Abstract
The increasing number of disease-causing mutations demands a simple, direct, and cost-effective diagnostic genotyping technique capable of detecting multiple mutations. This study validated the efficacy of a novel melting curve analysis–based genotyping assay (MeltPro HBB assay) for 24 β-thalassemia mutations in the Chinese population. The diagnostic potential of this assay was evaluated in 1022 pretyped genomic DNA samples, including 909 clinical cases of β-thalassemia minor or major, using a double-blind analysis in a multicenter validation study. Reproducibility of the assay was 100%, and the limit of detection was 10 pg per reaction. All 24 β-thalassemia mutations were accurately genotyped, and β-thalassemia genotypes were correctly determined in all 1022 samples, yielding overall sensitivity and specificity of 100%. The concordance rate was 99.4% between this assay and the reference method. It was concluded that the MeltPro HBB assay is useful for reliable genotyping of multiple β-thalassemia mutations in clinical settings and may have potential as a versatile method for rapid genotyping of known mutations because of its high throughput, accuracy, ease of use, and low cost.
See related Commentary on page 369
β-Thalassemia, an autosomal recessive form of hemoglobinopathy, is one of the most widespread life-shortening genetic diseases in humans.1,2 According to the World Health Organization,3 approximately 1.5% of the world population are carriers of this disease. Each year, approximately 40,000 infants positive for β-thalassemia major are reported in areas endemic for thalassemia, which include the Mediterranean countries, parts of north and west Africa, the Middle East, India, and southeast Asia including southern China.3,4 In China, the estimated incidence of carriers of β-thalassemia is as high as 7% in the Guangxi Province, and 1% to 6% in populations elsewhere in the endemic areas of southern China.5–8
β-Thalassemia is a severe disease, and its treatment requires many resources, both medical and financial. While many cities in China have facilities for diagnosis and treatment of this disease, free medical services are not provided by the government in most rural areas. Treatment is available to only the few patients who can afford it, although this situation is more favorable in some regions with better economy. To improve the overall situation, the Chinese government is instituting a population-based screening program with the objective of reducing the birth rate of fetuses with β-thalassemia in endemic regions, and one of its goals is to provide free testing. As part of this program, a simple, reliable, cost-effective genetic testing method is required for disease confirmation and to screen for carriers.
In a decades-long effort to establish an ethnic group–specific β-thalassemia genetic testing assay, we and our Chinese colleagues systematically investigated the types and frequency of the causative mutations of the hemoglobin-β gene (HBB) in the endemic regions of China. To date, 45 single-nucleotide mutations and small deletions in HBB have been detected in the Chinese population.5–10 Translation of the data into clinical practice requires a simple, reliable, cost-effective method that is able to detect all of these mutations. Among dozens of methods that have been developed for detecting β-thalassemia mutations,11–13 reverse dot blot (RDB) analysis is the only method approved in China.14 Despite its cost-effectiveness, broad coverage of mutations, and long-term adoption in clinical settings, its use in carrier screening is limited, primarily because of low throughput and subjective results. We previously developed and evaluated a denaturing high-performance liquid chromatography–based mutation screening method; however, it did not achieve widespread use because of its cost, tedious manipulations, and narrow detection of the mutation spectrum.15 This experience taught us that a method for routine β-thalassemia carrier screening must be inexpensive, easy to use, rapid, accurate, and robust.
In the present study, we validated a new β-thalassemia mutation detection kit specifically developed to screen for Chinese carriers based on the mutation spectrum. The MeltPro HBB assay (Xiamen Zeesan Biotech Co., Ltd., Xiamen, Fujian, China) is a qualitative in vitro diagnostic method designed to genotype a panel of 24 single-nucleotide mutations and small indels in the HBB gene that cause β-thalassemia or abnormal hemoglobin. The test kit is based on a proprietary, multicolored, self-quenched, probe-based melting curve analysis that is performed using a standard real-time PCR instrument, from which genotype information for each mutation is retrieved based on the melting temperature (Tm) or difference in Tm between wild-type and mutant (ΔTm) DNA samples. To assess the clinical value of this test, we conducted a multicenter validation study including 1022 pretyped genomic DNA samples collected from six hospitals in the Guangdong and Guangxi Provinces, with special attention to factors required for routine use.
Materials and Methods
DNA Samples
Genomic DNA (gDNA) samples were collected from November 2008 to April 2010, and met the following eligibility criteria: (1) all had a record of hematologic phenotypes determined via full blood cell counts and a hemoglobin test, as previously described,6 (2) all had HBB genotype information determined via RDB analysis or sequencing,14 and (3) the template concentration was >10 ng/μL, with a 260- to 280-nm absorption ratio in the range of 1.6 to 2.0.
In total, 1022 gDNA samples were collected from individuals with wild-type, heterozygous carriers, homozygotes, and compound heterozygotes. The latter three sample types contained a number of different genotypes (Table 1). Because these gDNA samples were obtained from different laboratories, the DNA extraction method also varied. Of the 1022 samples, 723 were extracted using the classic chloroform-phenol method, 41 using the QIAamp DNA Blood Mini Kit (Qiagen NV, Venlo, The Netherlands), 198 using the Lab-Aid 820 Automated Blood DNA Extraction System (Xiamen Zeesan Biotech Co., Inc., Xiamen, China), and 58 using the RelaxGene Blood DNA System (TianGen Biotech Co., Ltd., Beijing, China); and two amniotic fluid samples were extracted using the QIAamp DNA Blood Mini Kit. The quantity and quality of the extracted DNA were evaluated by measuring absorption at 260 and 280 nm using a spectrophotometer (ND-1000 UV-Vis; NanoDrop Technologies, Inc., Wilmington, DE).
Table 1.
Sample Types Detected
| Sample type | Case no. | Genotype no. | Allele no. |
|---|---|---|---|
| Wild-type | 113 | 1 | 226 |
| Heterozygous | 733 | 21 | 1466⁎ |
| Homozygous | 54 | 11 | 108† |
| Compound heterozygous | 122 | 31 | 244† |
| Total | 1022 | 64 | 2044 |
Includes 733 wild-type and 733 mutant alleles comprising 21 mutation types.
Mutant alleles comprising 15 mutation types.
Multicenter Validation Procedure
The multicenter validation study included six hospital laboratories from Guangdong and Guangxi Provinces, representing the regions in China most endemic for β-thalassemia. The study was performed in one central academic laboratory. DNA samples were renumbered by a technician (X.W.) who was solely in charge of the data collection and statistical analysis. The coded DNA samples from each hospital laboratory were analyzed by two technicians (X.C. consistently analyzed the DNA samples with the help of Q.X., T.Z., S.L., X.Y., Y.H., and Y.Q.) from the corresponding laboratories. The results obtained were reported to a third individual (F.X.), who checked the data and calculated the coincidence rate, sensitivity, and specificity.
MeltPro HBB Assay
The MeltPro HBB assay uses asymmetric PCR with two primers to generate excess single-stranded amplicons for probe hybridization followed by melting curve analysis using four-color real-time PCR (Figure 1). This assay is a one-step genotyping method, with PCR amplification and melting curve analysis performed using one program on one machine. As an alternative protocol, PCR and melting curve analysis can also be performed using separate machines; for example, amplification is performed using a standard thermal cycler, and melting analysis is performed using a real-time PCR instrument.
Figure 1.
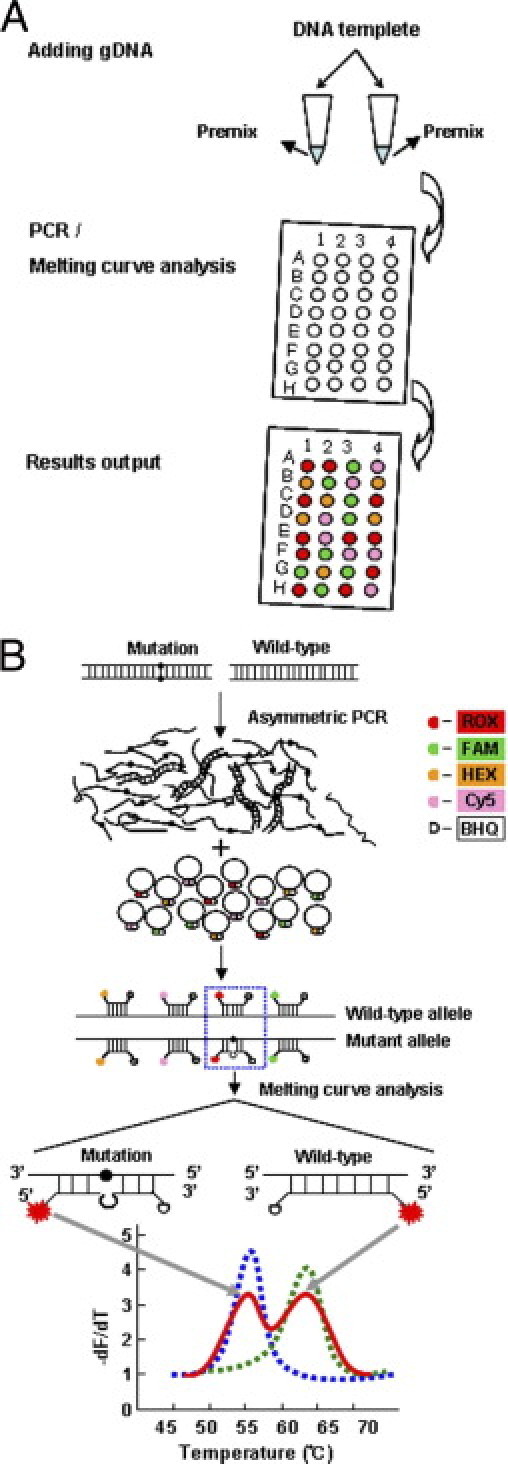
Proposed MeltPro HBB assay. A: Genotyping procedure. After DNA extraction, each sample tested is mixed with premixed PCR reagents in two separate tubes. PCR and the subsequent melting curve analysis were performed using a real-time PCR thermal cycler. Graphic output of genotyping results is produced. B: Principle of the MeltPro HBB assay. Asymmetric PCR is used to generate single-strand target sequences for hybridization using dual-labeled self-quenched probes, which are labeled at one of four different fluorophores at 5′-end and a quencher dye at 3′-end. One probe may detect one or several adjacent mutations, and it may also be possible for certain mutations to be detected by more than one probe. Examples shown are heterozygous (red line), wild-type (dashed green line), and homozygous mutant (dashed blue line) samples.
In each assay, extracted gDNA was added to two reaction tubes (5 μL gDNA per reaction tube) containing premixed PCR reagent. The assay protocol began with an amplification procedure consisting of denaturation for 3 minutes at 95°C; 50 amplification cycles of 15 seconds at 95°C, 10 seconds at 55°C, and 20 seconds at 76°C; followed by a melting analysis procedure consisting of denaturation for 1 minute at 95°C, hybridization for 5 minutes at 35°C, and incremental temperature increase (1°C per 5 seconds) from 40°C to 80°C. Fluorescence from FAM, HEX, ROX, and CY5 channels was recorded at each step during the melting analysis procedure. The evaluation experiments were performed using real-time PCR instruments (Rotor-Gene 3000 and 6000; Corbett Research Australia, Mortlake, NSW, Australia) with software that automatically identifies the mutation type and sample genotype based on Tm values.
Reproducibility Study
The reproducibility study was performed in two phases based on proficiency and performance. The proficiency phase was performed to ensure that each operator had the required expertise in use of the method so that meaningful data could be generated. For proficiency testing, two technicians from each of the six hospitals ran the same panel in duplicate. Data generated during this phase were used to demonstrate technician proficiency. Reproducibility testing for the performance phase was conducted using a panel of samples containing all of the types of mutations. Each technician ran the same panel in duplicate on five nonconsecutive days. The panel included one wild-type sample, 8 plasmid samples, and 24 prospectively collected clinical case samples. The Tm value of each genotype and the Tm difference between the wild-type and mutant samples were detected.
Limit of Detection Study
For the study of analytical sensitivity, wild-type human gDNA was extracted from whole blood using the QIAamps DNA Blood Mini Kit (Qiagen NV). The extracted DNA was quantified using UV and serially diluted 10-fold with water, yielding DNA concentrations ranging from 100 ng to 10 pg in a 5 μL reaction, which were analyzed using the MeltPro HBB assay. For each concentration, three replicates were analyzed using the MeltPro HBB assay. Water was used as no-template control.
Statistical Analysis
The χ2 test was used to evaluate whether a significant difference existed between the results obtained using the MeltPro HBB assay and RDB analysis or DNA sequencing. Statistical analysis was performed using commercially available software (STATA version 8.0; StataCorp LP, College Station, TX). P < 0.05 was considered significant.
Results
Reproducibility
The reproducibility study demonstrated 100% agreement for within-operator or within-day results, between days or within-operator results, between operators or within-hospital results, and between-hospital results (Figure 2 and Table 2). No Tm overlap was observed between wild-type and mutant alleles in any of the detection channels: ΔTm (3 SD) range, 1.7°C (0.35°C) (ROX channel in tube 1) to 20.0°C (0.45°C) (ROR channel in tube 2). Tm overlap was not observed between different alleles within the same detection channel; the smallest ΔTm difference (1.3°C between c.216-217insT and c.216-217insA of ROX channel, tube 2) was above the resolution level of 0.6°C (defined by 3 SD). Among the 24 mutant alleles included in the MeltPro HBB assay, three pairs of alleles, c.-80T>C and c.-81A>C, c.92 + 1G>T and c.92 + 5G>C, and c.113G>A and c.130G>T, are designated as degenerate mutant alleles because they are not differentiable within each pair, although all of them can be differentiated from wild-type and other mutant alleles.
Figure 2.
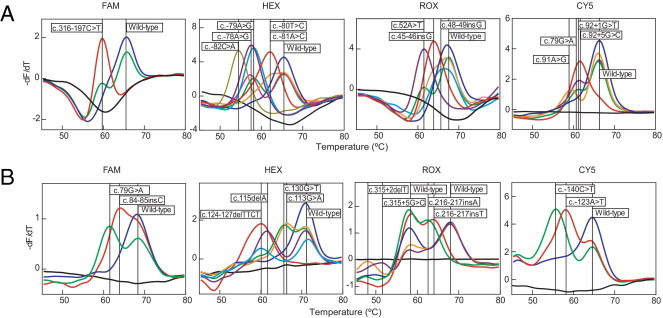
Identification of β-thalassemia genotypes using the MeltPro HBB assay. The negative derivative fluorescence over temperature versus temperature was plotted for each sample. Melting curves and corresponding genotypes (box) of the 24 mutations plus wild-type alleles are given according to tube 1 (A) and tube 2 (B). The four detection channels are shown by the corresponding fluorophores. Melting curves of HEX and CY5 channels in tube 1 and of HEX and ROX channels in tube 2 are separated into two panels. Black lines indicate no-template control.
Table 2.
Calibrated Tm of 24 Mutations Detected Using the MeltPro HBB Assay
| Tube | Color | Genotype | Tm (°C) | ΔTm (3 SD) (°C)⁎ |
|---|---|---|---|---|
| 1 | FAM | Wild-type | 65.9 | NA |
| c.316-197C>T | 59.9 | 6.0 (0.42) | ||
| HEX | Wild-type | 65.4 | NA | |
| c.-78A>G | 57.0 | 8.4 (0.39) | ||
| c.-79A>G | 58.4 | 7.0 (0.54) | ||
| c.-80T>C or c.-81A>C† | 62.1 | 3.3 (0.42) | ||
| c.-82C>A | 54.3 | 11.1 (0.39) | ||
| ROX | Wild-type | 67.7 | NA | |
| c.45-46insG | 66.0 | 3.1 (0.45) | ||
| c.48-49insG | 66.6 | 1.7 (0.35) | ||
| c.52A>T | 62.2 | 5.5 (0.54) | ||
| Cy5 | Wild-type | 67.1 | NA | |
| c.79G>A | 61.6 | 5.4 (0.31) | ||
| c.92 + 1G>T or c.92 + 5G>C† | 63.4 | 3.7 (0.33) | ||
| c.91A>G | 59.0 | 7.9 (0.51) | ||
| 2 | FAM | Wild-type | 68.4 | NA |
| c.79G>A | 61.4 | 7.0 (0.36) | ||
| c.84-85insC | 64.0 | 4.4 (0.45) | ||
| HEX | Wild-type | 71.0 | NA | |
| c.124-127delTTCT | 60.1 | 10.8 (0.54) | ||
| c.115delA | 61.3 | 9.7 (0.48) | ||
| c.130G>T or c.113G>A† | 66.3 | 4.7 (0.48) | ||
| ROX | Wild-type | 68.1 | NA | |
| c.216-217insT | 63.8 | 4.3 (0.54) | ||
| c.216-217insA | 62.5 | 5.6 (0.50) | ||
| c.315 + 2delT | 48.1 | 20.0 (0.45) | ||
| c.315 + 5G>C | 51.4 | 16.7 (0.24) | ||
| Cy5 | Wild-type | 64.6 | NA | |
| c.-123A>T | 58.1 | 6.5 (0.42) | ||
| c.-140C>T | 55.7 | 8.9 (0.33) |
NA, not applicable.
ΔTm = Tm (wild-type) − Tm (mutant).
These mutations are judged to be the same type within pairs using the MeltPro HBB assay.
Limit of Detection
Sources of clinical samples include whole blood, sputum, hair, and amniotic fluid, among others, and different sample types may yield various amounts of gDNA. It is, therefore, necessary to test the analytical sensitivity and concentration range of the assay. Using the 10-fold serially diluted gDNA ranging from 100 ng to 10 pg in a 5 μL reaction, the results demonstrated that the entire range could be detected repeatedly in all three replicates in both real-time PCR and melting curve analysis (Figure 3). The limit of detection was 10 pg gDNA per reaction, which is equivalent to roughly one diploid genome (6.7 pg gDNA) per reaction. The same analysis was also performed using gDNA obtained from the four extraction methods: chloroform-phenol method, QIAamp DNA Blood Mini Kit (Qiagen NV), Lab-Aid 820 Automated Blood DNA Extraction System (Xiamen Zeesan Biotech Co., Inc.), and RelaxGene Blood DNA System (TianGen Biotech Co., Ltd.). Identical limit of detection was obtained regardless of the extraction method.
Figure 3.
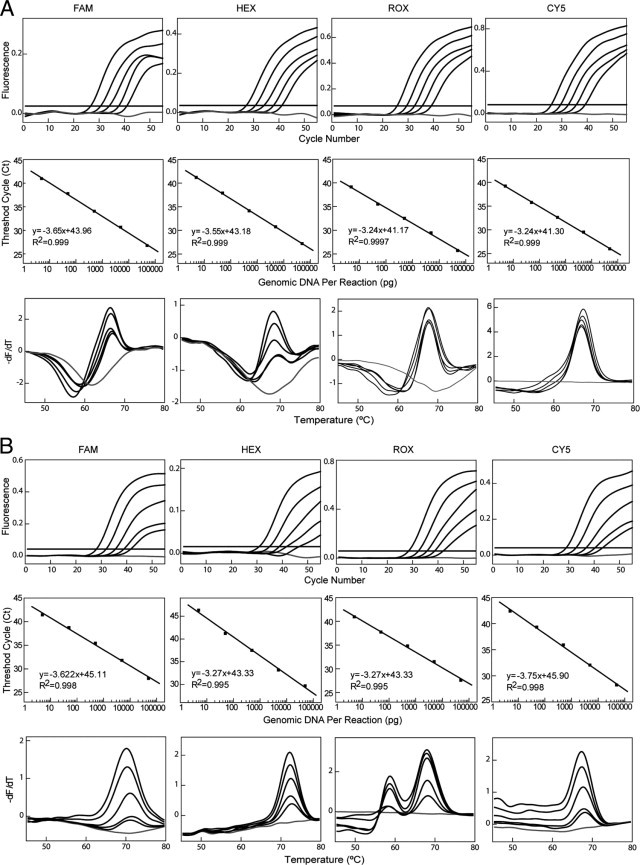
Analytical sensitivity analysis of the MeltPro HBB assay. Tenfold serial dilutions of wild-type DNA template ranging from 100 ng to 10 pg in a 25-μL reaction were analyzed using four detection channels (indicated by the labeling fluorophores) in reaction tube 1 (A) and tube 2 (B). Top panels: Real-time PCR amplification curves. Middle panels: Linear relationship between the number of threshold cycle values (Cq) and the logarithm of the mass of genomic DNA. Bottom panels: Corresponding melting curves. Water was used as the no-template control (gray lines).
Double-Blind Analysis of 1022 Samples
The 1022 samples were precharacterized using RDB analysis or bidirectional sequencing. Because RDB analysis detects 17 mutations, and 15 of these are detected by the MeltPro HBB assay, the other 9 mutations not detected using RDB analysis were determined using the sequencing method. Two mutations, c.2T>G and c.162delT, detected using RDB analysis but not the MeltPro HBB assay, were used as negative controls for the HBB assay. Of note, 45 β-thalassemia mutations have been reported in southern China. Among these, 8 mutations account for more than 95% of all HBB mutations in the Chinese population. These 8 common mutations and 7 mutations with medium prevalence are detected by both the MeltPro HBB assay and RDB analysis. The remaining 30 mutations, including the 2 detected using RDB analysis but not the MeltPro HBB assay, 9 mutations detected by the MeltPro assay but not RDB analysis, and 21 mutations not detected by either of the two methods, are all rare, and their omission does not pose a negative effect on the utility of an assay. Inasmuch as the 2 rare mutations detected using RDB analysis have not been detected in recent years, they have been removed from the MeltPro HBB assay, and 9 additional rare mutations recently detected have been added. Although the current version of the kit is designed to detect a limited number of mutations, the method can detect all mutations in the amplicon between the two primers.
Twenty-seven allele types were included in the validation study, including 24 mutant alleles (all detected using the MeltPro HBB assay), 1 wild-type allele, and 2 mutant alleles not detected using the MeltPro HBB assay. Except for the 2 mutant alleles unique to RDB analysis, all alleles were correctly detected using the MeltPro HBB assay. The allele types and numbers detected in the 1022 samples using the MeltPro HBB assay are given in Table 3, which demonstrates complete agreement between results of RDB analysis and DNA sequencing. The genotypes and numbers are listed in Table S1 (available at http://jmd.amjpathol.org). The double-blind assay demonstrated that the MeltPro HBB assay yielded 100% sensitivity and specificity for all included mutations. The concordance rate between the MeltPro HBB assay and RDB analysis or DNA sequencing was 99.4% because of the 2 mutations omitted from the MeltPro HBB assay.
Table 3.
Mutation Type and Number of Alleles Detected
| Number | Mutation type | MeltPro HBB assay | RDB | Sequencing | Phenotype |
|---|---|---|---|---|---|
| 1 | c.124-127delTTCT | 336 | 336 | ND | β0 |
| 2 | c.316-197C>T | 185 | 185 | ND | β+ |
| 3 | c.52A>T | 166 | 166 | ND | β0 |
| 4 | c.-78A>G | 127 | 127 | ND | β+ |
| 5 | c.79G>A | 77 | 77 | ND | HbE |
| 6 | c.216-217insA | 68 | 68 | ND | β0 |
| 7 | c.130G>T or c.113G>A⁎ | 26 | 23 | 3 | β0 |
| β0 | |||||
| 8 | c.92 + 1G>T or c.92 + 5G>C⁎ | 25 | 25 | ND | β0 |
| β+ (severe) | |||||
| 9 | c.-79A>G | 21 | 21 | ND | β+ |
| 10 | c.84-85insC | 17 | 17 | ND | 0 |
| 11 | c.315 + 5G>C | 9 | NT | 9 | β+ |
| 12 | c.45-46insG | 8 | 8 | ND | β0 |
| 13 | c.216-217insT | 2 | NT | 2 | β0 |
| 14 | c.48-49insG | 2 | NT | 2 | β0 |
| 15 | c.-140C>T | 1 | NT | 1 | β+ |
| 16 | c.-123A>T | 1 | NT | 1 | β+ |
| 17 | c.-82C>A | 2 | 2 | ND | β+ |
| 18 | c.-81A>C or c.-80T>C⁎ | 3 | 2 | 1 | β+ |
| β+ | |||||
| 19 | c.91A>G | 1 | NT | 1 | β0 |
| 20 | c.115delA | 1 | NT | 1 | β0 |
| 21 | c.315 + 2delT | 1 | NT | 1 | β0 |
| 22 | c.162delT† | NT | NT | 3 | Dominant |
| 23 | c.2T>G† | NT | 3 | ND | β0 |
| 24 | Wild type | 965 | 959 | ND | NA |
| Total | 2044 | 2019 | 25 | NA |
NA, not applicable; ND, not done; NT, not tested; RDB, reverse dot blot method.
These mutations are judged to be the same type within pairs using the MeltPro HBB assay.
Mutation detected using RDB but not the MeltPro HBB assay.
Comparison of Assay Throughput
The MeltPro HBB assay is a one-step homogenous assay. Overall analysis time was 2 hours 40 minutes using the four-color Rotor-Gene 3000 or 6000 real-time rotary analyzer. Compared with RDB analysis and DNA sequencing, overall decreased time using the MeltPro HBB assay was 38% and 57%, respectively (Figure 4). Decreased operative time was even more significant when considering that the MeltPro HBB assay is basically a single-step procedure with negligible hands-on operation, whereas both RDB analysis and DNA sequencing require multiple steps after PCR. In our hands, the overall MeltPro HBB assay time was further shortened to less than 2 hours when the amplification procedure was performed in a fast 96-well PCR thermal cycler (Black KingKong EDC-810 Gene Amplifier, Eastwin, Beijing, China) and the melting curve analysis was performed using real-time PCR. Although operative time was lengthened to 15 minutes because of the additional tube transfer step, the overall throughput was significantly increased.
Figure 4.
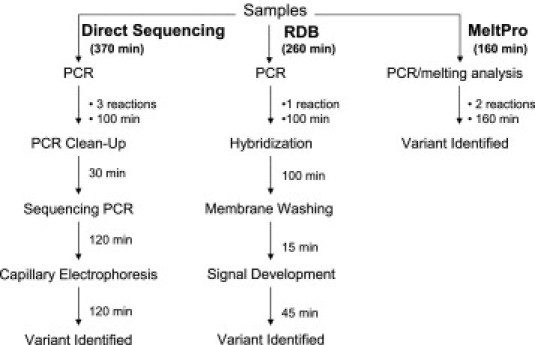
Comparison of the MeltPro HBB assay, RDB analysis, and sequencing insofar as time used for HBB variant genotyping.
Discussion
The MeltPro HBB assay was validated insofar as reproducibility, limit of detection, accuracy, and assay throughput. Reproducibility was 100% in all aspects, and limit of detection was 10 pg per reaction, which is close to that of a single-diploid genome per reaction. Overall accuracy was 99.4% for genotype and 99.7% for allele number, based on the multicenter study with 1022 clinical samples from six hospitals. Moreover, the assay has much higher throughput and requires less operative time than either RDB analysis or DNA sequencing. To our knowledge, the MeltPro HBB assay detects the largest number of mutations for β-thalassemia in China.
Insofar as clinical practice, the homogeneity of the MeltPro HBB assay provides advantages over heterogeneous methods such as RDB analysis and denaturing high-performance liquid chromatography. First, the MeltPro HBB assay requires no complex post-PCR processing, resulting in minimal risk of carryover contamination. In contrast, all heterogeneous methods involve heavy manipulations of PCR products, entailing stringent measures in laboratory management and personnel training to avert carryover contamination. Second, compared with heterogeneous methods, the MeltPro HBB assay has much higher throughput. The entire procedure involves a simple step of adding gDNA to two reaction tubes, and mutation detection can be automatically accomplished within 3 hours. When combined with use of a standard thermal cycler, overall throughput can be further improved, although an extra reaction tube transfer step is needed. This improvement was reflected in that the overall experimental part of this validation study was accomplished within 1 week. It is conceivable that by using one 4-color 96-well real-time PCR instrument supported by a sufficient number of standard thermal cyclers, more than 1000 samples can be analyzed within 1 workday. Such high throughput could afford large-scale population screening in a medium-sized genetic testing laboratory. In contrast, using standard RDB analysis, regardless of its intensive labor and long operative time, at least 10 workdays would be needed to process the same number of samples. Third, the readout of the MeltPro HBB assay is automatically generated, and the final report can be exported to the laboratory information management system via dedicated software. In contrast, many heterogeneous methods such as RDB analysis or denaturing high-performance liquid chromatography require manual reading, which may lead to ambiguous results and difficult information transfer. Fourth, the MeltPro HBB assay is extremely cost-effective. Its list price is approximately $2 per sample, compared with about $5 per sample for RDB analysis and about $10 per sample for DNA sequencing. Although a multicolor real-time PCR instrument can be a large investment, the overall operating expense is low when considering its minimal consumables cost, small labor cost, and short turnaround time. Thus, the data demonstrate that the MeltPro HBB assay outperforms existing heterogeneous methods and is suitable for routine confirmative diagnosis and for large-scale screening for carriers of β-thalassemia.
The merit of melting curve analysis for mutation detection has been well demonstrated by wide application of LightCycler technology (Roche Diagnostics Corp., Indianapolis, IN) in molecular diagnostics.16 Melting methods have also been developed for detection of HBB mutations using LightCycler technology with dual hybridization probes; however, the limited number of mutations detected by these methods hinder their use in clinical diagnostics.17,18 Because one probe can detect more than one allele in melting curve analysis, these reports showed that more mutations could be detected in one reaction than with real-time PCR. This advantage is also reflected in the MeltPro HBB assay because 24 mutations can be detected in two reactions. To our knowledge, this is the largest number of mutations detectable in one assay based on melting curve analysis. The MeltPro HBB assay uses self-quenched probes for melting curve analysis. Unlike the dual-hybridization probes, which are best used with the LightCycler system for multicolor detection, the MeltPro HBB assay can be used in any real-time PCR instrument equipped with four detection channels. This enhanced cross-platform compatibility is an additional benefit of the MeltPro HBB assay in clinical settings equipped with different models of real-time PCR instruments.
Many simple methods for genotyping HBB mutations have been developed including restriction fragment-length polymorphism analysis,19 the amplification refractory mutation system,20 single-strand conformation polymorphism analysis,21 denaturing gradient gel electrophoresis,22,23 and direct DNA sequencing.24 Although these research-orientated methods can detect HBB mutations on different levels, they cannot be readily adopted for routine use, primarily because of low throughput. Recent advances in genotyping technology have enabled high throughput screening of multiple mutations in a large number of samples. For example, allele-specific arrayed primer extension has been designed for simultaneous detection of 15 nondeletion α-globin gene defects and 23 β-globin gene mutations commonly observed in countries in southeast Asia.25 Multiplex minisequencing has also been developed in combination with subsequent detection using a variety of platforms including capillary electrophoresis,26 denaturing high-performance liquid chromatography,27,28 and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.29,30 However, these high-end techniques are best suited for use in a well-equipped central laboratory because of their expense and the high level of expertise required.
The high prevalence and severity of β-thalassemia necessitate an effective diagnostic and prevention program. Although phenotype determination is widely used in population screening, genetic testing is often considered a confirmative diagnostic tool. Therefore, genetic testing is often used for rapid mutation screening in carriers of reproductive age and in confirmatory diagnostic testing of prenatal samples. However, the technical complexity inherent in genetic testing often hinders its widespread application, especially in developing countries. Because of its accuracy, robustness, ease of use, low cost, and high throughput, the MeltPro HBB assay can be recommended as an alternative test to screen for β-thalassemia carriers, and can be used routinely in laboratories with access to real-time PCR. In addition, use of the MeltPro HBB assay may be extended to detection of mutations in ethnic populations other than Chinese.
Footnotes
Supported by grant U0632005 from the National Science Foundation of China–Guangdong Joint Fund and by grant 2008A030201018 from the Project of Technology of Guangdong Province.
F.X. and Q.H. contributed equally to this work.
Supplemental material for this article can be found at http://jmd.amjpathol.org or at doi:10.1016/j.jmoldx.2011.03.005.
Contributor Information
Qingge Li, Email: qgli@xmu.edu.cn.
Xiangmin Xu, Email: gzxuxm@pub.guangzhou.gd.cn.
Supplemental data
References
- 1.Cao A., Galanello R. Beta-thalassemia. Genet Med. 2010;12:61–76. doi: 10.1097/GIM.0b013e3181cd68ed. [DOI] [PubMed] [Google Scholar]
- 2.Weatherall D.J. The inherited diseases of hemoglobin are an emerging global health burden. Blood. 2010;115:4331–4336. doi: 10.1182/blood-2010-01-251348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modell B., Darlison M. Global epidemiology of haemoglobin disorders and derived service indicators. Bull World Health Organ. 2008;86:480–487. doi: 10.2471/BLT.06.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galanello R., Origa R. Beta-thalassemia. Orphanet J Rare Dis. 2010;5:11. doi: 10.1186/1750-1172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng Y., Huang S. The studies of hemoglobinopathies and thalassemia in China: the experiences in Shanghai Institute of Medical Genetics. Clin Chim Acta. 2001;313:107–111. doi: 10.1016/s0009-8981(01)00660-x. [DOI] [PubMed] [Google Scholar]
- 6.Xu X.M., Zhou Y.Q., Luo G.X., Liao C., Zhou M., Chen P.Y., Lu J.P., Jia S.Q., Xiao G.F., Shen X., Li J., Chen H.P., Xia Y.Y., Wen Y.X., Mo Q.H., Li W.D., Li Y.Y., Zhuo L.W., Wang Z.Q., Chen Y.J., Qin C.H., Zhong M. The prevalence and spectrum of alpha and beta thalassaemia in Guangdong Province: implications for the future health burden and population screening. J Clin Pathol. 2004;57:517–522. doi: 10.1136/jcp.2003.014456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan H.F., Long G.F., Li Q., Feng Y.N., Lei Z.Y., Wei H.W., Huang Y.Y., Huang J.H., Lin N., Xu Q.Q., Ling S.Y., Chen X.J., Huang T. Current status of thalassemia in minority populations in Guangxi: China. Clin Genet. 2007;71:419–426. doi: 10.1111/j.1399-0004.2007.00791.x. [DOI] [PubMed] [Google Scholar]
- 8.Xiong F., Sun M., Zhang X., Cai R., Zhou Y., Lou J., Zeng L., Sun Q., Xiao Q., Shang X., Wei X., Zhang T., Chen P., Xu X. Molecular epidemiological survey of haemoglobinopathies in the Guangxi Zhuang autonomous region of southern China. Clin Genet. 2010;78:139–148. doi: 10.1111/j.1399-0004.2010.01430.x. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y.T., Huang S.Z. Disorders of haemoglobin in China. J Med Genet. 1987;24:578–583. doi: 10.1136/jmg.24.10.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee H.H., Chang J.G., Chen R.T., Yang M.L., Choo K.B. Prenatal diagnosis of beta-thalassemic mutations in Chinese by multiple restriction fragment–single strand conformation polymorphism analysis. Proc Natl Sci Counc Repub China B. 1994;18:112–117. [PubMed] [Google Scholar]
- 11.Old J.M. Screening and genetic diagnosis of haemoglobinopathies. Scand J Clin Lab Invest. 2007;67:71–86. doi: 10.1080/00365510601046466. [DOI] [PubMed] [Google Scholar]
- 12.Battistella S., Damin F., Chiari M., Delgrosso K., Surrey S., Fortina P., Ferrari M., Cremonesi L. Genotyping beta-globin gene mutations on copolymer-coated glass slides with the ligation detection reaction. Clin Chem. 2008;54:1657–1663. doi: 10.1373/clinchem.2008.107870. [DOI] [PubMed] [Google Scholar]
- 13.Cousens N.E., Gaff C.L., Metcalfe S.A., Delatycki M.B. Carrier screening for beta-thalassaemia: a review of international practice. Eur J Hum Genet. 2010;18:1077–1083. doi: 10.1038/ejhg.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai S.P., Wall J., Kan Y.W., Chehab F.F. Reverse dot blot probes for the screening of β-thalassemia mutations in Asians and American blacks. Hum Mutat. 1994;3:59–63. doi: 10.1002/humu.1380030110. [DOI] [PubMed] [Google Scholar]
- 15.Wu G., Hua L., Zhu J., Mo Q.H., Xu X.M. Rapid, accurate genotyping of beta-thalassaemia mutations using a novel multiplex primer extension/denaturing high-performance liquid chromatography assay. Br J Haematol. 2003;122:311–316. doi: 10.1046/j.1365-2141.2003.04431.x. [DOI] [PubMed] [Google Scholar]
- 16.Lyon E., Wittwer C.T. LightCycler technology in molecular diagnostics. J Mol Diagn. 2009;11:93–101. doi: 10.2353/jmoldx.2009.080094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrmann M.G., Dobrowolski S.F., Wittwer C.T. Rapid β-globin genotyping by multiplexing probe melting temperature and color. Clin Chem. 2000;46:425–428. [PubMed] [Google Scholar]
- 18.Vrettou C., Traeger-Synodinos J., Tzetis M., Malamis G., Kanavakis E. Rapid screening of multiple beta-globin gene mutations by real-time PCR on the LightCycler: application to carrier screening and prenatal diagnosis of thalassemia syndromes. Clin Chem. 2003;49:769–776. doi: 10.1373/49.5.769. [DOI] [PubMed] [Google Scholar]
- 19.Old J.M., Petrou M., Modell B., Weatherall D.J. Feasibility of antenatal diagnosis of beta thalassaemia by DNA polymorphisms in Asian Indian and Cypriot populations. Br J Haematol. 1984;57:255–263. [PubMed] [Google Scholar]
- 20.Fortina P., Dotti G., Conant R., Monokian G., Parrella T., Hitchcock W., Rappaport E., Schwartz E., Surrey S. Detection of the most common mutations causing beta-thalassemia in Mediterraneans using a multiplex amplification refractory mutation system (MARMS) PCR Methods Appl. 1992;2:163–166. doi: 10.1101/gr.2.2.163. [DOI] [PubMed] [Google Scholar]
- 21.Chinchang W., Viprakasit V., Pung-Amritt P., Tanphaichitr V.S., Yenchitsomanus P.T. Molecular analysis of unknown beta-globin gene mutations using polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) technique and its application in Thai families with beta-thalassemias and beta-globin variants. Clin Biochem. 2005;38:987–996. doi: 10.1016/j.clinbiochem.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Ghanem N., Girodon E., Vidaud M., Martin J., Fanen P., Plassa F., Goossens M. A comprehensive scanning method for rapid detection of beta-globin gene mutations and polymorphisms. Hum Mutat. 1992;1:229–239. doi: 10.1002/humu.1380010310. [DOI] [PubMed] [Google Scholar]
- 23.Losekoot M., Fodde R., Harteveld C.L., van Heeren H., Giordano P.C., Bernini L.F. Denaturing gradient gel electrophoresis and direct sequencing of PCR amplified genomic DNA: a rapid and reliable diagnostic approach to beta thalassaemia. Br J Haematol. 1990;76:269–274. doi: 10.1111/j.1365-2141.1990.tb07883.x. [DOI] [PubMed] [Google Scholar]
- 24.Wong C., Dowling C.E., Saiki R.K., Higuchi R.G., Erlich H.A., Kazazian H.H., Jr Characterization of beta-thalassaemia mutations using direct genomic sequencing of amplified single copy DNA. Nature. 1987;330:384–386. doi: 10.1038/330384a0. [DOI] [PubMed] [Google Scholar]
- 25.Chan K., Wong M.S., Chan T.K., Chan V. A thalassaemia array for Southeast Asia. Br J Haematol. 2004;124:232–239. doi: 10.1046/j.1365-2141.2003.04758.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang W., Kham S.K., Yeo G.H., Quah T.C., Chong S.S. Multiplex minisequencing screen for common Southeast Asian and Indian beta-thalassemia mutations. Clin Chem. 2003;49:209–218. doi: 10.1373/49.2.209. [DOI] [PubMed] [Google Scholar]
- 27.Su Y.N., Lee C.N., Hung C.C., Chen C.A., Cheng W.F., Tsao P.N., Yu C.L., Hsieh F.J. Rapid detection of beta-globin gene (HBB) mutations coupling heteroduplex and primer-extension analysis by DHPLC. Hum Mutat. 2003;22:326–336. doi: 10.1002/humu.10265. [DOI] [PubMed] [Google Scholar]
- 28.Yip S.P., Pun S.F., Leung K.H., Lee S.Y. Rapid, simultaneous genotyping of five common Southeast Asian beta-thalassemia mutations by multiplex minisequencing and denaturing HPLC. Clin Chem. 2003;49:1656–1659. doi: 10.1373/49.10.1656. [DOI] [PubMed] [Google Scholar]
- 29.Liao H.K., Su Y.N., Kao H.Y., Hung C.C., Wang H.T., Chen Y.J. Parallel minisequencing followed by multiplex matrix-assisted laser desorption/ionization mass spectrometry assay for beta-thalassemia mutations. J Hum Genet. 2005;50:139–150. doi: 10.1007/s10038-005-0234-z. [DOI] [PubMed] [Google Scholar]
- 30.Thongnoppakhun W., Jiemsup S., Yongkiettrakul S., Kanjanakorn C., Limwongse C., Wilairat P., Vanasant A., Rungroj N., Yenchitsomanus P.T. Simple, efficient, and cost-effective multiplex genotyping with matrix assisted laser desorption/ionization time-of-flight mass spectrometry of hemoglobin beta gene mutations. J Mol Diagn. 2009;11:334–346. doi: 10.2353/jmoldx.2009.080151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


