of signal intensities of several transcripts with increasing cryopreservation time. Small but significant changes were also observed when analyzing the present call rates, with a pronounced decrease in present call rates over time. Moreover, unsupervised gene selection approaches, combined with hierarchical clustering, revealed a tendency to distinct separation of the long-term cryopreserved samples corresponding to their cryopreservation time periods, further corroborating cryopreservation time-dependent changes in the gene expression profile. The time-dependent alterations of GEP in long-term cryopreserved PBMCs are in line with the frequent observation that PBMC samples cryopreserved in liquid nitrogen for longer time periods behave differently in functional assays, compared with unfrozen samples.10–14 Together, these findings support the view that the transcriptome of viable PBMCs is not stabilized during the freezing period at all.
Because cryopreservation in liquid nitrogen was shown to be an unfavorable method for storage of PBMCs prior to GEP, we evaluated the freezing effect on GEP of peripheral blood samples that were directly drawn and stored in an RNA stabilizing agent. PAXgene blood collection tubes were stored at −20°C for up to 12 months. This approach was encouraged by recent findings demonstrating that freezing of PAXgene-stabilized blood samples at −70°C had no major effect on subsequent GEP.19,21 However, the stability of GEP of PAXgene blood samples in the context of long-term storage hitherto had not been addressed. In our longitudinal storage approach, no obvious effects on GEP were observed in PAXgene-stabilized blood samples, not even after prolonged cryopreservation time periods. In fact, even intraindividual differences were conserved over time. This indicates that gene expression profiles of whole blood samples remain highly stabilized also during prolonged freezing periods. In addition, samples that were directly processed after 24 hours incubation at room temperature showed high correlation with their frozen counterparts. These findings are corroborated by the absence of any significant transcriptional changes across the PAXgene data set. Within the time period of up to 12 months, we did not detect any significant changes in gene expression between immediately processed samples and samples stored at −20°C. This is in stark contrast to the freezing procedure used for preservation of PBMCs (Figure 1).
Altogether, we have provided evidence that cryopreservation of PBMCs in liquid nitrogen during short and long-term periods induces significant alterations to the blood-based transcriptome over time. Therefore, the use of adequate RNA stabilizing devices (such as the PAXgene RNA blood collection tube) is of advantage for preservation of clinical blood samples prior to GEP. The implementation of such procedures into standard tissue bank protocols for preservation of blood samples would be desirable. In any event, it is essential to establish an obligatory standard sample collection procedure before initiation of the respective GEP trial. This is important to avoid the introduction of technical bias that would render the resulting data ultimately not evaluable. In case of functional investigation of PBMCs, the use of cryopreserved PBMCs is still indispensable. However, one must keep in mind that the transcriptional and functional cryopreservation-induced changes might distort experimental outcome and data interpretation. Our GEP data set of cryopreserved PBMCs might help investigators to estimate in advance any cryopreservation-induced changes for their genes of interest before further investigation.
Footnotes
Supported in part by a Sofja-Kovalevskaja award from the Alexander von Humboldt-Foundation (J.L.S.) and a grant from the Deutsche Krebshilfe (S.D.-P.).
Supplemental material for this article can be found at http://jmd.amjpathol.org or at doi: 10.1016/j.jmoldx.2011.03.006.
Supplementary data
A: Distribution of signal intensities of cluster II and cluster IV genes in the different study groups (cryopreservation time period groups 1 to 4). B: For cluster II and cluster IV genes, a pairwise comparison of groups 2, 3, and 4 against group 1 as baseline was performed, and distribution of quantitative changes in mean expression for these comparisons were plotted. Minimum and maximum changes for each comparison are indicated. The boundary of the box closest to zero indicates the 25th percentile, the line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. Whiskers above and below the box indicate the 90th and 10th percentiles. Outliers are plotted as dots.
References
- 1.Ginsburg G.S., Willard H.F. Genomic and personalized medicine: foundations and applications. Transl Res. 2009;154:277–287. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Pene F., Courtine E., Cariou A., Mira J.P. Toward theragnostics. Crit Care Med. 2009;37(1 Suppl):S50–S58. doi: 10.1097/CCM.0b013e3181921349. [DOI] [PubMed] [Google Scholar]
- 3.Bates S. Progress towards personalized medicine. Drug Discov Today. 2010;15:115–120. doi: 10.1016/j.drudis.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Rockett J.C., Burczynski M.E., Fornace A.J., Herrmann P.C., Krawetz S.A., Dix D.J. Surrogate tissue analysis: monitoring toxicant exposure and health status of inaccessible tissues through the analysis of accessible tissues and cells. Toxicol Appl Pharmacol. 2004;194:189–199. doi: 10.1016/j.taap.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Staratschek-Jox A., Classen S., Gaarz A., Debey-Pascher S., Schultze J.L. Blood-based transcriptomics: leukemias and beyond. Expert Rev Mol Diagn. 2009;9:271–280. doi: 10.1586/erm.09.9. [DOI] [PubMed] [Google Scholar]
- 6.Campo Dell'Orto M., Zangrando A., Trentin L., Li R., Liu W.M., te Kronnie G., Basso G., Kohlmann A. New data on robustness of gene expression signatures in leukemia: comparison of three distinct total RNA preparation procedures. BMC Genomics. 2007;8:188. doi: 10.1186/1471-2164-8-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohlmann A., Haschke-Becher E., Wimmer B., Huber-Wechselberger A., Meyer-Monard S., Huxol H., Siegler U., Rossier M., Matthes T., Rebsamen M., Chiappe A., Diemand A., Rauhut S., Johnson A., Liu W.M., Williams P.M., Wieczorek L., Haferlach T. Intraplatform reproducibility and technical precision of gene expression profiling in 4 laboratories investigating 160 leukemia samples: the DACH study. Clin Chem. 2008;54:1705–1715. doi: 10.1373/clinchem.2008.108506. [DOI] [PubMed] [Google Scholar]
- 8.Debey S., Schoenbeck U., Hellmich M., Gathof B.S., Pillai R., Zander T., Schultze J.L. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J. 2004;4:193–207. doi: 10.1038/sj.tpj.6500240. [DOI] [PubMed] [Google Scholar]
- 9.Madabusi L.V., Latham G.J., Andruss B.F. RNA extraction for arrays. Methods Enzymol. 2006;411:1–14. doi: 10.1016/S0076-6879(06)11001-0. [DOI] [PubMed] [Google Scholar]
- 10.Owen R.E., Sinclair E., Emu B., Heitman J.W., Hirschkorn D.F., Epling C.L., Tan Q.X., Custer B., Harris J.M., Jacobson M.A., McCune J.M., Martin J.N., Hecht F.M., Deeks S.G., Norris P.J. Loss of T cell responses following long-term cryopreservation. J Immunol Methods. 2007;326:93–115. doi: 10.1016/j.jim.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkord E. Frequency of human T regulatory cells in peripheral blood is significantly reduced by cryopreservation. J Immunol Methods. 2009;347:87–90. doi: 10.1016/j.jim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Costantini A., Mancini S., Giuliodoro S., Butini L., Regnery C.M., Silvestri G., Montroni M. Effects of cryopreservation on lymphocyte immunophenotype and function. J Immunol Methods. 2003;278:145–155. doi: 10.1016/s0022-1759(03)00202-3. [DOI] [PubMed] [Google Scholar]
- 13.Disis M.L., dela Rosa C., Goodell V., Kuan L.Y., Chang J.C., Kuus-Reichel K., Clay T.M., Kim Lyerly H., Bhatia S., Ghanekar S.A., Maino V.C., Marcher H.T. Maximizing the retention of antigen specific lymphocyte function after cryopreservation. J Immunol Methods. 2006;308:13–18. doi: 10.1016/j.jim.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Weinberg A., Zhang L., Brown D., Erice A., Polsky B., Hirsch M.S., Owens S., Lamb K. Viability and functional activity of cryopreserved mononuclear cells. Clin Diagn Lab Immunol. 2000;7:714–716. doi: 10.1128/cdli.7.4.714-716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mollinedo F., López-Pérez R., Gajate C. Differential gene expression patterns coupled to commitment and acquisition of phenotypic hallmarks during neutrophil differentiation of human leukaemia HL-60 cells. Gene. 2008;419:16–26. doi: 10.1016/j.gene.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Graziano R.F., Ball E.D., Fanger M.W. The expression and modulation of human myeloid-specific antigens during differentiation of the HL-60 cell line. Blood. 1983;61:1215–1221. [PubMed] [Google Scholar]
- 17.Foell J.L., Volkmer I., Giersberg C., Kornhuber M., Horneff G., Staege M.S. Loss of detectability of Charcot-Leyden crystal protein transcripts in blood cells after treatment with dimethyl sulfoxide. J Immunol Methods. 2008;339:99–103. doi: 10.1016/j.jim.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Reimann K.A., Chernoff M., Wilkening C.L., Nickerson C.E., Landay A.L. Preservation of lymphocyte immunophenotype and proliferative responses in cryopreserved peripheral blood mononuclear cells from human immunodeficiency virus type 1-infected donors: implications for multicenter clinical trials: The ACTG Immunology Advanced Technology Laboratories. Clin Diagn Lab Immunol. 2000;7:352–359. doi: 10.1128/cdli.7.3.352-359.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Øvstebø R., Lande K., Kierulf P., Haug K.B. Quantification of relative changes in specific mRNAs from frozen whole blood - methodological considerations and clinical implications. Clin Chem Lab Med. 2007;45:171–176. doi: 10.1515/CCLM.2007.035. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy L., Vass J.K., Haggart D.R., Moore S., Burczynski M.E., Crowther D., Miele G. Hematopoietic lineage transcriptome stability and representation in PAXgene collected peripheral blood utilising SPIA single-stranded cDNA probes for microarray. Biomark Insights. 2008;3:403–417. doi: 10.4137/bmi.s938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vartanian K., Slottke R., Johnstone T., Casale A., Planck S.R., Choi D., Smith J.R., Rosenbaum J.T., Harrington C.A. Gene expression profiling of whole blood: comparison of target preparation methods for accurate and reproducible microarray analysis. BMC Genomics. 2009;10:2. doi: 10.1186/1471-2164-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rainen L., Oelmueller U., Jurgensen S., Wyrich R., Ballas C., Schram J., Herdman C., Bankaitis-Davis D., Nicholls N., Trollinger D., Tryon V. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–1890. [PubMed] [Google Scholar]
- 23.Thurner B., Röder C., Dieckmann D., Heuer M., Kruse M., Glaser A., Keikavoussi P., Kämpgen E., Bender A., Schuler G. Generation of large numbers of fully mature and stable dendritic cells from leukapheresis products for clinical application [Erratum appeared in J Immunol Methods 1999, 224:211] J Immunol Methods. 1999;223:1–15. doi: 10.1016/s0022-1759(98)00208-7. [DOI] [PubMed] [Google Scholar]
- 24.Feezor R.J., Baker H.V., Mindrinos M., Hayden D., Tannahill C.L., Brownstein B.H., Fay A., MacMillan S., Laramie J., Xiao W., Moldawer L.L., Cobb J.P., Laudanski, Miller-Graziano C.L., Maier R.V., Schoenfeld D., Davis R.W., Tompkins R.G., Inflammation and Host Response to Injury, Large-Scale Collaborative Research Program Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol Genomics. 2004;19:247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- 25.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., Hornik K., Hothorn T., Huber W., Iacus S., Irizarry R., Leisch F., Li C., Maechler M., Rossini A.J., Sawitzki G., Smith C., Smyth G., Tierney L., Yang J.Y., Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultze J.L., Eggle D. IlluminaGUI: graphical user interface for analyzing gene expression data generated on the Illumina platform. Bioinformatics. 2007;23:1431–1433. doi: 10.1093/bioinformatics/btm101. [DOI] [PubMed] [Google Scholar]
- 27.Backes C., Keller A., Kuentzer J., Kneissl B., Comtesse N., Elnakady Y.A., Muller R., Meese E., Lenhof H.P. GeneTrail–advanced gene set enrichment analysis. Nucl Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debey S., Zander T., Brors B., Popov A., Eils R., Schultze J.L. A highly standardized, robust, and cost-effective method for genome-wide transcriptome analysis of peripheral blood applicable to large-scale clinical trials. Genomics. 2006;87:653–664. doi: 10.1016/j.ygeno.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Classen S., Muth C., Debey-Pascher S., Eggle D., Beyer M., Mallmann M.R., Rudlowski C., Zander T., Pölcher M., Kuhn W., Lahn M., Schultze J.L., Staratschek-Jox A. Application of T cell-based transcriptomics to identify three candidate biomarkers for monitoring anti-TGFbetaR therapy. Pharmacogenet Genomics. 2010;20:147–156. doi: 10.1097/FPC.0b013e328335731c. [DOI] [PubMed] [Google Scholar]
- 30.Julià A., Barceló M., Erra A., Palacio C., Marsal S. Identification of candidate genes for rituximab response in rheumatoid arthritis patients by microarray expression profiling in blood cells. Pharmacogenomics. 2009;10:1697–1708. doi: 10.2217/pgs.09.99. [DOI] [PubMed] [Google Scholar]
- 31.Younossi Z.M., Baranova A., Afendy A., Collantes R., Stepanova M., Manyam G., Bakshi A., Sigua C.L., Chan J.P., Iverson A.A., Santini C.D., Chang S.Y. Early gene expression profiles of patients with chronic hepatitis C treated with pegylated interferon-alfa and ribavirin. Hepatology. 2009;49:763–774. doi: 10.1002/hep.22729. [DOI] [PubMed] [Google Scholar]
- 32.Burczynski M.E., Twine N.C., Dukart G., Marshall B., Hidalgo M., Stadler W.M., Logan T., Dutcher J., Hudes G., Trepicchio W.L., Strahs A., Immermann F., Slonim D.K., Dorner A.J. Transcriptional profiles in peripheral blood mononuclear cells prognostic of clinical outcomes in patients with advanced renal cell carcinoma. Clin Cancer Res. 2005;11:1181–1189. [PubMed] [Google Scholar]
- 33.Twine N.C., Stover J.A., Marshall B., Dukart G., Hidalgo M., Stadler W., Logan T., Dutcher J., Hudes G., Dorner A.J., Slonim D.K., Trepicchio W.L., Burczynski M.E. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res. 2003;63:6069–6075. [PubMed] [Google Scholar]
- 34.Aarøe J., Lindahl T., Dumeaux V., Saebø S., Tobin D., Hagen N., Skaane P., Lönneborg A., Sharma P., Børresen-Dale A.L. Gene expression profiling of peripheral blood cells for early detection of breast cancer. Breast Cancer Res. 2010;12:R7. doi: 10.1186/bcr2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma P., Sahni N.S., Tibshirani R., Skaane P., Urdal P., Berghagen H., Jensen M., Kristiansen L., Moen C., Sharma P., Zaka A., Arnes J., Sauer T., Akslen L.A., Schlichting E., Børresen-Dale A.L., Lönneborg A. Early detection of breast cancer based on gene-expression patterns in peripheral blood cells. Breast Cancer Res. 2005;7:R634–R644. doi: 10.1186/bcr1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Showe M.K., Vachani A., Kossenkov A.V., Yousef M., Nichols C., Nikonova E.V., Chang C., Kucharczuk J., Tran B., Wakeam E., Yie T.A., Speicher D., Rom W.N., Albelda S., Showe L.C. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with nonsmall cell lung cancer from patients with nonmalignant lung disease. Cancer Res. 2009;69:9202–9210. doi: 10.1158/0008-5472.CAN-09-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berry M.P., Graham C.M., McNab F.W., Xu Z., Bloch S.A., Oni T., Wilkinson K.A., Banchereau R., Skinner J., Wilkinson R.J., Quinn C., Blankenship D., Dhawan R., Cush J.J., Mejias A., Ramilo O., Kon O.M., Pascual V., Banchereau J., Chaussabel D., O'Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaussabel D., Allman W., Mejias A., Chung W., Bennett L., Ramilo O., Pascual V., Palucka A.K., Banchereau J. Analysis of significance patterns identifies ubiquitous and disease-specific gene-expression signatures in patient peripheral blood leukocytes. Ann N Y Acad Sci. 2005;1062:146–154. doi: 10.1196/annals.1358.017. [DOI] [PubMed] [Google Scholar]
- 39.Chaussabel D., Quinn C., Shen J., Patel P., Glaser C., Baldwin N., Stichweh D., Blankenship D., Li L., Munagala I., Bennett L., Allantaz F., Mejias A., Ardura M., Kaizer E., Monnet L., Allman W., Randall H., Johnson D., Lanier A., Punaro M., Wittkowski K.M., White P., Fay J., Klintmalm G., Ramilo O., Palucka A.K., Banchereau J., Pascual V. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batliwalla F.M., Li W., Ritchlin C.T., Xiao X., Brenner M., Laragione T., Shao T., Durham R., Kemshetti S., Schwarz E., Coe R., Kern M., Baechler E.C., Behrens T.W., Gregersen P.K., Gulko P.S. Microarray analyses of peripheral blood cells identifies unique gene expression signature in psoriatic arthritis. Mol Med. 2005;11:21–29. doi: 10.2119/2006-00003.Gulko. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grunblatt E., Bartl J., Zehetmayer S., Ringel T.M., Bauer P., Riederer P., Jacob C.P. Gene expression as peripheral biomarkers for sporadic Alzheimer's disease. J Alzheimers Dis. 2009;16:627–634. doi: 10.3233/JAD-2009-0996. [DOI] [PubMed] [Google Scholar]
- 42.Gaarz A., Debey-Pascher S., Classen S., Eggle D., Gathof B., Chen J., Fan J.B., Voss T., Schultze J.L., Staratschek-Jox A. Bead array-based microRNA expression profiling of peripheral blood and the impact of different RNA isolation approaches. J Mol Diagn. 2010;12:335–344. doi: 10.2353/jmoldx.2010.090116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asare A.L., Kolchinsky S.A., Gao Z., Wang R., Raddassi K., Bourcier K., Seyfert-Margolis V. Differential gene expression profiles are dependent upon method of peripheral blood collection and RNA isolation. BMC Genomics. 2008;9:474. doi: 10.1186/1471-2164-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min J.L., Barrett A., Watts T., Pettersson F.H., Lockstone H.E., Lindgren C.M., Taylor J.M., Allen M., Zondervan K.T., McCarthy M.I. Variability of gene expression profiles in human blood and lymphoblastoid cell lines. BMC Genomics. 2010;11:96. doi: 10.1186/1471-2164-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Distribution of signal intensities of cluster II and cluster IV genes in the different study groups (cryopreservation time period groups 1 to 4). B: For cluster II and cluster IV genes, a pairwise comparison of groups 2, 3, and 4 against group 1 as baseline was performed, and distribution of quantitative changes in mean expression for these comparisons were plotted. Minimum and maximum changes for each comparison are indicated. The boundary of the box closest to zero indicates the 25th percentile, the line within the box marks the median, and the boundary of the box farthest from zero indicates the 75th percentile. Whiskers above and below the box indicate the 90th and 10th percentiles. Outliers are plotted as dots.


