Abstract
Mathematical models that estimate the proportion of foodborne illnesses attributable to food commodities at specific points in the food chain may be useful to risk managers and policy makers to formulate public health goals, prioritize interventions, and document the effectiveness of mitigations aimed at reducing illness. Using human surveillance data on laboratory-confirmed Salmonella infections from the Centers for Disease Control and Prevention and Salmonella testing data from U.S. Department of Agriculture Food Safety and Inspection Service's regulatory programs, we developed a point-of-processing foodborne illness attribution model by adapting the Hald Salmonella Bayesian source attribution model. Key model outputs include estimates of the relative proportions of domestically acquired sporadic human Salmonella infections resulting from contamination of raw meat, poultry, and egg products processed in the United States from 1998 through 2003. The current model estimates the relative contribution of chicken (48%), ground beef (28%), turkey (17%), egg products (6%), intact beef (1%), and pork (<1%) across 109 Salmonella serotypes found in food commodities at point of processing. While interpretation of the attribution estimates is constrained by data inputs, the adapted model shows promise and may serve as a basis for a common approach to attribution of human salmonellosis and food safety decision-making in more than one country.
Introduction
Attribution of the burden of foodborne illness to specific foods and settings is one of the Centers for Disease Control and Prevention's (CDC) Foodborne Diseases Active Surveillance Network (FoodNet) four objectives. Foodborne illness attribution can assist risk managers and policy makers as they formulate public health goals, develop and prioritize interventions, and assess the effectiveness of efforts to reduce illness and improve public health (Batz et al., 2005). Ideally, foodborne illness attribution can be performed using data collected at different points in the food production and distribution system—the reservoir, at processing, and food preparation and retail levels—and provide insight into points where food safety interventions may be most effective. However, the lack of current and robust estimates of the prevalence and microbial concentration of specific human pathogens across the U.S. domestic food supply limits our ability to attribute observed human illnesses to all potential food sources. Attribution of human enteric illnesses to specific foodborne exposure pathways is also complicated by the wide variety of both food and nonfood sources of infection. Specific sources of foodborne illness are rarely identified outside the outbreak setting. Consequently, stochastic attribution models that incorporate data from a variety of research programs or surveillance and regulatory systems can be useful to estimate the proportion of human cases of illness resulting from specific food exposure pathways. These mathematical models can collectively provide information not available from a single data source.
There are important reasons to estimate the attribution of salmonellosis to various sources in the United States. Salmonella enterica is the most common bacterial pathogen reported from population-based, active laboratory surveillance in FoodNet. The overall incidence of laboratory-confirmed Salmonella infection in 2009 was 15.2 cases per 100,000 persons, an incidence more than twice that listed as the national health objective for the year 2010 (6.8 cases per 100,000 persons) (U.S. DHHS, 2000; CDC, 2010). Despite a regulatory and public health focus on reducing the incidence of Salmonella infection, the burden of illness has remained relatively unchanged in recent years.
We focused on adapting the Bayesian attribution model methodology developed by Hald et al. (2004) to U.S. data sources. In Denmark, the model has been used to attribute sporadic human salmonellosis to specific animal reservoirs and food commodities, and has facilitated the implementation of commodity-specific interventions that have reduced the incidence of foodborne salmonellosis (Pires and Hald, 2010). The goal of our study was to determine if the Danish model could be directly applied to U.S. data sources and to assess the potential for the resulting model to be the basis of human illness estimates attributable to specific U.S.-origin food commodities.
This article presents the first U.S. effort to develop a functional Bayesian stochastic model that attributes sporadic, domestically acquired cases of human salmonellosis to the consumption of specific animal-product food commodities.
Methods
Model overview
The basic model attribution equation as described by Hald et al. (2004) is
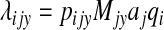 |
(1) |
where λijy is the expected number of sporadic human cases of Salmonella serotype i (i.e., not associated with a recognized outbreak) that are acquired domestically (i.e., not related to travel outside the United States) and attributed to food product j in year y. The prevalence of Salmonella serotype i in food product j in year y is represented by the term pijy, and Mjy is the amount of food product j available for consumption in year y. Since the U.S. regulatory verification sampling system is not designed to estimate pathogen prevalence in food commodities, the term “prevalence” is used here to refer to the probability of isolating Salmonella serotype i from a commodity sample at the point of processing during regulatory food safety inspection testing.
The proportionality factors aj and qi are used to estimate the relative roles of both known and unknown food-source-dependent and bacteria-dependent factors, respectively, on the expected number of human cases of salmonellosis attributed to each source. Food-source-dependent factors reflect relative differences in food commodities that influence their ability to serve as vehicles of Salmonella. Examples of food-source-dependent factors are characteristics of foods or their processing that affect the distribution or growth of bacteria in the commodity, differences between the U.S. Department of Agriculture's Food Safety and Inspection Service (FSIS) sampling programs applied to different commodities that affect the observed estimates of Salmonella presence in a commodity, or postprocessing wholesale, retail, and consumer handling practices that influence the probability of pathogen survival in the commodity at the time of consumption. Bacteria-dependent factors reflect relative differences between the Salmonella serotypes, such as the survival characteristics of a Salmonella serotype in food commodities, virulence or dose-related properties, other modes of transmission not related to commodities in the model, or human pathogenicity.
The underlying assumption of the model is that the annual number of domestically acquired sporadic cases of each Salmonella serotype attributed to a given food is proportional to the amount of food consumed that is contaminated with each serotype, such that λijy μ pijy Mjy. Since there is no estimate of microbial load or dose per serving in this model, the pijy Mjy terms do not estimate the expected numbers of infectious exposures. Thus, the total number of human cases of serotype i in a given year, Niy, is approximated by the sum of commodity-specific results obtained from the right hand side of equation 1, using Bayesian estimates of aj and qi for each commodity and serotype, respectively.
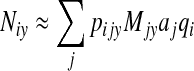 |
(1a) |
Description of U.S. data sources
Domestically acquired sporadic cases of Salmonella serotype i per year
The annual number of domestically acquired sporadic cases for each Salmonella serotype i was estimated using data on culture-confirmed human Salmonella infections obtained from the National Salmonella Surveillance System (NSSS) for the study years 1998 through 2003. NSSS data include Salmonella serotypes isolated from human clinical specimens reported to CDC by U.S. public health laboratories. Data from Mississippi, Florida, and Texas were excluded because isolate serotype was not consistently reported during the study period. The NSSS database does not contain epidemiologic information about reported illnesses. However, since 2004 each case of salmonellosis ascertained in the FoodNet surveillance system has epidemiologic information regarding whether travel- or outbreak-related (“Yes,” “No,” or “Unknown”). Therefore, data collected from FoodNet in 2004 were used to estimate the probability that an individual case of serotype i reported to NSSS was both domestically acquired and sporadic. Table 1 illustrates the matrix that was used to classify cases obtained from FoodNet surveillance data to estimate the relative proportions of human illnesses caused by serotype i in each epidemiologic category. The total numbers of serotype-specific culture-confirmed Salmonella infections in the NSSS database that were domestically acquired and sporadic were modeled stochastically using the methods outlined in Table 2. For this model we assumed that the underlying U.S. proportions of Salmonella cases of serotype i that were outbreak- or travel-related were constant from 1998 to 2003 and equal to that of the FoodNet catchment in 2004.
Table 1.
Foodborne Diseases Active Surveillance Network Classification Matrix Used to Estimate the Total Number of Domestically Acquired (DA+) and Sporadic Cases of Laboratory-Confirmed Salmonellosis
| International travel | ||||
|---|---|---|---|---|
| Outbreak | Yes | No | Unknown | |
| Yes | a | koi | ||
| No | kti | ksi | bi | |
| Unknown | ai | Ui | ||
Cell parameters reflect variable designations used in Bayesian models outlined in Table 2 to estimate the unknown portions of cases (gray cells) that were either DA+, sporadic, or both.
This cell had very few cases in the FoodNet database. Since the proportion of cases assigned to this category was negligible compared to the other categories, and since these cases were likely misclassified, these cases were re-classified as sporadic, international travel-related (kti).
FoodNet, Foodborne Diseases Active Surveillance Network.
Table 2.
Estimation of the Annual Number of Domestically Acquired Sporadic Cases of Salmonellosis
| Notation | Description | Estimation |
|---|---|---|
| Cases per Salmonella serotype | ||
| I | Subscript for Salmonella serotype associated with a reported case of salmonellosis | |
| Y | Subscript for study year: 1998–2003 | |
| sssyi | Observed cases of i reported to national surveillance system in year y | NSSS Data |
| kti | Observed cases of i that were known to be unrelated to an outbreak (sporadic) but were international travel-related (also includes cases that were classified as both outbreak- and international travel-related) | FoodNet Data |
| ksi | Observed cases of i that were known to be sporadic and not travel-related (domestically acquired) | FoodNet Data |
| αuti | Probability that a case of i that is known to be unrelated to an outbreak but is missing travel information is international travel-related | Beta(kti + 1; ksi + 1) |
| bi | Observed cases of i that were sporadic but had no information regarding travel | FoodNet Data |
| λti | Estimated expected number of international travel-related cases of i among those cases known to be unrelated to outbreaks but missing travel information | bi x αuti |
| xi | Expected number of international travel-related cases of i among those cases known to be unrelated to outbreaks but missing travel information | Poisson(λti)) x I[xi < bi] |
| ntspi | Estimated number of sporadic cases of i with no known travel history that were domestically acquired | bi - xi |
| koi | Observed cases of serotype i that were known to be domestically acquired and outbreak-related | FoodNet Data |
| αobi | Probability that a case of serotype i that was known to be domestically acquired but had no outbreak history was outbreak-related | Beta(koi + 1; ksi + 1) |
| ai | Observed cases of serotype i that were known to be domestically acquired but had an unknown outbreak history | FoodNet Data |
| λobi | Estimated expected number of cases of serotype i that were outbreak-related among those cases that were domestically acquired but lacked outbreak information | ai x αobi |
| oi | Expected number of outbreak-related cases of i among those cases known to be domestically acquired but lacking outbreak information | Poisson(λobi) x I[oi < ai] |
| nospi | Estimated number of cases of i that were sporadic among those known to be domestically acquired but had an unknown outbreak history | ai - oi |
| αspoi αdobi αtrspoi | Probability that a randomly selected FoodNet case of i was domestically acquired, sporadic (αspoi), domestically acquired, outbreak-related (αdobi) and international travel-related, sporadic cases (αtrspoi). All three estimated probabilities sum to 1. | Dirichlet(ksi + 1, koi + 1, kti + 1) |
| Ui | Observed number of FoodNet cases of i designated as unknown for both international travel and outbreak. | FoodNet Data |
| si | Estimated number of domestically acquired, sporadic cases of i that were designated as unknown for both international travel and outbreak in FoodNet Data (estimated numbers of domestically acquired, outbreak-related and international travel-related, sporadic cases were also calculated) | Multinomial (Ui, αspoi, αdobi, αtrspoi) |
| Sporadic | Total estimated number of domestically acquired, sporadic cases of i reported by FoodNet sites | ksi + ntspi + nospi + si |
| αfnspoi | Proportion of FoodNet cases of i that were domestically acquired, sporadic after allocating all cases with partial and completely unknown data (estimated proportions were also calculated for domestically acquired, outbreak-related [αfnoutyi] and international travel-related, sporadic categories [αfntravyi]) | Gamma(Sporadic; 1) Σ Gamma(Sporadic; 1) |
| ssspoyi | Estimated number of sporadic cases of i in year y reported to Salmonella Surveillance System (estimated case numbers were also calculated for outbreak [ssoutyi] and travel-related cases [sstravyi]) | pfnspoi x sssyi |
Adapted from Hald et al. (2004).
NSSS, National Salmonella Surveillance System.
Salmonella serotype prevalence in food commodities
The distributions of Salmonella serotypes in ground beef, intact beef, chicken, turkey, pork, and FSIS-regulated egg products were estimated using data collected by FSIS regulatory sampling programs for raw products during the years 1998–2003.1 These data describe the number of samples obtained from each food commodity type, the number of samples that yielded Salmonella, and the serotype of each isolate. The distribution of Salmonella serotypes in shell eggs was estimated based on data from the Salmonella Enteritidis Pilot Project, a mid-1990s project designed to monitor Salmonella Enteritidis in laying flocks and eggs in Pennsylvania (Schlosser et al., 1999).
The estimates for the prevalence of each Salmonella serotype (Si) in each food commodity or product, (Pj), was computed using Equation 2
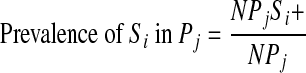 |
(2) |
where NPjSi + is the number of samples that were positive for serotype i in product j and NPj is the number of samples of product j tested for Salmonella.
Because there were no FSIS regulatory sampling program data for turkey carcasses during the period of study, we assumed that the ratio of Salmonella prevalence for ground turkey to whole turkey is the same as the ratio for ground chicken to broilers.
FSIS collects Salmonella data on cows and bulls and on steers and heifers. Consequently, the estimates for intact beef were generated using both sources and the following equation:
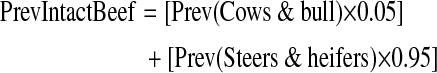 |
(3) |
The weighting factors 0.05 and 0.95 are the relative proportions of each animal source in intact beef estimated by experts from the American Meat Institute (AMI).
Information about methods of sample collection and enrichment and isolation of Salmonella from FSIS regulated commodities is available in the FSIS Microbiology Laboratory Guidebook.2
Food consumption
Food consumption data were obtained from the U.S. Department of Agriculture Economic Research Service Food Consumption Data System3 and the AMI. The Economic Research Service estimates domestic food consumption by calculating the annual per capita disappearance of domestically available food. It divides the total pounds of food commodity annually available for domestic consumption by the yearly U.S. population estimate, adjusting for losses at the retail/institutional level, nonedible portions, as well as cooking losses at the consumer level. Data for the yearly food disappearance at retail for the study years 1998 through 2003 were obtained for each of the following food categories: chicken, beef, turkey, egg products, pork, and shell eggs. Estimates for ground beef consumption obtained from the AMI (Jim Hodges, pers. comm., December 13, 2005) were used to determine the proportion of total beef consumed that was ground. Use of these data requires the assumption that all food estimated to be available for domestic consumption (after adjustment for losses) is consumed. In addition, these data do not differentiate between domestic and imported sources of food, so we assumed that during our study period the amount of imported food in our commodity categories was insignificant compared with the amount of domestic food available in the U.S. inventory.
Implementation of the attribution model
Model code, written in WinBUGS, was supplied by Hald et al. (2004). Adaptation of the code included multiple programming loops to estimate the annual number of illnesses due to serotype i for multiple years (1998–2003). Multiple independent Markov chains of over 40,000 iterations each were used for each model run. Values for the qi and aj parameters were initially estimated using the same continuous uniform distributions described previously (Hald et al., 2004). Likewise, estimated qi values were parameterized by using the method described by Hald of setting the q for serotype Enteritidis at 1. Implementation of the Bayesian model (outlined in Table 2) to estimate the number of domestically acquired, sporadic cases was implemented concurrently with the estimation of qi and aj parameter values. The model code is available from the corresponding author of this report.
All observed cases estimated to be domestically acquired and sporadic are assumed to be foodborne and attributable to the food commodities in the model, so serotypes were excluded if they were not identified in both the food commodity and the human case databases to limit spurious attribution of serotypes. One hundred nine Salmonella serotypes were identified in both NSSS human surveillance and FSIS regulatory raw product sampling databases. Initial modeling efforts included all 109 individual serotypes individually parameterized; however, the resultant model was not stable. Therefore, serially evaluated models were run to determine the maximum number of serotypes that could be parameterized (i.e., qi value estimated) in a stable model, and an “other” serotype category was added to include data for all serotypes not individually specified in the model. Sequential model runs included the most frequently occurring serotypes in NSSS data specified individually. It was noted that serotype nomenclature was not consistent across study years and between datasets, so nomenclature rationalization (e.g., Oranienburg was recorded as a combination of Oranienburg and Oranienburg Var. 14+, formerly Thielalee) was performed before selection of a stable model.
Inclusion of 30 serotypes (plus the additional category “other” containing the data for the remaining 79 serotypes) was the upper limit for model stability, accounting for >95% of all reported cases of salmonellosis among the 109 study serotypes reported during 1998–2003 (Supplementary Figs. S1 and S2 illustrate the distribution of the 30 serotypes in human illness and modeled food commodities; Supplementary Data are available online at www.liebertonline.com/fpd). Once a stable model was identified, the model properties and estimated outputs under different model conditions were evaluated, and a model was identified for the estimation of the proportion of illnesses attributable to each commodity.
Results
Figures illustrating the model-estimated qi (i.e., bacteria-dependent factor) and aj (i.e., food-source-dependent factor) values are available online (Supplementary Figs. S3 and S4). Because the qi of Salmonella Enteritidis was held at 1, qi estimates for the other Salmonella serotypes are interpreted relative to this value. Newport and Javiana had the largest median qi values (9.76 and 9.67, respectively; prior distribution range: 0–10) in the 30-serotype model. Since this parameter is a multiplier, higher estimated values reflect a relatively disproportionate number of observed human cases of serotype i relative to the distribution of the serotype in the modeled food commodities. Recognizing this, data for Salmonella Javiana were excluded from subsequent models because human infection with Javiana has not been epidemiologically associated with the food commodities in this model. Clustering of qi estimates at the upper limit of the specified prior uniform distributions was noted for several serotypes in the 30-serotype model. To address this, the number of individually specified serotypes was reduced to 15 and 10, and the uniform distributions were dynamically adjusted for individual serotypes.
The relative ranking of estimated qi values for Salmonella serotypes was fairly stable in both the 15- and 10-serotype models that included shell eggs (Supplementary Fig. S3). Even with dynamic adjustment of the prior distributions, the estimated qi values for Oranienburg and Newport continued to cluster near the upper limit of the specified distribution, suggesting that the Danish approach of using Enteritidis as the baseline serotype may not be as applicable to our data. None of the serotypes in the 15- or 10-serotype models that included shell eggs had estimated qi values that suggested that they were less likely to cause disease than Salmonella Enteritidis when present in a food commodity. Although the estimated qi values for serotypes Agona and Montevideo were closest to Enteritidis, the estimated values were still more than four times higher than the Enteritidis baseline.
In the 30-serotype model, consumption of a given amount (in pounds) of shell eggs was associated with an estimated probability of salmonellosis nearly 200 times higher than consumption of the same amount of ground beef. However, as the 30-serotype model was modified to examine the stability of estimated qi values, it became clear that inclusion of the shell egg commodity was contributing to overall model instability. These data were not comparable to the data used for the other commodities in the model in that they were not nationally representative and were collected several years before the study period. Consequently, these data were removed with the shell egg commodity from the 15- and 10-serotype models. Removing the shell egg data changed the model rankings of some of the remaining food commodities. Specifically, egg products, ranked fifth in the 30-serotype model, had the highest aj estimates in the 15- and 10-serotype models. Intact beef was ranked the least risky of the seven commodities in the 30-serotype model but was fourth (out of six commodities) in both reduced serotype models. Removing the shell egg commodity also significantly reduced the credibility intervals for the aj estimates for egg products n the 15- and 10-serotype models (Supplementary Fig. S4).
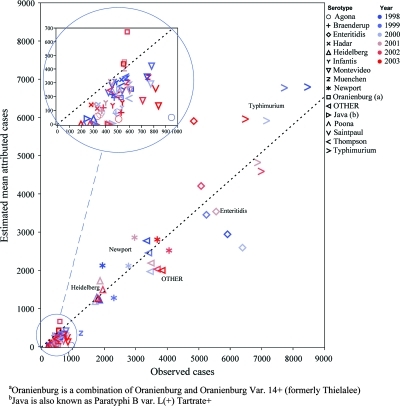
We determined the 15-serotype model without the shell egg commodity to be our best current model for estimating the attribution of salmonellosis to food commodities. There were 160,000 laboratory-reported cases of salmonellosis included in the Bayesian model. The model estimated that 106,000 of these illnesses were domestically acquired, sporadic, and were attributed to the modeled food commodities. Figure 1 illustrates the number of observed cases that the model estimated to be domestically acquired, sporadic for each serotype. A plot of the best-fitting line reveals that the estimated number of domestically acquired sporadic illnesses was ∼0.73 times the observed number of laboratory-reported illnesses during the study period. The modeled estimates of serotypes more likely to be reported by FoodNet sites to be of unknown travel or outbreak status—such as Enteritidis, Agona, and Braenderup—reduced the overall proportion of observed cases estimated to be domestically acquired, sporadic below the 2004 FoodNet estimate of 85% (FoodNet includes only those with known epidemiologic data) (CDC, 2006).
FIG. 1.
Estimated mean annual attributed Salmonella cases versus observed annual attributable cases by serotype and year of study. The dotted line represents a least squares fit constrained to pass through the point (0, 0). The slope of the fitted line is 0.73, which suggests that, at the level of serotype and year, 73% of observed cases are attributed to commodity. The inset reveals a lack of fit with respect to this relationship and the data, with serotypes having lower counts showing a generally lower percentage of observed cases attributed to commodity. Color images are available online at www.liebertonline.com/fpd
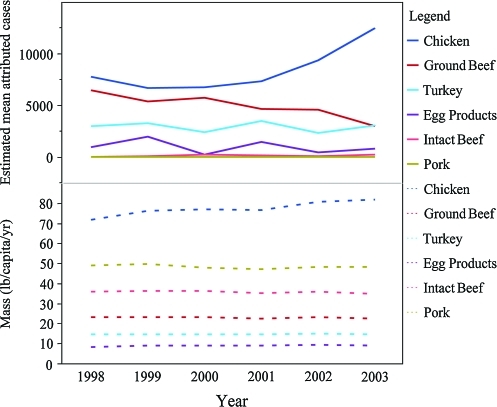
Figure 2 illustrates the annual consumption data and mean number of estimated Salmonella cases attributed to each commodity. Of the reported salmonellosis cases attributed by this model to the included commodities, 48% were attributed to chicken, 28% to ground beef, 17% to turkey, 6% to egg products, 1% to intact beef, and <1% to pork. These proportions were stable and did not change significantly as inputs were varied during sensitivity analyses, which included excluding the “other” serotype category, modifying estimated serotype distributions in the commodities, and varying the estimated numbers of outbreak- and travel-related cases (results not shown).
FIG. 2.
Annual commodity consumption data as estimated via disappearance (mass in pounds per capita per year) and estimated mean attributed Salmonella cases versus year by commodity for six commodities. Color images are available online at www.liebertonline.com/fpd
Discussion
In this study we sought to adapt an attribution model for foodborne salmonellosis (Hald et al., 2004) to use with U.S. data. This adaptation of the model retained much of the original methodology and represents a potential opportunity to inform food safety efforts and develop a common understanding of foodborne disease attribution in multiple countries. The high estimated consumption of chicken relative to the other modeled commodities during the period of study, as well as the distribution of positive Salmonella samples from chicken at the point of processing, resulted in the highest model-estimated proportion of illnesses being attributed to this commodity. Thus, the model seems to provide reasonable relative attribution estimates for included commodities based upon domestic consumption and the probability of Salmonella presence in food sources. Although data availability limited the number of food commodities that were included in the model, the estimated relative proportions of Salmonella illness attribution across the commodities were robust to changes in data inputs and model constraints. This is an important feature for future applications, as shell eggs were initially estimated to be the most risky food vehicle per pound of consumption, but inclusion of this data led to model instability and could not be used to attribute Salmonella illnesses to commodities. Removal of the shell egg commodity resulted in a shift to egg products and improved stability of the food-source-dependent factor estimate for this commodity. This suggests that it may be possible to include specific food commodities that lack robust data by using data from an alternative model commodity in a “what if” exploration of foodborne disease attribution.
Differences between the two countries' data inputs result in distinct interpretations of the estimated values for food-source-dependent (aj) and bacteria-dependent (qi) factors. All of our data on Salmonella presence in food commodities were obtained from the point of processing, whereas both preharvest surveillance data and end-product samples were used in the Hald model. Because the estimated food-source parameter reflects the cumulative effect of all processes in the food chain between the point of observed Salmonella presence in the food source and consumption, our variable estimates reflect a smaller range of food production-specific factors that may influence Salmonella presence at the point of human consumption. In addition, our model did not incorporate Salmonella phage typing data; we also included serotypes found in many food commodities that have been sources of human infection but were not included in our final attribution model, such as shell eggs, produce, milk, and fish (Lynch et al., 2006), as well as sources of infection not associated with food, such as direct contact with animals (Sato et al., 2000; NASPHV, 2005; Milstone et al., 2006) and household environmental exposures (Barker and Bloomfield, 2000). The modeled bacteria-dependent parameters are dependent upon the assumption that the food commodities included in the model are the only reservoirs of human infection for the included serotypes. Consequently, the observed clustering of estimated serotype qi values for serotypes such as Javiana and Newport toward the upper limit of the specified prior distribution likely reflects the presence of additional exposure pathways not included in our model rather than a higher intrinsic likelihood of the serotype causing disease.
All of the model iterations in our study involved the estimation of more parameters than the originally described model and subsequent adaptations (Hald et al., 2004; Mullner et al., 2009; Little et al., 2010). Estimating a larger number of parameters likely contributed to some of our difficulties with model convergence. Mullner et al. (2009) noted that a major limitation of this attribution model is the high number of estimated food-source-dependent and bacteria-dependent factors compared with the limited number of data points used to generate these, and used a hierarchical approach to generate random values from a hypothetical distribution of bacteria-dependent factors (Mullner et al., 2009). While this approach simplifies the parameterization of the model and improves its convergence properties, it makes the comparison of bacteria-dependent factors among Salmonella types problematic. This limitation may not be desirable when considering the potential value of these outputs to food safety programs.
Incomplete human illness data limit the validity of the estimated attribution outputs. Laboratory data from three states had to be excluded from the model because of incomplete reporting. Exclusion of these cases may have resulted in an underestimation of the burden of human salmonellosis attributable to the modeled commodities. In addition, the geographic distribution of serotypes included in this model is not uniform (CDC, 2003); thus, the inclusion of cases from these states might have had significant impacts on estimated bacteria-dependent factors if the serotype distributions in these populations were known. Our model also excluded all model-estimated outbreak-related cases of Salmonella infection from attribution. Since our study used epidemiologic data reported to FoodNet in 2004 to estimate the total number of outbreak-associated cases among the 1998–2003 NSSS cases, we were not able to identify individual outbreaks in our study. Consequently, we chose to exclude all model-estimated outbreak cases to avoid the introduction of a highly uncertain estimated fraction in our attribution. The annual frequency and size of reported outbreaks, as well as the availability of data that distinguish outbreak and sporadic illnesses, drive the choice of how outbreak cases are used in source attribution. As the 2010 multi-state outbreak of Salmonella Enteritidis highlights (CDC, 2010), a single, large foodborne disease outbreak can double the burden of illness associated with a single serotype within a specific time frame and significantly impact the annual attribution to an individual commodity. Likewise, the number of submitted isolates that are directly attributable to the outbreak source is unknown. With additional data, regional differences in attribution as well as the potential role of outbreaks on the number of annually reported Salmonella cases may be evaluated, and allow us to better represent the overall disease process.
Another major limitation of our model was the absence of data for Salmonella presence among shell egg samples—a commodity well known to be a source of human infection. Because of the currently very low incidence of Salmonella contamination of eggs (estimates suggest that somewhere between 1 in 10,000 to 1 in 20,000 eggs are contaminated [Schlosser et al., 1999; Ebel and Schlosser, 2000]), it is difficult to generate statistically robust sampling data for Salmonella in eggs. Given the importance of shell eggs as vehicles of Salmonella infection, a new, nationally representative survey of Salmonella in and on shell eggs should be considered to inform future attribution models.
Collectively, foodborne disease attribution efforts raise the question of how much human salmonellosis is due to animal product food sources. Hald et al. attribute ∼75% of estimated sporadic, domestic cases to animal product food sources. Mullner et al. attributed all estimated cases of salmonellosis to six food animal commodity categories. We also attributed all of the 106,000 cases estimated by our model to be domestically acquired and sporadic to our six animal product food commodities. This approach likely over-estimates the burden of foodborne salmonellosis attributed to the commodities included in the model.
Conclusions
Our adaptation of the Hald model was focused on exploring model behavior and characteristics using U.S. data while retaining as many parameters as possible. Other adaptations have mathematically simplified the model to make it more broadly applicable to a wide variety of data resolutions, data sources, and nonfoodborne exposure pathways (Mullner et al., 2009; Little et al., 2010). We believe that by generating additional and robust Salmonella surveillance data from reported human illnesses and food commodities and through continued refinement of the number of parameters included in the model, we can develop increasingly useful results to help guide development, formulation, and implementation of sound mitigation strategies to reduce salmonellosis and protect public health.
Supplementary Material
Footnotes
Disclosure Statement
No competing financial interests exist.
References
- Barker J. Bloomfield SF. Survival of Salmonella in bathrooms and toilets in domestic homes following salmonellosis. J Appl Microbiol. 2000;89:137–144. doi: 10.1046/j.1365-2672.2000.01091.x. [DOI] [PubMed] [Google Scholar]
- Batz MB. Doyle MP. Morris G., Jr. Painter J. Singh R. Tauxe RV. Taylor MR. Lo Fo Wong DM Food Attribution Working Group. Attributing illness to food. Emerg Infect Dis. 2005;11:993–999. doi: 10.3201/eid1107.040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. FoodNet Surveillance Report for 2004 (Final Report) Jun, 2006. www.cdc.gov/FoodNet/annual/2004/Report.pdf. [Feb 9;2010 ]. www.cdc.gov/FoodNet/annual/2004/Report.pdf (Online.)
- [CDC] Centers for Disease Control and Prevention. Investigation update: multistate outbreak of human Salmonella Enteritidis infections associated with shell eggs. 2010. Atlanta, Georgia: US Department of Health and Human Services, CDC; 2010. [Oct 10;2010 ]. (Online.) [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. Morb Mortal Wkly Rep. 2010;59:418–422. [PubMed] [Google Scholar]
- [CDC] Centers for Disease Control and Prevention. Salmonella surveillance: annual summary. Atlanta, Georgia: US Department of Health and Human Services, CDC; 2003. [Oct 5;2010 ]. 2004. (Online.) [Google Scholar]
- Ebel E. Schlosser W. Estimating the annual fraction of eggs contaminated with Salmonella enteritidis in the United States. Int J Food Microbiol. 2000;61:51–62. doi: 10.1016/s0168-1605(00)00375-5. [DOI] [PubMed] [Google Scholar]
- Hald T. Vose D. Wegener HC. Koupeev T. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 2004;24:255–269. doi: 10.1111/j.0272-4332.2004.00427.x. [DOI] [PubMed] [Google Scholar]
- Little CL. Pires SM. Gillespie IA. Kathie G. Nichols GL. Attribution of human Listeria monocytogenes infections in England and Wales to ready-to-eat food sources placed on the market: adaptation of the Hald Salmonella source attribution model. Foodborne Pathog Dis. 2010;7:749–756. doi: 10.1089/fpd.2009.0439. [DOI] [PubMed] [Google Scholar]
- Lynch M. Painter J. Woodruff R. Braden C Centers for Disease Control and Prevention. Surveillance for foodborne-disease outbreaks—United States, 1998–2002. MMWR Morb Mortal Wkly Rep. 2006;55:1–42. [PubMed] [Google Scholar]
- Milstone AM. Agwu AG. Angulo FJ. Alerting pregnant women to the risk of reptile-associated salmonellosis. Obstet Gynecol. 2006;107:516–518. doi: 10.1097/01.AOG.0000187950.37065.87. [DOI] [PubMed] [Google Scholar]
- Mullner P. Jones G. Noble A. Spencer SE. Hathaway S. French N. Source attribution of food-borne zoonoses in New Zealand: a modified hald model. Risk Anal. 2009;29:970–984. doi: 10.1111/j.1539-6924.2009.01224.x. [DOI] [PubMed] [Google Scholar]
- [NASPHV] National Association of State Public Health Veterinarians IN. Compendium of measures to prevent disease associated with animals in public settings, 2005. MMWR Morb Mortal Wkly Rep. 2005;54:1–12. [PubMed] [Google Scholar]
- Pires SM. Hald T. Assessing the differences in public health impact of Salmonella subtypes using a Bayesian microbial subtyping approach for source attribution. Foodborne Pathog Dis. 2010;7:143–151. doi: 10.1089/fpd.2009.0369. [DOI] [PubMed] [Google Scholar]
- Sato Y. Mori T. Koyama T. Nagase H. Salmonella Virchow infection in an infant transmitted by household dogs. J Vet Med Sci. 2000;62:767–769. doi: 10.1292/jvms.62.767. [DOI] [PubMed] [Google Scholar]
- Schlosser WD. Henzler DJ. Mason J. Kradel D. Shipman L. Trock S. Hurd SH. Hogue AT. Sischo W. Ebel ED. The Salmonella enterica serovar Enteritidis Pilot Project. In: Saeed AM, editor; Gast RK, editor; Potter ME, editor; Wall PG, editor. Salmonella enterica serovar Enteritidis in Humans and Animals: Epidemiology, Pathogenesis, and Control. Ames: Iowa State University Press; 1999. pp. 353–366. [Google Scholar]
- [U.S. DHHS] U.S. Department of Health and Human Services. Healthy people 2010. Washington, DC: US Department of Health and Human Services; 2000. [May 5;2008 ]. (Online.) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.