Abstract
RREB1 is an alternatively spliced transcription factor implicated in Ras signaling and cancer. Little is known about the expression of RREB1 isoforms in cell lines or human tumors, or about the clinical relevance of the latter. We have developed tools for IHC of RREB1 protein isoform-specific amplification of RREB1 mRNA and selective knockdown of RREB1 isoforms and use these to provide new information by characterizing RREB1 expression in bladder and prostate cancer cell lines and human tissue samples. Previously described splice variants RREB1α, RREB1β, RREB1γ, and RREB1δ were identified, as well as the novel variant RREB1ε. Total and isoform-specific mRNA expression was lower in most but not all tumors, compared with normal tissues. RREB1 IHC performed on a bladder cancer TMA did not indicate a relationship between total RREB1 expression and overall survival after radical cystectomy for invasive bladder cancer. In contrast, in vitro proliferation studies using the UMUC-3 bladder cancer cell line after selective isoform-specific knockdown of expression indicate that RREB1α is not necessary for proliferation, but that RREB1β may be required. These contributions should accelerate progress in the nascent RREB1 field by providing new reagents while also providing clues to the role of RREB1 isoforms in human cancer and raising the possibility of isoform-specific roles in human carcinogenesis and progression.
The Ras family of GTPases, with their regulators and effectors, have been implicated in tumor progression.1–7 We recently found that the Ral (Ras-like) GTPase pathway downstream of Ras plays an important role in bladder cancer cell migration.8,9 Notably, the RREB1 (Ras-responsive element binding protein 1) transcription factor was identified as a putative Ral-regulated gene through batch analysis of promoter sequences in Ral target genes.10 Experiments confirmed that Ral manipulation affects RREB1 reporter activity in bladder cancer cells.10
The significance of RREB1 continues to be elucidated as studies have found it to function in either the induction or repression of gene expression. Genes induced by RREB1 include those encoding calcitonin,11 FSH,12 MT-IIA,13 p53,14 and secretin15; genes repressed by RREB1 include those encoding angiotensinogen,16 HLA-G,17 hZIP1,18 p16,19 and PSA.20 RREB1 has also been found to bind nuclear proteins, such as CtBP,21 NeuroD,15 and androgen receptor (AR).20
The initial study of RREB1 found that the gene product bound the calcitonin promoter in medullary thyroid carcinomas in response to Ras.11 In bladder cancer, the tumor suppressor p16 is commonly lost as an early event in tumorigenesis, and RREB1 was found to bind and repress transcription of the Cdkn2a locus.19 Depletion of RREB1 by siRNA slows cell migration and cell spreading in breast cell lines,22 whereas in breast cancer and osteosarcoma cells RREB1 binds the p53 promoter and transactivates p53 expression on DNA damage.14 The RREB1 gene is a locus of viral integration for hepatitis B virus in hepatocellular carcinoma.23 Furthermore, RREB1 was identified as a potential oncogene in Moloney murine leukemia virus (MuLV) infected p19ARF and p53 knockout mice.24 Finally, the human RREB1 locus has been found to be amplified in melanoma and is currently an area of intense investigation for its potential in molecular diagnostic testing.25–30 In prostate cancer, RREB1 binds the PSA promoter only in association with AR to repress transcription.20 In summary, the current literature on RREB1 suggests context-dependent phenotypes that may suppress or promote carcinogenesis and tumor progression.
RREB1 is a transcription factor containing between 13 and 15 zinc finger domains, depending on alternative splicing.16 It was initially described as a 755-amino-acid C2H2 zinc finger protein (RREB-1),11 although subsequent analyses in chicken and human cells indicated that RREB1 encodes a longer protein of 1656 (Finb) amino acids (AA) in humans.13,31 A second variant encoding 1397 amino acids [Finb (cl-32)] with a unique C-terminus was also identified.13 Two additional RREB1 C-terminal isoforms exhibiting addition or removal of cassette exons were isolated and designated Finb188 (1742AA) and Finb159 (1476AA). These isoforms exhibit a translation start site 57 bp upstream of the earlier described Finb and Finb (cl-32) isoforms (Figure 1A).16 The authors also discovered that Finb contains regions without homology to consensus protein sequence of Finb188. cDNA sequence alignment of Finb and Finb188 reveals that the former does not conform to sequences for the human genome, potentially because of cloning artifacts.16 Thus, there is a critical need for consensus on the RREB1 proteins.
Figure 1.
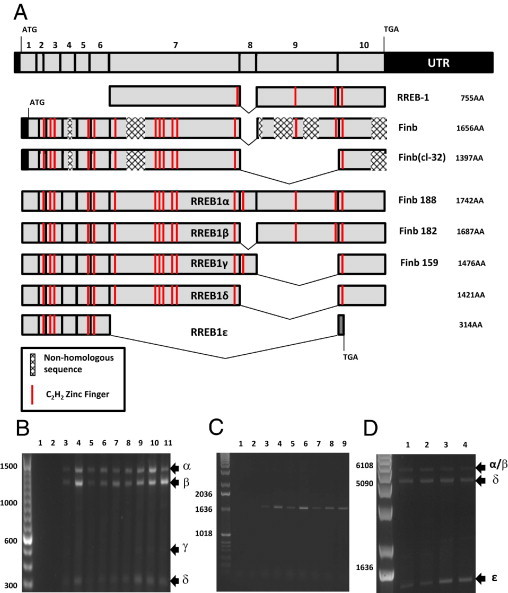
RREB1 alternative splicing. A: 10 coding exons of RREB and previously described RREB1 sequences were aligned. Finb and Finb (cl-32) contained several regions of frame shifts in the cDNA that resulted in nonhomologous protein sequence. These sequences also described an alternative translation start site that results in a protein without the N-terminal 19 amino acids. We propose using Greek lettering to describe RREB1 alternative splicing: RREB1α, RREB1β, RREB1γ, RREB1δ, and RREB1ε. B: Primers designed to span the region of known RREB1 alternative splicing (exon 7 to exon 10) were interrogated on the following bladder and prostate cancer cell lines: 1, negative PCR control; 2, negative RT control; 3, TERT; 4, 293T; 5, UMUC-3; 6, LUL2; 7, UMUC13D; 8, J82; 9, 1A6; 10, PC3; and 11, LNCAP. C: Primers designed to span from exon 1 to exon 7 of RREB1 were used to amplify cDNA from the following samples: 1, negative PCR control; 2, negative RT control; 3, TERT; 4, 293T; 5, UMUC-3; 6, LUL2; 7, UMUC13D; 8, J82; and 9, 1A6. D: Primers designed to span exon 1 to exon 10 of RREB1 were used to amplify cDNA from the following samples: 1, UMUC-3; 2, LUL2; 3, LNCaP; and 4, J82. Arrows and Greek letters indicate the RREB1 splice variants.
Given the number and diversity of known targets of RREB1 signaling, and that splice variants encoding four protein isoforms have already been described, it is likely that RREB1 signaling is mechanistically and phenotypically complex and that expression of isoforms in various proportions could contribute to this complexity. Surprisingly, there has been no comprehensive evaluation of RREB1 isoforms in human cancer cell lines and normal or tumor tissues to date. Furthermore, no study has shown evidence suggestive of isoform-specific phenotypes.
With the present study, we developed tools to evaluate RREB1 isoforms in human bladder and prostate cell lines and human tumors at the RNA and immunohistochemical levels. We then used these tools to evaluate the expression patterns and their clinical relevance in human samples. Here, we present evidence that isoforms have different functional effects on in vitro bladder cancer cell proliferation.
Materials and Methods
Cell Lines and Human Tissues
Human cancer cell lines were grown as described previously.32 TERT cells33 were a gift from Margaret A. Knowles. LUL2 cells were isolated from lung tumors by successive passages of the UMUC-3 cell line by tail vein injection into nude mice (unpublished data). With strict observance of NIH and University of Virginia guidelines and with approval of the Institutional Review Board, deidentified flash-frozen and archival tissues of human bladder and prostate cancer and adjacent non-neoplastic epithelia, procured by the University of Virginia Biorepository and Tissue Research Facility, were obtained for analysis of RREB1 expression. The tumor tissue was macrodissected to approximately 80% purity, as previously described.34 The bladder TMA described here has been reported previously.35
RNA Isolation, Reverse Transcription, and PCR
RNA isolation was performed using an RNeasy mini kit (Qiagen, Valencia, CA) and cDNA synthesis using iScript (Bio-Rad, Hercules, CA). Quantitative RT-PCR (qRT-PCR) was performed on cDNA using iQ SYBR Green Supermix (Bio-Rad). RNA quantification was performed as previously described.36 The following primers were used with annealing temperatures: RREB1 (total) forward 5′-CTTCCTATAACTGCCCCC-3′, reverse 5′-ATGAGTGGTCGGCTCCTCC-3′; RREB1α forward 5′-TGGATCCCATGATAGCACAGAC-3′, reverse 5′-TGCTCTCTGTCCCGTGAGG-3′; RREB1β forward 5′-CACATGCTCACACACACTGACA-3′, reverse 5′-CCGACGGCTGCTCTCTGT-3′; RREB1δ forward 5′-ACCAACTGCCTGCAGAAGATCA-3′, reverse 5′-GTATGGCCTTTCCCCAGTGTGT-3′; RREB1ε forward 5′-TACAGAACAACCCTTCAATTCCT-3′, reverse 5′-TATGGCCTTTCCCCTGAG-3′. The PCR for the 5′ and 3′ ends of RREB1 was performed using AccuPrime SuperMix II (Invitrogen, Carlsbad, CA) for 35 cycles with the following primers: RREB1-5′ forward 5′-TCGGATTGGCAGAAGGAA-3′, reverse 5′-CAGGCTCAGCAGGTTGGT-3′; RREB1 3′ forward 5′-CGGAACTCGTACACCAACTG-3′, reverse 5′-CGCTGTGGGTGGACTCATTC-3′; RREB1 full length forward 5′-GATCAAGCTTACGTCAAGTTCGCCCGCT-3′, reverse 5′-GATCCTCGAGTCACTCCATCCCCACGAG-3′.
Cloning of RREB1
RREB1δ and RREB1ε were cloned out of UMUC-3 cells. Sequences were submitted to GenBank: RREB1δ (HM369361) and RREB1ε (HM369360). Total RNA was isolated as described above and cDNA synthesis using SuperScript III first-strand synthesis supermix (Invitrogen) with 50 μmol/L random hexamers and 1 μg total RNA. The PCR reaction also included 10× PfuUltra HF reaction buffer (Agilent Technologies, Foster City, CA), 100 mmol/L dNTP mix (Agilent), 5% dimethyl sulfoxide, 1 mol/L betaine solution (Sigma-Aldrich, St. Louis, MO). The primers used were as follows: RREB1 cloning forward 5′-GATCATCGATATGACGTCAAGTTCGCCC-3′, reverse 5′-GATCTCTAGACTCCATCCCCACGAGCTG-3′. PCR products were isolated in a 1% agarose gel and purified using a QIAquick gel extraction kit (Qiagen). Isolated products were digested at 37°C by the enzymes ClaI and XbaI (New England Biolabs, Ipswich, MA) and were ligated into the p3XFLAG-CMV-14 expression vector (Sigma-Aldrich). RREB1α and RREB1β,16 kindly provided by Dr. Akiyoshi Fukamizu (University of Tsukuba, Tsukuba, Japan), were subcloned into the p3XFLAG-CMV-14.
Antibodies, Immunoblotting, and IHC
The following antibodies were used in detection of RREB1: anti-RREB1 from GenWay Biotech (cat. no. 18-732-2922332; Biotech, San Diego, CA), from Cosmo Bio (cat. no: CBX-CBX00717; Carlsbad, CA; Tokyo, Japan), and from Sigma-Aldrich (cat. no. HPA001756), as well as anti-FLAG (cat. no: F1804; Sigma-Aldrich), anti-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-TBP (TFIID) (Santa Cruz Biotechnology). Nuclear and cytoplasmic isolates were made with an NE-PER extraction kit (Thermo Scientific, Rockford, IL). Immunoblotting and detection were performed as described previously.8 Immunohistochemistry (IHC) was performed on a Dako Autostainer instrument (Dako, Glostrup, Denmark) with the following protocol: antigen retrieval (125°C, Dako TRS9 buffer, 30 seconds), Dako dual endogenous enzyme block (10 minutes), RREB1 antibody (Sigma-Aldrich; 1:100 in Dako antibody diluent, 30 minutes), detection (Dako Envision dual link anti-rabbit, 30 minutes), chromogen (Dako diaminobenzidine Dab+ substrate, 10 minutes), and counterstain (hematoxylin, 5 minutes). Staining of RREB1 was scored semiquantitatively as negative (absence of staining), low/focal (a blush or positivity of cells <10%), moderate (clearly detectable nuclear staining pattern in up to 50% of cells), or high (a strong nuclear and/or cytoplasmic positivity in >50% of cells).
Transfection and siRNA
Transient vector transfection was performed using FuGENE 6 transfection reagent (Roche, Basel, Switzerland) according to the manufacturer's instructions. Stable expression was achieved by cutting RREB1-expressing p3XFLAG-CMV-14 vector with the ScaI restriction enzyme (New England Biolabs) and transfected with FuGENE 6. Selection was performed for 14 days in 800 μg/mL of G418 (Geneticin, Invitrogen). Oligofectamine (Invitrogen) was used to transiently transfect siRNA according to the manufacturer's instructions. All siRNA was transfected at a final concentration of 25 nmol/L: RREB1 total 5′-GGAGUUUGUUUGCAAGUAU-3′ and 5′-GUUCAGACCUAUCUUCCAU-3′ (used in combination at 12.5 nmol/L), GL2 5′-CGUACGCGGAAUACUUCGAdTdT-3′, RREB1 exon 8-1 5′-CCUGAGAAGAAACGGGCUUUU-3′, RREB1 exon 8-2 5′-CGCAAACACGGAGUUACCACCUGUU-3′, RREB1 exon 8-3 5′-GAUGUUGGAUCCCAUGAUAUU-3′, RREB1 exon 9-1 5′-CAGAGAAGAGCGACGAUGAdTdT-3′, RREB1 9-2 5′-CCACCAAGCUCAUGGACUUUU-3′, and RREB1 exon 9-3 5′-GGAAGAAGGUCUGCAGCGUdTdT-3′.
In Vitro Cell Growth Assays
In vitro cell growth assays were performed by using Alamar Blue (Invitrogen) fluorescence emission as described previously.37 Briefly, Alamar Blue fluorescence was measured at 96 hours after siRNA depletion in UMUC-3 cells.
Results
Alternative Splicing Creates a Unique RREB1 Splice Variant
Using bladder and prostate cancer cell lines as models, primers were designed to interrogate expression of variants in the last four coding exons of RREB1, a region in alternative splicing has been described (Figure 1A).13,16 Predicted PCR product sizes for the four RREB1 isoforms characterized thus far were 1237 bp for Finb188, 1072 bp for Finb182 (RREB-1, Finb), 439 bp for Finb159, and 348 bp for Finb (cl-32). In nine cell lines examined, bands corresponding to all four splice variants were identified (Figure 1B). Each band was sequenced to confirm the PCR products corresponded to the predicted variant. To examine whether the first six identified coding exons of RREB1 transcripts underwent alternative splicing, primers spanning from the first to the seventh coding exon were tested on seven cell lines. Each cell line showed a single band, leading us to conclude that alternative splicing of RREB1 occurs predominantly in the exon 6 to exon 10 region of the RNA (Figure 1C).
To search for novel splice variants, primers were designed to interrogate all 10 coding exons (Figure 1B). PCR products for Finb188/182 and Finb (cl-32) were identified, as well as a novel variant migrating at a much smaller size (Figure 1D). Sequencing of this band revealed it to be a novel isoform of RREB1, lacking coding exons 7, 8, and 9 and causing a frame shift in exon 10, resulting in a loss of a C2H2 zinc finger and a new translation stop site. Given these findings, we propose the following nomenclature for the RREB1 splice variants: RREB1α, RREB1β, RREB1γ, RREB1δ, and RREB1ε (Figure 1A).
RREB1 mRNA Expression in Bladder and Prostate Cell Lines
To evaluate RREB1 isoform expression, we developed primers for qRT-PCR that allow specific amplification of each isoform. These primers were used in conjunction with primers spanning the second and third exons, the area we found not to be involved in differential splicing, to detect total RREB1 mRNA expression. We examined one telomerase (TERT) immortalized urothelial cell line,33 five urothelial bladder cancer cell lines, and two prostate cancer cell lines. The data presented in Figure 1B, as well as the use of isoform-specific primers to estimate relative abundance of isoforms, demonstrated that RREB1α and RREB1β comprised the vast majority of total RREB1 mRNA (see Supplemental Figure S1 at http://ajp.amjpathol.org). Expression of RREB1γ was so low as to not be reliably quantifiable. Expression of RREB1δ typically was only 1% to 4%, and RREB1ε expression was <1% of total RREB1. For further studies, therefore, we focused on total RREB1, RREB1α, and RREB1β. The expression levels for total RREB1, RREB1α, and RREB1β in cell lines were variable (Figure 2A). Notably, the total RREB1 expression is higher in the nontransformed TERT urothelial cell line than in all of the urothelial (bladder) cancer cell lines.
Figure 2.
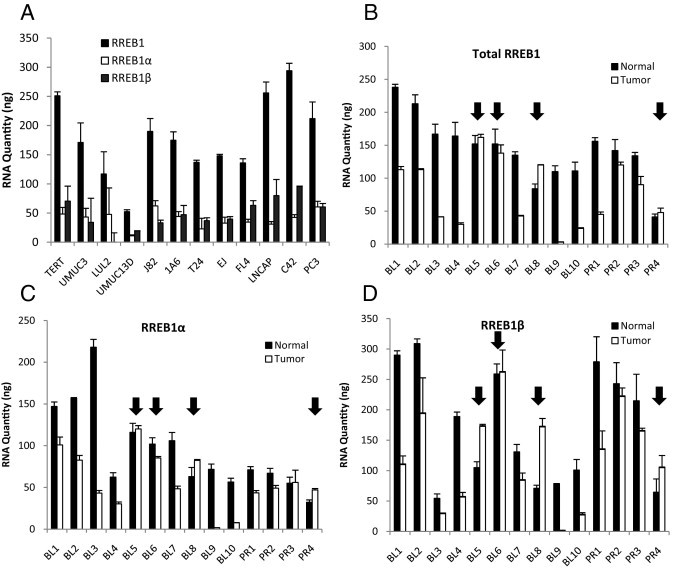
RREB1 transcripts in bladder tumors and cancer cell lines. A: mRNA expression of total RREB1, RREB1α, and RREB1β was measured in bladder and prostate cancer cell lines. RNA quantities cannot be compared among total RREB1, RREB1α, and RREB1β, because the levels were calculated on unique standard curves for each primer pair. B: Total RREB1 expression was measured in paired normal and cancer (tumor) urothelial bladder (BL) and prostate (PR) tissues, using primers that span exons 2 and 3 of RREB1, an area lacking alternative splicing. Arrows indicate samples in which RREB1 expression is greater in the tumor than in the paired normal tissue. C and D: RNA expression of the RREB1α and RREB1β splice variants was measured using primers targeting base pairs at unique splice junctions.
Next we examined paired human normal and cancerous bladder (n = 10) and prostate (n = 4) tissues for total RREB1 (Figure 2B), RREB1α (Figure 2C), and RREB1β (Figure 2D) mRNA expression. Of 10 bladder cancers, 7 samples had lower total RREB1 expression, as well as lower expression of the specific α and β isoforms, than their normal counterparts (P = 0.005), whereas 3 samples (indicated by arrows in Figure 2) demonstrated similar or higher total RREB1, RREB1α, and RREB1β isoform expression. Of the four prostate cancer samples, three demonstrated a decrease in total RREB1 expression and one had an increase in total RREB1 expression, compared with its normal counterpart (P = 0.22).
Detection and Cellular Localization of RREB1 Protein
The expression of RREB1 splice variants at the protein level has not been examined previously. A major obstacle appears to be the lack of a well-characterized antibody. To define the sensitivity and specificity of three commercial RREB1 antibodies, we cloned RREB1α, RREB1β, RREB1δ, and RREB1ε into a C-terminal 3X-FLAG tagged vector. (RREB1γ was excluded because it was undetectable in the cell lines and tissues we examined.) These constructs were transfected into 293T cells and their expression was confirmed by anti-FLAG antibody (Figure 3A). The three RREB1 antibodies were evaluated on these lysates. Two antibodies (from Cosmo Bio and GenWay) detected RREB1α and RREB1β, the GenWay antibody also detecting RREB1δ. The Cosmo Bio antibody suffered from low sensitivity in detecting endogenous RREB1, and the GenWay antibody had higher nonspecific binding (see Supplemental Figure S2 at http://ajp.amjpathol.org). The Sigma-Aldrich antibody had robust detection of RREB1α, RREB1β, and RREB1δ tagged transgenes (Figure 3A). Support for the specificity of the antibody was provided by observing decreases in endogenous bands of appropriate sizes to the splice variants RREB1α, RREB1β, and RREB1δ on treatment with siRNA to total RREB1 in UMUC-3 cells (Figure 3B).
Figure 3.
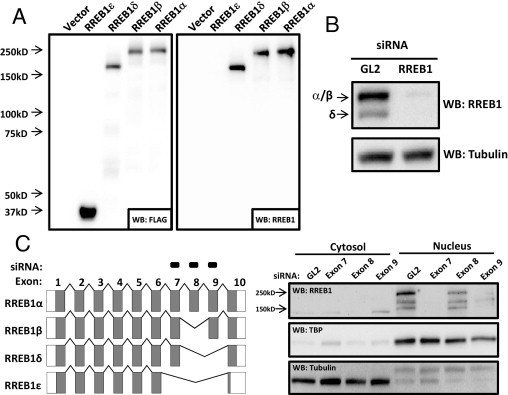
Detection of RREB1 protein isoforms in human bladder cancer cells. A: RREB1 isoforms were either isolated from UMUC-3 cells (Isoforms δ and ε) or received as a gift from Dr. Akiyoshi Fukamizu (University of Tsukuba, Tsukuba, Japan) (isoforms α and β). The splice variants were cloned into a C-terminal 3XFLAG-tagged expression construct and expressed in 293T cells. Detection with anti-FLAG (1:1000) or anti-RREB1 (1:1000) (Sigma-Aldrich) antibody is shown. B: RREB1 (total) siRNA, designed to knock down all splice variants, was transfected into UMUC-3 cells. RREB1 antibody (Sigma-Aldrich) detection (1:1000) on Western blot is shown. C: siRNAs targeting exons 7, 8, or 9 were transfected into UMUC-3 cells. Cytoplasmic and nuclear fractions were isolated after 96 hours and RREB1 protein splice variants were detected on Western blot (1:1000) (Sigma-Aldrich).
To determine which protein species detected by the antibody corresponded to each RREB1 splice variant, we designed siRNAs targeting specific RREB1 exons. In parallel, the nuclei and cytosol were fractionated to examine the subcellular localization of specific variants (because this had not been shown previously).13 RREB1 seems to localize primarily in the nucleus (Figure 3C). siRNA to exon 8 knocks down the RREB1α and RREB1γ splice variants. Because RREB1γ mRNA is undetectable, all measurable protein depleted by siRNA to exon 8 represents RREB1α. Exon 8 siRNA decreased the intensity of the largest band (Figure 3C), which, based on size (∼250 kDa), is likely RREB1α. siRNA to exon 9, which depletes RREB1β in addition to RREB1α, resulted in an elimination of the largest RREB1 band, which (given only a 3% difference in amino acid content between the splice variants) likely represents both RREB1α and RREB1β. Surprisingly, a second band at ∼150 kDa was also knocked down with exon 9 siRNA. This band could represent a unique RREB1 isoform, which is either generated by an unknown pre-mRNA splicing event that has yet to be identified or is an alternative translation product from the RREB1β isoform. Finally, siRNA to exon 7, which knocks down RREB1α, RREB1β, and RREB1δ, eliminated the upper band of RREB1α and RREB1β and the second largest band (∼200 kDa), which migrates at the same size as the cloned RREB1δ mRNA. The presence of the cytosolic band at ∼140 kDa that appears to disappear with siRNA to exon 8 may also represent another unique RREB1 isoform, one that has yet to be identified. The Sigma-Aldrich RREB1 antibody showed the greatest sensitivity and specificity in detecting the exogenously and endogenous isoforms, and therefore it was used in all subsequent experiments.
Expression RREB1 Protein in Normal and Cancerous Bladder and Prostate Tissues
Next we optimized the Sigma-Aldrich RREB1 antibody for immunohistochemical staining to determine whether the expression patterns and subcellular localization observed in cells reflected those found in human tissue. After optimization, IHC was performed on two samples of bladder cancer and two samples of prostate cancer and adjacent normal tissue. We found a general decrease in RREB1 staining in the malignant tissue, compared with adjacent benign tissue (Figure 4A), a finding consistent with qRT-PCR analysis (Figure 4B). In general, RREB1 staining was strongest in the nucleus (although, as we observed in cell line fractionation experiments, some cytoplasmic expression was also detected).
Figure 4.
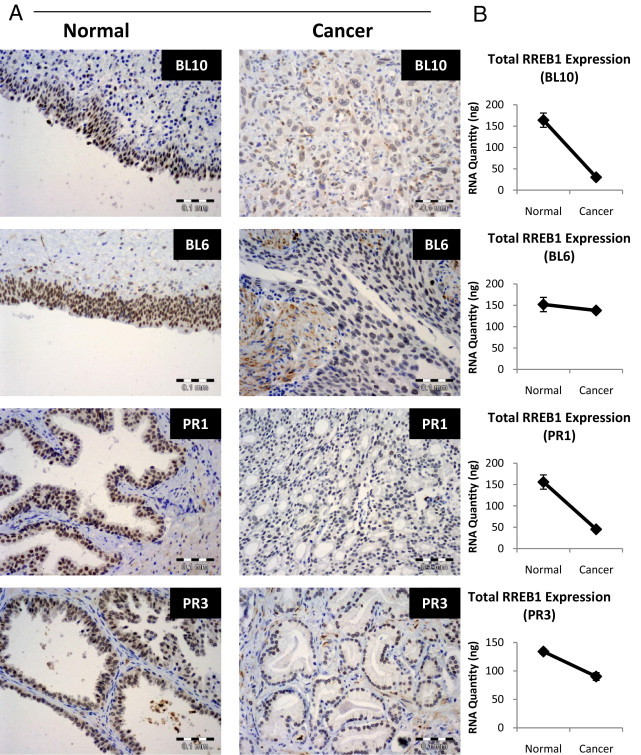
IHC of RREB1 in human carcinoma. A: RREB1 IHC (1:100) (Sigma-Aldrich) of paired normal and cancer tissues for urothelial bladder (BL10 and BL6) and prostate (PR1 and PR3). B: Total RREB1 RNA levels by qRT-PCR from the same tissue as in A. Scale bars: 0.1 mm.
RREB1β Is Necessary for UMUC-3 Proliferation
To study a possible RREB1 isoform-specific phenotype, we investigated the effect of total RREB1 knockdown versus specific knockdown of RREB1α or RREB1α and RREB1β together on proliferation of the UMUC-3 urothelial cancer cell line. It is not possible to knock down RREB1β specifically, because of a lack of a siRNA sequence in the unique splice junction site. Knockdown of RREB1 (total) using a siRNA targeting a region common to all isoforms inhibited proliferation of UMUC-3 cells (Figure 5A). To determine the isoform specificity of the RREB1 growth phenotype, we tested isoform-specific siRNAs in depletion experiments in UMUC-3. siRNA to exon 8 (exon 8-1), which targets RREB1α, showed no decrease in UMUC-3 growth in vitro (Figure 5A). qRT-PCR for RREB1α was performed to judge the extent of knockdown and showed >80% loss of expression (Figure 5B). To determine whether this effect was specific, we designed two additional siRNAs to separate regions within exon 8 (exon 8-2 and exon 8-3). The additional siRNAs had similar effects on UMUC-3 growth (Figure 5A), while maintaining efficient knockdown of RREB1α (Figure 5B). However, siRNA to exon 9 (exon 9-1), which knocks down RREB1α and RREB1β together, decreased growth similar to that of siRNAs targeting all RREB1 splice variants (Figure 5C). Isoform-specific qRT-PCR showed that RREB1α and RREB1β were robustly depleted in these cells (Figure 5D). Two additional siRNAs were designed to unique regions within exon 9 to confirm the specificity of these phenotypic effects. The additional siRNAs targeted to exon 9 (exon 9-2 and 9-3) showed identical results on UMUC-3 growth (Figure 5C).
Figure 5.

Depletion of RREB1β decreases UMUC-3 growth in vitro. A: UMUC-3 cells were treated with 25 nmol/L siRNA of control (GL2), RREB1 (all isoforms), and three unique siRNAs targeting exon 8 (8-1, 8-2, and 8-3). Growth was measured 96 hours after siRNA depletion using Alamar Blue and normalized to the GL2 control. B: Real-time PCR for the RREB1α splice variant was performed for the experiment described in panel A to confirm knockdown of RREB1α. RNA levels were normalized to the GL2 control siRNA. C: UMUC-3 cells were treated with 25 nmol/L siRNA of control (GL2), RREB1 (all isoforms), and three unique siRNAs targeting exon 9 (9-1, 9-2, and 9-3). D: RREB1α and RREB1β-specific real-time PCR was used to confirm RREB1α and RREB1β knockdown.
RREB1 Protein Expression Does Not Predict Overall Survival in Bladder Cancer
With the knowledge that RREB1 is necessary for proliferation of the bladder cancer cell line UMUC-3, we asked whether expression of RREB1 in bladder cancer tissue is predictive of overall survival. We performed IHC on a 142-sample TMA35 from patients with bladder cancer who underwent cystectomy. The intensity of RREB1 staining was scored as negative, low/focal, moderate, and high (Figure 6A). A Kaplan-Meier analysis of overall survival for stratified RREB1 expression groupings was plotted (Figure 6B). None of the four RREB1 staining patterns were predictive of survival (P = 0.86). Furthermore, RREB1 staining did not correlate with pathological tumor grade (P = 0.62), stage (pT) (P = 0.43), or nodal metastasis (pN) (P = 0.69) (see Supplemental Table S1 at http://ajp.amjpathol.org).
Figure 6.
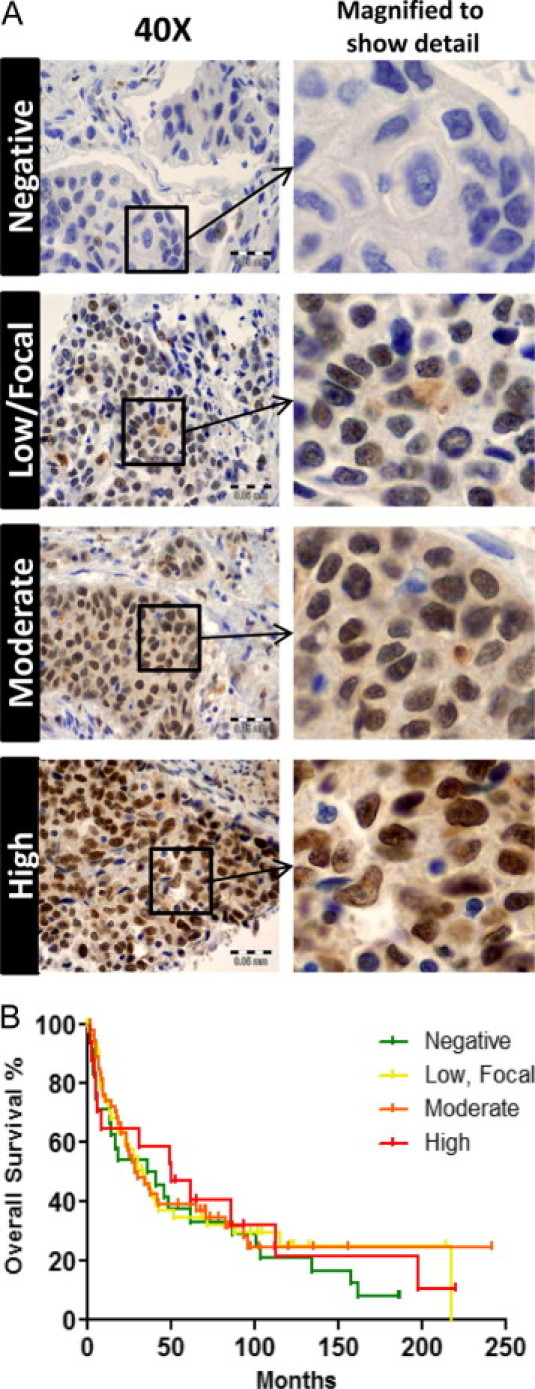
Total RREB1 expression in bladder cancer does not predict overall survival. A: A TMA of archived bladder cancers (n = 142) was used to assay for RREB1 expression. Representative examples of RREB1 expression grading are shown, as described under Materials and Methods (negative, low/focal, moderate, and high). B: Survival data of patients whose tumors comprised the bladder cancer TMA was used to generate a Kaplan-Meier analysis comparing RREB1 staining versus overall survival. RREB1 staining did not significantly correlate with this clinical parameter (P = 0.86, log rank).
Discussion
Despite its implication in Ras, Ral, p16, p53, and androgen receptor10,11,14,19,20 signaling pathways, sparse data exist regarding the nature, expression, or function of RREB1 mRNA and protein in cancer cells. This is the first study to address this gap by describing RREB1 expression in human cancers, as well as the first to examine isoform expression in human tissues and cell lines. We confirmed expression of four previously described mRNA splice variants, clarified prior sequencing artifacts,13,16 and identified a novel RREB1 splice variant. For the sake of clarity within the field, we propose a new nomenclature for RREB1 splice variants, ordered from the largest to the smallest protein coding sequence: RREB1α, RREB1β, RREB1γ, RREB1δ, and RREB1ε.
Characterization of currently available RREB1 antibodies failed to detect the new RREB1ε isoform, or to specifically detect any single RREB1 isoform. However, we could readily detect proteins corresponding to the most abundant RREB1 mRNA species. The comparison of RREB1 expression as measured by IHC and its correlation with clinical outcome did not reveal a relationship to disease-free survival for bladder cancer patients. The antibody used for the TMA cannot differentiate between expression of RREB1α and RREB1β, and indeed no such antibodies exist at this time. Given our results with isoform-specific qRT-PCR in bladder cancers, and the possibility of differential effects of RREB1 in subsets of tumors and isoform-specific effects, it is possible that expression of a specific RREB1 isoform in a particular subset of patients will be predictive of disease survival. These findings indicate the need for further reagent development as hypotheses for RREB1 function become more sophisticated.
Given its associations with cancer signaling, we suspected that RREB1 expression would be altered in tumors. By using qRT-PCR for the most abundant isoforms, namely RREB1α and RREB1β, as well as IHC for RREB1 protein, we found that RREB1 expression was lower in tumors, compared with histologically similar but untransformed tissue from the same patient. This suggests that loss of RREB1 expression is associated with transformation.
Although most of the cancers examined by qRT-PCR showed decreased RREB1 expression, there were exceptions. In tumors in which total RREB1 mRNA expression was seen to increase compared with normal tissue, we observed that the RREB1β mRNA increased more than the RREB1α isoform. Although the number of samples examined is small, these observations at least suggest the possibility that RREB1β is preferentially increased in some tumors, relative to RREB1α. This allowed us to hypothesize that RREB1 may have a growth-promoting effect in some subset of tumors, and that this phenotype may be largely mediated by the β isoform. Our in vitro experiments with selective RREB1 knockdown in UMUC-3 bladder cancer cells support this hypothesis. In UMUC-3 cells, RREB1α was not required for cell proliferation, whereas knockdown of both α and β inhibited proliferation. The most parsimonious explanation of this result is that RREB1β is necessary for proliferation but that RREB1α is dispensable, at least in the context of UMUC-3 cells. This result also implies that UMUC-3 cells are more representative of a subset of tumors in which RREB1 expression could promote tumor growth.
The recently reported interaction of RREB1 with AR in prostate cancer cells may provide a clue as to why decreased RREB1 expression is associated with this particular tumor type. Mukhopadhyay et al20 reported that RREB1 binds AR and inhibits its function as a transcription factor. In addition, RREB1 binding and inhibition of AR activity was relieved by activated Ras signaling. Because increased androgen signaling is thought to play a central role in prostate cancer tumorigenesis, the decreased RREB1 expression we observe in prostate cancer specimens may result in heightened AR activity. Ours is the first report of diminished RREB1 expression in prostate cancer clinical samples, and the findings suggest that the molecular interactions described by Mukhopadhyay et al20 from in vitro studies may be operational in actual human prostate tumors.
Other signaling targets may account for decreased RREB1 expression in tumor types in which AR is not an obvious contributing factor. Liu et al14 reported that transcription of the p53 tumor suppressor gene in response to genotoxic stress is dependent on RREB1 binding to the p53 promoter in a variety of cancer cell lines, including osteosarcoma, breast and colon cancer cells. Apoptosis induced by p53 in response to DNA damage was also RREB1-dependent. It is possible, therefore, to hypothesize that a decrease in RREB1 in tumors would result in resistance to DNA-damaging chemotherapy agents used in treating urothelial tumors. This hypothesis can now be tested in bladder and other cancers using the tools we have developed.
The work presented here provides both new tools for investigating RREB1 expression and insights into its function. Our observations also provide interesting and testable hypotheses regarding the role of RREB1 and AR in prostate cancer progression, as well as that of RREB1 in resistance to chemotherapy. This work will also, we hope, provide an impetus for future studies to differentiate between isoform-specific effects in the nascent field of RREB1 signaling in cancer.
Acknowledgments
We thank Dr. Christopher Moskaluk and the University of Virginia Biorepository and Tissue Research Facility (BTRF) for their assistance in obtaining deidentified human clinical specimens for evaluation.
Footnotes
Supported by NIH grant CA143971 (D.T.).
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.03.038.
Current address of S.C.S., University of Michigan, Ann Arbor, Michigan.
Supplementary data
RREB1α and RREB1β account for the majority of RREB1 expression in bladder and prostate cancer cell lines. RNA expression of RREB1 splice variants were measured in the indicated cell lines using primers designed to the unique splice junctions. RREB1γ was not included, because of sub quantifiable expression. Isoform expression levels were calculated using the Pfaffl method (Pfaffl MW: A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res 2001, 29:e45).
RREB1 isoform detection by commercial RREB1 antibodies. RREB1 isoforms were isolated from UMUC-3 cells (isoforms δ and ε) or a gift from Dr. Akiyoshi Fukimizu (isoforms α and β). The splice variants were cloned into a C-terminal 3XFLAG tagged expression construct and expressed in 293T cells. Detection with anti-FLAG (1:1000), anti-RREB1 (1:500) (Cosmo Bio, Carlsbad, CA; Tokyo, Japan), or anti-RREB1 (1:1000) (GenWay Biotech, San Diego, CA) is shown.
References
- 1.Traynor P., McGlynn L.M., Mukhergee R., Grimsley S.J., Bartlett J.M., Edwards J. An increase in N-Ras expression is associated with development of hormone refractory prostate cancer in a subset of patients. Dis Markers. 2008;24:157–165. doi: 10.1155/2008/918087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erlich S., Tal-Or P., Liebling R., Blum R., Karunagaran D., Kloog Y., Pinkas-Kramarski R. Ras inhibition results in growth arrest and death of androgen-dependent and androgen-independent prostate cancer cells. Biochem Pharmacol. 2006;72:427–436. doi: 10.1016/j.bcp.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Weber M.J., Gioeli D. Ras signaling in prostate cancer progression. J Cell Biochem. 2004;91:13–25. doi: 10.1002/jcb.10683. [DOI] [PubMed] [Google Scholar]
- 4.Min J., Zaslavsky A., Fedele G., McLaughlin S.K., Reczek E.E., DE Raedt T., Guney I., Strochlic D.E., Macconaill L.E., Beroukhim R., Bronson R.T., Ryeom S., Hahn W.C., Loda M., Cichowski K. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oxford G., Theodorescu D. The role of Ras superfamily proteins in bladder cancer progression. J Urol. 2003;170:1987–1993. doi: 10.1097/01.ju.0000088670.02905.78. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen P.L., Swanson P.E., Jaszcz W., Aeppli D.M., Zhang G., Singleton T.P., Ward S., Dykoski D., Harvey J., Niehans G.A. Expression of epidermal growth factor receptor in invasive transitional cell carcinoma of the urinary bladder: A multivariate survival analysis. Am J Clin Pathol. 1994;101:166–176. doi: 10.1093/ajcp/101.2.166. [DOI] [PubMed] [Google Scholar]
- 7.Theodorescu D., Cornil I., Fernandez B.J., Kerbel R.S. Overexpression of normal and mutated forms of HRAS induces orthotopic bladder invasion in a human transitional cell carcinoma. Proc Natl Acad Sci USA. 1990;87:9047–9051. doi: 10.1073/pnas.87.22.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oxford G., Owens C.R., Titus B.J., Foreman T.L., Herlevsen M.C., Smith S.C., Theodorescu D. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- 9.Gildea J.J., Harding M.A., Seraj M.J., Gulding K.M., Theodorescu D. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002;62:982–985. [PubMed] [Google Scholar]
- 10.Oxford G., Smith S.C., Hampton G., Theodorescu D. Expression profiling of Ral-depleted bladder cancer cells identifies RREB-1 as a novel transcriptional Ral effector. Oncogene. 2007;26:7143–7152. doi: 10.1038/sj.onc.1210521. [DOI] [PubMed] [Google Scholar]
- 11.Thiagalingam A., De Bustros A., Borges M., Jasti R., Compton D., Diamond L., Mabry M., Ball D.W., Baylin S.B., Nelkin B.D. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol Cell Biol. 1996;16:5335–5345. doi: 10.1128/mcb.16.10.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xing W., Sairam M.R. Cross talk of two Krupple transcription factors regulates expression of the ovine FSH receptor gene. Biochem Biophys Res Commun. 2002;295:1096–1101. doi: 10.1016/s0006-291x(02)00812-4. [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto-Nishiyama A., Ishii S., Matsuda S., Inoue J., Yamamoto T. A novel zinc finger protein, Finb, is a transcriptional activator and localized in nuclear bodies. Gene. 1997;195:267–275. doi: 10.1016/s0378-1119(97)00172-8. [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Hew H.C., Lu Z.G., Yamaguchi T., Miki Y., Yoshida K. DNA damage signalling recruits RREB-1 to the p53 tumour suppressor promoter. Biochem J. 2009;422:543–551. doi: 10.1042/BJ20090342. [DOI] [PubMed] [Google Scholar]
- 15.Ray S.K., Nishitani J., Petry M.W., Fessing M.Y., Leiter A.B. Novel transcriptional potentiation of BETA2/NeuroD on the secretin gene promoter by the DNA-binding protein Finb/RREB-1. Mol Cell Biol. 2003;23:259–271. doi: 10.1128/MCB.23.1.259-271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Date S., Nibu Y., Yanai K., Hirata J., Yagami K., Fukamizu A. Finb, a multiple zinc finger protein, represses transcription of the human angiotensinogen gene. Int J Mol Med. 2004;13:637–642. [PubMed] [Google Scholar]
- 17.Flajollet S., Poras I., Carosella E.D., Moreau P. RREB-1 is a transcriptional repressor of HLA-G. J Immunol. 2009;183:6948–6959. doi: 10.4049/jimmunol.0902053. [DOI] [PubMed] [Google Scholar]
- 18.Milon B.C., Agyapong A., Bautista R., Costello L.C., Franklin R.B. Ras responsive element binding protein-1 (RREB-1) down-regulates hZIP1 expression in prostate cancer cells. Prostate. 2010;70:288–296. doi: 10.1002/pros.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S., Qian X., Redman C., Bliskovski V., Ramsay E.S., Lowy D.R., Mock B.A. p16 INK4a gene promoter variation and differential binding of a repressor, the ras-responsive zinc-finger transcription factor, RREB. Oncogene. 2003;22:2285–2295. doi: 10.1038/sj.onc.1206257. [DOI] [PubMed] [Google Scholar]
- 20.Mukhopadhyay N.K., Cinar B., Mukhopadhyay L., Lutchman M., Ferdinand A.S., Kim J., Chung L.W., Adam R.M., Ray S.K., Leiter A.B., Richie J.P., Liu B.C., Freeman M.R. The zinc finger protein ras-responsive element binding protein-1 is a coregulator of the androgen receptor: implications for the role of the Ras pathway in enhancing androgenic signaling in prostate cancer. Mol Endocrinol. 2007;21:2056–2070. doi: 10.1210/me.2006-0503. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y., Sawada J., Sui G., Affar el B., Whetstine J.R., Lan F., Ogawa H., Luke M.P., Nakatani Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 22.Melani M., Simpson K.J., Brugge J.S., Montell D. Regulation of cell adhesion and collective cell migration by hindsight and its human homolog RREB1. Curr Biol. 2008;18:532–537. doi: 10.1016/j.cub.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 23.Tamori A., Yamanishi Y., Kawashima S., Kanehisa M., Enomoto M., Tanaka H., Kubo S., Shiomi S., Nishiguchi S. Alteration of gene expression in human hepatocellular carcinoma with integrated hepatitis B virus DNA. Clin Cancer Res. 2005;11:5821–5826. doi: 10.1158/1078-0432.CCR-04-2055. [DOI] [PubMed] [Google Scholar]
- 24.Uren A.G., Kool J., Matentzoglu K., de Ridder J., Mattison J., van Uitert M., Lagcher W., Sie D., Tanger E., Cox T., Reinders M., Hubbard T.J., Rogers J., Jonkers J., Wessels L., Adams D.J., van Lohuizen M., Berns A. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morey A.L., Murali R., McCarthy S.W., Mann G.J., Scolyer R.A. Diagnosis of cutaneous melanocytic tumours by four-colour fluorescence in situ hybridisation. Pathology. 2009;41:383–387. doi: 10.1080/00313020902915875. [DOI] [PubMed] [Google Scholar]
- 26.Gerami P., Wass A., Mafee M., Fang Y., Pulitzer M.P., Busam K.J. Fluorescence in situ hybridization for distinguishing nevoid melanomas from mitotically active nevi. Am J Surg Pathol. 2009;33:1783–1788. doi: 10.1097/PAS.0b013e3181ba6db6. [DOI] [PubMed] [Google Scholar]
- 27.Pouryazdanparast P., Newman M., Mafee M., Haghighat Z., Guitart J., Gerami P. Distinguishing epithelioid blue nevus from blue nevus-like cutaneous melanoma metastasis using fluorescence in situ hybridization. Am J Surg Pathol. 2009;33:1396–1400. doi: 10.1097/PAS.0b013e3181a92cbc. [DOI] [PubMed] [Google Scholar]
- 28.Gerami P., Mafee M., Lurtsbarapa T., Guitart J., Haghighat Z., Newman M. Sensitivity of fluorescence in situ hybridization for melanoma diagnosis using RREB1, MYB, Cep6, and 11q13 probes in melanoma subtypes. Arch Dermatol. 2010;146:273–278. doi: 10.1001/archdermatol.2009.386. [DOI] [PubMed] [Google Scholar]
- 29.Busam K.J., Fang Y., Jhanwar S.C., Pulitzer M.P., Marr B., Abramson D.H. Distinction of conjunctival melanocytic nevi from melanomas by fluorescence in situ hybridization. J Cutan Pathol. 2010;37:196–203. doi: 10.1111/j.1600-0560.2009.01488.x. [DOI] [PubMed] [Google Scholar]
- 30.Gaiser T., Kutzner H., Palmedo G., Siegelin M.D., Wiesner T., Bruckner T., Hartschuh W., Enk A.H., Becker M.R. Classifying ambiguous melanocytic lesions with FISH and correlation with clinical long-term follow up. Mod Pathol. 2010;23:413–419. doi: 10.1038/modpathol.2009.177. [DOI] [PubMed] [Google Scholar]
- 31.Miyake J.H., Szeto D.P., Stumph W.E. Analysis of the structure and expression of the chicken gene encoding a homolog of the human RREB-1 transcription factor. Gene. 1997;202:177–186. doi: 10.1016/s0378-1119(97)00491-5. [DOI] [PubMed] [Google Scholar]
- 32.Lee J.K., Havaleshko D.M., Cho H., Weinstein J.N., Kaldjian E.P., Karpovich J., Grimshaw A., Theodorescu D. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci USA. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman E.J., Hurst C.D., Pitt E., Chambers P., Aveyard J.S., Knowles M.A. Expression of hTERT immortalises normal human urothelial cells without inactivation of the p16/Rb pathway. Oncogene. 2006;25:5037–5045. doi: 10.1038/sj.onc.1209513. [DOI] [PubMed] [Google Scholar]
- 34.Titus B., Frierson H.F., Jr, Conaway M., Ching K., Guise T., Chirgwin J., Hampton G., Theodorescu D. Endothelin axis is a target of the lung metastasis suppressor gene RhoGDI2. Cancer Res. 2005;65:7320–7327. doi: 10.1158/0008-5472.CAN-05-1403. [DOI] [PubMed] [Google Scholar]
- 35.Smith S.C., Nicholson B., Nitz M., Frierson H.F., Jr, Smolkin M., Hampton G., El-Rifai W., Theodorescu D. Profiling bladder cancer organ site-specific metastasis identifies LAMC2 as a novel biomarker of hematogenous dissemination. Am J Pathol. 2009;174:371–379. doi: 10.2353/ajpath.2009.080538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitz M.D., Harding M.A., Theodorescu D. Invasion and metastasis models for studying RhoGDI2 in bladder cancer. Methods Enzymol. 2008;439:219–233. doi: 10.1016/S0076-6879(07)00417-X. [DOI] [PubMed] [Google Scholar]
- 37.Havaleshko D.M., Cho H., Conaway M., Owens C.R., Hampton G., Lee J.K., Theodorescu D. Prediction of drug combination chemosensitivity in human bladder cancer. Mol Cancer Ther. 2007;6:578–586. doi: 10.1158/1535-7163.MCT-06-0497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RREB1α and RREB1β account for the majority of RREB1 expression in bladder and prostate cancer cell lines. RNA expression of RREB1 splice variants were measured in the indicated cell lines using primers designed to the unique splice junctions. RREB1γ was not included, because of sub quantifiable expression. Isoform expression levels were calculated using the Pfaffl method (Pfaffl MW: A new mathematical model for relative quantification in real-time RT-PCR. Nucl Acids Res 2001, 29:e45).
RREB1 isoform detection by commercial RREB1 antibodies. RREB1 isoforms were isolated from UMUC-3 cells (isoforms δ and ε) or a gift from Dr. Akiyoshi Fukimizu (isoforms α and β). The splice variants were cloned into a C-terminal 3XFLAG tagged expression construct and expressed in 293T cells. Detection with anti-FLAG (1:1000), anti-RREB1 (1:500) (Cosmo Bio, Carlsbad, CA; Tokyo, Japan), or anti-RREB1 (1:1000) (GenWay Biotech, San Diego, CA) is shown.


