Abstract
An important challenge in prostate cancer research is to develop effective predictors of tumor recurrence following surgery to determine whether immediate adjuvant therapy is warranted. To identify biomarkers predictive of biochemical recurrence, we isolated the RNA from 70 formalin-fixed, paraffin-embedded radical prostatectomy specimens with known long-term outcomes to perform DASL expression profiling with a custom panel that we designed of 522 prostate cancer–relevant genes. We identified a panel of 10 protein-coding genes and two miRNA genes (RAD23B, FBP1, TNFRSF1A, CCNG2, NOTCH3, ETV1, BID, SIM2, LETMD1, ANXA1, miR-519d, and miR-647) that could be used to separate patients with and without biochemical recurrence (P < 0.001), as well as for the subset of 42 Gleason score 7 patients (P < 0.001). We performed an independent validation analysis on 40 samples and found that the biomarker panel was also significant at prediction of biochemical recurrence for all cases (P = 0.013) and for a subset of 19 Gleason score 7 cases (P = 0.010), both of which were adjusted for relevant clinical information including T-stage, prostate-specific antigen, and Gleason score. Importantly, these biomarkers could significantly predict clinical recurrence for Gleason score 7 patients. These biomarkers may increase the accuracy of prognostication following radical prostatectomy using formalin-fixed specimens.
Prostate cancer remains the most common noncutaneous cancer diagnosed for U.S. males, and ranks second among tumor site–specific mortality, with estimates for 2009 at over 192,000 new cases and 27,000 deaths.1 The majority of patients with prostate cancer are clinically asymptomatic with early-stage, organ-confined disease, and in fact, more than 50% of men who reach the age of 80 develop clinically insignificant prostate cancer. However, a subpopulation of prostate cancer patients progress to highly aggressive, androgen-independent metastatic disease, which is inevitably fatal. One of the important challenges in current prostate cancer research is to develop effective methods to determine whether a patient is likely to progress to aggressive, metastatic disease, to aid clinicians in deciding on the appropriate course of treatment. Biomarker assays that could predict progression and metastasis for prostate cancer patients would be of great utility in aiding clinical management of this large patient population. An important challenge in prostate cancer research is to develop effective predictors of tumor recurrence following surgery to determine whether immediate adjuvant therapy is warranted. Thus, biomarkers that could predict the likelihood of success for surgical therapies would be of great clinical significance.
In the past few years, enormous progress has been made in developing technologies to exploit formalin-fixed, paraffin-embedded (FFPE) tumor tissue samples for gene expression analysis. The DASL (cDNA-mediated Annealing, Selection, extension and Ligation) assay is a unique expression profiling platform that is based on massively multiplexed RT-PCR applied in a microarray format, that allows for the determination of expression of RNA isolated from 96 FFPE tumor tissue samples in a high throughput format.2,3
Here, we have identified biomarkers predictive of recurrence by expression profiling archived FFPE tumor samples using both a custom panel of prostate cancer–associated mRNA genes and a panel of microRNA (miRNA) genes. These biomarkers were developed on a training set of 70 patients (29 with biochemical recurrence and 41 controls) and validated on an independent set of 40 samples (13 with biochemical recurrence and 27 controls) and were able to significantly discriminate between patients with and without biochemical recurrence following radical prostatectomy. Moreover, these biomarkers were able to discriminate biochemical recurrence in patients with Gleason score 7, for whom outcome is particularly difficult to predict.
Materials and Methods
Patient Samples
In the initial training set, 70 prostate cancer cases were used (29 with biochemical recurrence and 41 controls without recurrence), 45 patients from Sunnybrook Health Science Center (Toronto, ON), and 25 patients from Emory University (Atlanta, GA). The 45 cases of paraffin-embedded tissue samples from Toronto were drawn from men who underwent radical prostatectomy as the sole treatment for clinically localized prostate cancer between 1998 and 2006. The clinical data include multiple clinicopathological variables such as prostate-specific antigen (PSA) levels, histological grade (Gleason score), tumor stage (pathological stage category, for example; organ confined, pT2; or with extra-prostatic extension, pT3a; or with seminal vesicle invasion, pT3b), and biochemical recurrence rates. For the cases from Emory University, both the training set (25 cases) and validation set (40 cases) FFPE samples were also selected from a screen of over a thousand patients through an institutional review board–approved retrospective study at Emory University of men who had undergone radical prostatectomy between 1990 and 1994. Those who were included met specific inclusion criteria, had available tissue specimens, had documented long-term follow-up, and consented to participate or were included by institutional review board waiver. The cases were assigned prostate identification numbers to protect their identities. These patients did not receive neoadjuvant or concomitant hormonal therapy. Their demographic, treatment, and long-term clinical outcome data have been collected and recorded in an electronic database. Clinical data recorded include PSA measurements, radiological studies and findings, clinical findings, tissue biopsies, and additional therapies that the subjects may have received. Clinical data associated with the samples used in this study are given in Supplemental Table S1 (available at http://ajp.amjpathol.org).
RNA Preparation
Tissue cores (1 mm) were used for RNA preparation rather than sections because of the heterogeneity of samples and the opportunity for obtaining cores with a very high percentage tumor content. H&E-stained slides were reviewed by a board-certified urologic pathologist (A.O.O.) to identify regions of cancer to select corresponding areas for cutting of cores from paraffin blocks. Total RNA was prepared at the Emory Biomarker Service Center from FFPE cores as previously described,4 using the Ambion Recoverall MagMax methodology in 96-well format on a MagMax 96 Liquid Handler Robot (Life Technologies, Carlsbad, CA). FFPE RNA was quantitated using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE), and tested for RNA integrity and quality by TaqMan analysis of the RPL13a ribosomal protein on a HT7900 real-time PCR instrument (Applied Biosystems, Foster City, CA). Samples with sufficient yield (>500 ng), A260/A280 ratio >1.8, and RPL13a CT values less than 30 cycles were used for miRNA and DASL profiling.
Custom Prostate Cancer DASL Assay Pool
The DASL assay enables quantitation of expression of up to 1536 probes using RNA isolated from archived FFPE tumor tissue samples in a high throughput format.2,3 Data from multiple publicly available gene expression datasets,5–8 along with genes involved in prostate cancer progression based on current understanding of the disease,6,9 were distilled to develop a highly predictive set of 522 genes for use in the DASL assay. Due to specific probe design considerations, this panel had three probes for 497 genes, two probes for 20 genes, and a single probe for five genes, two of which were specific to TMPRSS2-ERG and TMPRSS2-ETV1 fusion transcripts. The unique combination of genes was optimized for performance in the DASL assay using stringent criteria that predicts excellent performance of the primer sets. The panel includes genes found to be correlated with Gleason score in Liu et al,10 Bibikova et al,11 True et al,12 Lapointe et al,7 and/or Singh et al.13 It also includes prognostic markers from Dhanasekaran et al5 and Yu et al,14 and genes associated with metastasis in Varambally et al.6 In addition, a number of genes known from other studies to be critical in prostate cancer such as NKX3.1, PTEN, and the androgen receptor are all included in the panel. Other genes that play important roles in the Wnt, Hedgehog, TGFβ, Notch, MAPK, and PI3K pathways are also present in this gene set. Finally, primer sets that detect chromosomal translocations in ERG,9 ETV1,15 and ETV416 are also included in this panel. The custom prostate cancer panel list of 522 candidate genes (see Supplemental Table S2 at http://ajp.amjpathol.org) was submitted to Illumina for synthesis (Illumina, San Diego, CA). The optimal oligonucleotide sequence for each of the 1536 gene probes was determined using an oligonucleotide scoring algorithm. The oligonucleotide pool or DASL Assay Pool was synthesized by Illumina for use with the 96-well Universal Array Matrix.
The DASL Assay
The DASL assay was performed with our 522-gene custom-designed human prostate cancer panel using 200 ng of input RNA at the Emory Biomarker Service Center, Emory University, according to the manufacturer's protocols. Samples, including technical replicates (two, three, or four) were hybridized on Universal Array Matrices, and scanned using the BeadStation 500 Instrument (Illumina). For miRNA DASL assays, the human miRNA v2 DASL panel (Illumina), which allows for the determination of expression of 1146 human miRNAs (>97% coverage of miRBase release 12), was used. These data are available in the Gene Expression Omnibus (GEO) database under accession number GSE26367.
Data Analysis
DASL fluorescent intensities were interpreted in GenomeStudio, quantile normalized, and exported for meta-analysis. Average signal intensity, genes detected (P value = 0.01), background, and noise (SD of background) were analyzed for trends by plate, row, and column. The two endpoints of interest were postoperative biochemical recurrence, defined as two detectable PSA readings (>0.2 ng/mL), and clinical recurrence, defined as evidence of local or metastatic disease. The primary outcome of interest was time to biochemical recurrence following surgery. A local recurrence was defined as recurrence of cancer in the prostatic bed that was detected by either a palpable nodule on digital rectal examination and subsequently verified by a positive biopsy, and/or a positive imaging study [ProstaScint (EUSA Pharma, Langhorne, PA) or computed tomography scan] accompanied by a detectable postoperative PSA result and lack of evidence for metastases. Also, patients whose PSA levels decreased following adjuvant pelvic radiation therapy for elevated postoperative PSA were considered as local recurrence cases. A recurrence with metastases was defined as a positive imaging study indicating presence of a tumor outside of the prostatic bed.
To identify important biomarkers and build and evaluate prediction models for prostate cancer recurrence, we adopted the following strategy. In the training step, the prediction model was built based on the time to biochemical recurrence. Specifically, we first fit a univariate Cox proportional hazard (PH) model for each individual oligonucleotide probe using the training dataset, and a set of important mRNA and miRNA probes were then preselected based on a false discovery rate threshold of 0.30. Next, to identify the optimal prediction score based on the preselected probes, we fit a lasso Cox PH model17,18 using the training dataset, where the tuning parameter for lasso was selected using a leave-one-out cross-validation technique.18 The lasso Cox PH model was fitted first using the set of preselected mRNA probes only and then using the complete set of preselected mRNA and miRNA probes, resulting in an optimal mRNA panel and an optimal combined mRNA/miRNA panel, respectively. Based on each biomarker panel, a final prediction model for recurrence was built to also incorporate relevant clinical biomarkers, namely, T-stage, PSA, and Gleason score, through fitting Cox PH models. For comparison, we also built a prediction model using only clinical information, namely, T-stage, PSA, and Gleason score, through fitting a Cox PH model.
To evaluate and validate the final prediction models obtained from the training phase, 79 samples from 40 patients were used, and replicate samples from the same patient were again averaged to generate a single average signal for each patient. Each prediction model from the training phase was used to generate a predictive score for each subject in the validation dataset, and subjects were subsequently divided into high and low scoring groups using the median predictive score. Kaplan-Meier analysis was performed to compare the time to biochemical recurrence, between high (poor score) and low (good score) risk groups, and the statistical significance was determined using the log-rank test. Similarly, we also evaluated the final model that uses the combined mRNA/miRNA panel for predicting time to clinical recurrence in both training and validation datasets.
Missing data are present in this study, in particular, for clinical recurrence, PSA, and T-stage data. We adopted the available-case approach19 in our analyses, and the sample sizes used in each step of building and evaluating prediction models may be less than the total sample size.
Results
Custom Prostate DASL Profiling
We performed DASL expression profiling with our custom-designed prostate cancer panel (see the Materials and Methods section) and the Illumina DASL microRNA panel on 70 prostatectomy patient samples to identify biomarkers predictive of recurrence. An independent validation profiling experiment was performed on 40 additional samples. miRNA probes were filtered to retain only those that were present on the miRNA microarrays used for both the training and validation sets, reducing the total number of probes examined to 403 miRNA probes. The training set included 29 cases with observed biochemical PSA recurrence (median time to recurrence = 19 months), and 41 cases censored, ie, without observed recurrence during follow-up (median follow-up time = 83.0 months). A summary of the clinical characteristics of the training and validation sets of samples is provided in Table 1. The complete dataset for the combined mRNA and miRNA data are provided in Supplemental Table S3 for the training set and Supplemental Table S4 for the validation set (available at http://ajp.amjpathol.org).
Table 1.
A Summary of the Clinical Characteristics of the Training and Validation Sets of Patient Samples
| Training set (total) | Training set (no BCR) | Training set (BCR) | Validation set (total) | Validation set (no BCR) | Validation set (BCR) | |
|---|---|---|---|---|---|---|
| Number cases | 70 | 41 | 29 | 40 | 27 | 13 |
| Clinical recurrence | 8 | 0 | 8 | 11 | 0 | 11 |
| No clinical recurrence | 57 | 41 | 16 | 29 | 27 | 2 |
| Median time F/U (months) | 84 | 83 | 81 | 74 | 75 | 73 |
| Median time to BCR (months) | 19 | NA | 19 | 14 | NA | 14 |
| Median time no BCR (months) | 48 | 83 | 19 | 34.5 | 56 | 14 |
| Gleason score (avg ± SD) | 6.9 ± 0.6 | 6.7 ± 0.6 | 7.0 ± 0.6 | 7.0 ± 0.8 | 6.8 ± 0.7 | 7.4 ± 1 |
| PSA (avg ± SD) | 9.2 ± 5.4 | 8.7 ± 6.4 | 9.9 ± 3.8 | 12.7 ± 8.4 | 12.4 ± 9.9 | 13.1 ± 5.3 |
| Age (avg ± SD) | 61.9 ± 7.7 | 61.2 ± 7.7 | 62.9 ± 7.8 | 63.6 ± 8.4 | 63.5 ± 8.3 | 64 ± 8.9 |
BCR, biochemical recurrence; F/U, follow up; NA, not applicable; PSA, prostate specific antigen; SD, standard deviation.
Integrated DASL Biomarker Analysis
After fitting a univariate Cox proportional hazard model for each individual probe using the training data, a set of 27 important probes were preselected based on a false discovery rate threshold of 0.30 (see Supplemental Table S5 at http://ajp.amjpathol.org). Next, to identify the optimal prediction score based on the preselected probes, we fit a lasso Cox proportional hazard model17,18 first using the set of 25 preselected mRNA probes only, resulting in a panel of nine protein-coding genes shown in Table 2 (RAD23B, FBP1, TNFRSF1A, NOTCH3, ETV1, BID, SIM2, ANXA1, and BCL2). A final prediction model was then built to include the predictive score based on this panel of nine mRNA biomarkers as well as the relevant clinical biomarkers including T-stage, PSA, and Gleason score, which could be used to predict recurrence following radical prostatectomy. Kaplan-Meier analysis (Figure 1A) demonstrated that these probes could significantly discriminate patients at higher and lower risk of recurrence by the log-rank test (P < 0.001). We next applied the final predictive model developed on the training set to the validation set, a separate, independent DASL profiling experiment performed on a different day. Kaplan-Meier analysis (Figure 1B) on this validation set determined that the model could discriminate patients at higher and lower risk of recurrence (P = 0.010).
Table 2.
Nine-Gene Predictor of Prostate Cancer Recurrence Following Surgery
| Symbol | Description | Coefficient | References |
|---|---|---|---|
| RAD23B | RAD23 homolog B | 0.152 | 20, 21 |
| FBP1 | Fructose-1,6-bisphosphatase 1 | 0.310 | 7, 10, 33 |
| TNFRSF1A | Tumor necrosis factor receptor superfamily, member 1A | −0.560 | 23, 24 |
| NOTCH3 | Notch homolog 3 | 0.426 | 25, 26 |
| ETV1 | Ets variant gene 1 (ETV1) | 0.157 | 9, 15 |
| BID | BH3 interacting domain death agonist (BID) | 0.248 | 27, 28 |
| SIM2 | Single-minded homolog 2 | 0.043 | 29–32 |
| ANXA1 | Annexin A1 | −0.185 | 33–36 |
| BCL2 | B-cell CLL/lymphoma 2 | 0.028 | 37–38 |
Coefficient is derived from the lasso Cox proportion hazards model and was used for computing the predictive score. Positive coefficients indicate a positive association with recurrence, and negative coefficients indicate a negative association with recurrence.
Figure 1.
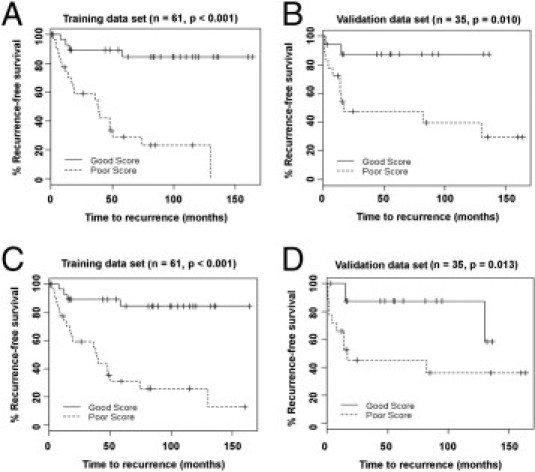
Prediction of biochemical recurrence in all prostate cancer patients using two biomarker panels, adjusted for clinical information. A: Kaplan-Meier analysis of the training set patients that were separated based on the mRNA panel described in Table 2. B: Kaplan-Meier analysis on the validation cases using the mRNA panel. C: Kaplan-Meier analysis of the training set using the combined mRNA and miRNA panel described in Table 3. D: Kaplan-Meier analysis of the validation set using the combined mRNA and miRNA panel.
Subsequently, we repeated the above training procedure using the complete set of 27 preselected mRNA and miRNA probes, and we identified an optimal panel of 10 mRNAs and two microRNAs (Table 3) and built a final prediction model for prostate cancer biochemical recurrence, which again included relevant clinical biomarkers. Kaplan-Meier analysis and the log-rank test determined that this panel could also significantly discriminate patients at higher and lower risk of recurrence both in the training set (P < 0.001, Figure 1C) and in the validation set (P = 0.013, Figure 1D).
Table 3.
Twelve-Gene Predictor of Prostate Cancer Recurrence Following Surgery Using 10 mRNAs and Two microRNAs
| Symbol | Description | Coefficient | References |
|---|---|---|---|
| RAD23B | RAD23 homolog B | 0.070 | 20, 21 |
| FBP1 | Fructose-1,6-bisphosphatase 1 | 0.251 | 7, 10, 22 |
| TNFRSF1A | Tumor necrosis factor receptor superfamily, member 1A | −0.588 | 23, 24 |
| CCNG2 | Cyclin G2 | 0.008 | 39–41 |
| hsa-miR-647 | hsa-miR-647 | −0.318 | |
| LETMD1 | LETM1 domain containing 1 | 0.063 | 42–44, 45 |
| NOTCH3 | Notch homolog 3 | 0.367 | 25, 26 |
| ETV1 | ETS variant gene 1 (ETV1) | 0.179 | 9, 15 |
| hsa-miR-519 days | hsa-miR-519 days | 0.551 | 46 |
| BID | BH3 interacting domain death agonist (BID) | 0.128 | 27, 28 |
| SIM2 | Single-minded homolog 2 | 0.124 | 29–32 |
| ANXA1 | Annexin A1 | −0.143 | 33–36 |
Coefficient is derived from the lasso Cox proportion hazards model and was used for computing the predictive score. Positive coefficients indicate a positive association with recurrence, and negative coefficients a negative association with recurrence.
Prediction of Cases with a Gleason Score 7
Prediction of recurrence for patients with a Gleason score 7 is particularly difficult. To address this issue, we applied the biomarker panels to the subset of cases in the training and validation sets that had a Gleason score 7. The prediction model based on the nine-mRNA panel was significant at discriminating biochemical recurrence in Gleason score 7 cases in both the training set (P < 0.001, Figure 2A) and the validation set (P = 0.027, Figure 2B). For the prediction model based on the combined panel of 10 mRNAs and two miRNAs in Table 3, the predictive value was again significant for both the training set (P = < 0.001, Figure 2C) and the validation set (P = 0.010, Figure 2D). A summary of the P values for predicting biochemical recurrence is given in Table 4. In all cases, the prediction models that use one of the two gene biomarker panels plus clinical information outperforms the prediction model using only clinical information.
Figure 2.
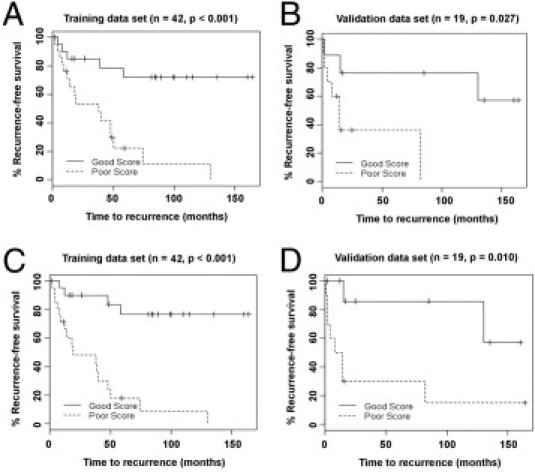
Prediction of biochemical recurrence in prostate cancer patients with a Gleason score 7 using two biomarker panels, adjusted for clinical information. A: Kaplan-Meier analysis of the training set of Gleason score 7 cases using the mRNA panel described in Table 2. B: Kaplan-Meier analysis of the Gleason score 7 cases in the validation set using the mRNA panel. C: Kaplan-Meier analysis of the Gleason score 7 cases in the training set using the combined mRNA and miRNA panel described in Table 3. D: Kaplan-Meier analysis of the Gleason score 7 cases in the validation set using the combined mRNA and miRNA panel.
Table 4.
Summary of P Values (Log-Rank Test) of Prediction of Biochemical Recurrence on Training and Validation Sets
| Dataset | Combined |
Clinical information only | |
|---|---|---|---|
| mRNA panel | mRNA/miRNA panel | ||
| Training | |||
| All cases (n = 61) | <0.001 | <0.001 | 0.096 |
| Gleason score 7 (n = 42) | <0.001 | <0.001 | 0.641 |
| Validation | |||
| All cases (n = 35) | 0.010 | 0.013 | 0.020 |
| Gleason score 7 (n = 19) | 0.027 | 0.010 | 0.028 |
Prediction of biochemical recurrence for the entire dataset and the subset of Gleason score 7 cases was made using two biomarker panels adjusted for T-stage, PSA, and Gleason score, or using clinical information only. Significant P values are indicated in bold.
Analysis of Clinical Recurrence
Although most patients who have clinical recurrence following prostatectomy also have biochemical recurrence, there is a significant population of patients with biochemical recurrence who do not have clinically significant recurrences observed during their follow-ups. To evaluate our biomarker panel of biochemical recurrence for predicting the clinical recurrence, we tested the prediction model based on the combined mRNA/miRNA panel in the same training and validation samples using their clinical recurrence outcome data. Unfortunately, clinical recurrence data were lacking on some of the samples, and the total number of samples used in the training set was reduced. In the training data, the combined mRNA/miRNA panel was highly significant for predicting clinical recurrence in all patients (P = 0.002) as well as in the subset of patients with a Gleason score 7 (P = 0.004); in the validation data, it was also significant for predicting recurrence in patients with a Gleason score 7 (P = 0.023) and trended toward significance in all patients (P = 0.078). A summary of the P values for predicting clinical recurrence is given in Table 5. In all cases, the prediction model that uses the combined mRNA and miRNA panel plus the clinical information, again, outperforms the prediction model that uses only the clinical information.
Table 5.
Summary of P Values (Log-Rank Test) of Prediction of Clinical Recurrence on Training and Validation Sets
| Dataset | Combined mRNA/miRNA panel | Clinical information only |
|---|---|---|
| Training | ||
| All cases (n = 56) | 0.002 | 0.262 |
| Gleason score 7 (n = 37) | 0.004 | 0.136 |
| Validation | ||
| All cases (n = 35) | 0.078 | 0.193 |
| Gleason score 7 (n = 19) | 0.023 | 0.080 |
Prediction of clinical recurrence for the entire dataset and the subset of Gleason score 7 cases was made using the combined mRNA/miRNA panel, adjusted for T-stage, PSA, and Gleason score, or using clinical information only. Significant P values are indicated in bold.
We also performed an analysis to construct a predictive set of biomarkers based on the clinical recurrence data instead of biochemical recurrence. Only three probes passed the initial preselection step for the univariate Cox PH modeling, all corresponding to the ETV1 gene, which is likely due to the considerably fewer number of clinical recurrences in the training set as well as the smaller total sample size. Furthermore, the prediction model built on this set of gene biomarkers did not perform as well as the models built on biochemical recurrence (data not shown).
Discussion
In the past few years, enormous progress has been made in developing technologies to exploit FFPE tumor tissue samples for gene expression and proteomic analysis. The use of FFPE tissues as a starting material is attractive because this approach should make biomarkers identified in this way much easier to translate into widespread clinical practice. DASL profiling makes it possible to define gene sets using FFPE prostate cancer tissues that could have potential prognostic and predictive value. For example, the DASL assay has been used recently to identify a 16-gene set that correlates with prostate cancer relapse.11 There was no overlap between our panel of 10 mRNA and two miRNA biomarkers described here and the previously described 16-gene panel even though 10 of the genes in the 16-gene panel previously reported were included in our 522 custom prostate DASL panel. When we analyzed the performance of the probes corresponding to those 10 mRNAs in our dataset, we found that they were not able to significantly discriminate patients at higher or lower risk of recurrence. In this previous study, the gene signature selection and prediction model building were performed in separate steps, and the signature selection was based on the correlation between the gene expression and Gleason score rather than between the gene expression and time to biochemical recurrence; our analytic approach overcomes these limitations. Specifically, our approach of building (training) prediction models takes advantage of recent advancement in regularized regression models for survival outcomes17,18; regularized regression models can achieve simultaneous feature selection and model estimation and avoid model overfitting, leading to better prediction performance. Our use of a preselection step is similar to the recently proposed sure independence screening methods,47,48 which have been shown to achieve better performance in the presence of high-dimensional data for survival analysis compared to regularized regression without a preselection step.49
Two other recent studies have used DASL profiling to prostate cancer, but have not detected any signature that improved on clinical models in validation sets.50,51 Although these studies used large cohorts with long-term follow-up, they examined different panels of mRNA transcripts and did not include probes corresponding to miRNA genes. Moreover, these earlier studies suggested that tumor heterogeneity may play an important role in confounding signature identification. For our study of prostatectomy specimens, we identified the most prominent tumor lesion, and used a tissue core sample from that region to minimize stromal contributions and tumor heterogeneity.
In our 12-gene predictive biomarker panel, nine of the genes are positively associated with recurrence, and three are negatively associated with recurrence. The nine genes positively associated with recurrence included miR-519d, Notch homolog 3 (Notch3), Fructose-1,6-bisphosphatase 1 (FBP1), ETS variant gene 1 (ETV1), BH3 interacting domain death agonist (BID), Single-Minded homolog 2 (SIM2), RAD23 homolog B (RAD23B), LETM1 domain containing 1 (LETMD1), and Cyclin G2 (CCNG2). Little is known about miR-519d other than it may be associated with obesity.46 NOTCH3 is one of four Notch family receptors in humans, and Notch signaling has been shown to be important for prostate cancer cell growth, migration, and invasion,25,26 as well as normal prostate development.52,53 FBP1 is expressed in the prostate and is involved in gluconeogenesis.22 The identification of this metabolic enzyme as a biomarker of recurrence is initially surprising, but given the recent identification of isocitrate dehydrogenase 1 (IDH1) mutations in glioblastoma,54 and the fact that FBP1 was overexpressed in independent microarray analyses of prostate cancers,7,10 the potential of FBP1 as a biomarker should not be underestimated. ETV1 is well established as one of the commonly recurrent translocations found in prostate cancers,9,15 and has been used in clinical models of recurrence following prostatectomy.55 BID is a proapoptotic protein that binds to BCL2 and potentiates apoptotic responses on cleavage in response to tumor necrosis factor α (TNFα) and other death receptors.27,28 SIM2 was identified as a potential biomarker of prostate cancer in 200229 and later independently confirmed by Halvorsen et al30 and Arredouani et al.31 SIM2 functions as a transcription factor that represses the proapoptotic gene BNIP3.56 RAD23B plays a critical role in DNA damage recognition and nucleotide excision repair,20 as well as inhibiting MDM2-mediated degradation of the p53 tumor suppressor.21 LETMD1 (also known as HCCR) is an oncogene that is induced by Wnt42 and PI3K/AKT signaling,43 inhibits p53 function,44 and is a biomarker for hepatocellular57 and breast45 cancers. Cyclin G2 is an atypical cyclin that is induced by DNA damage39 in a p53-independent manner, as well as by PI3K/AKT/FOXO signals,40 and induces p53-dependent cell cycle arrest.41
The three genes in the predictive biomarker panel negatively associated with recurrence were miR-647, the TNFα receptor (TNFRSF1A), and annexin A1 (ANXA1). Although little is known about miR-647, TNFRSF1A (also known as TNFR1) mediates proapoptotic responses to TNFα ligand.23,24 Annexin A1 expression is reduced in early onset prostate cancer33 and high-grade prostatic intraepithelial neoplasia.34 ANXA1 plays important roles in vesicle trafficking and reduced ANXA1 promotes EMT and metastasis,35 and up-regulates autocrine IL-6 signaling.36 Thus, as a whole, this panel of biomarkers appears to reflect changes in DNA stability, PI3K signaling, p53 activity, apoptosis, and differentiation consistent with more aggressive disease.
Although this study goes beyond a pilot study, enhanced by selection of samples from multiple institutions, the number of specimens tested is still relatively small. Re-analysis of our data using only the Emory samples for the training set did not identify any significant probes, likely due to the substantially smaller sample size. Thus, although the performance of our panel of biomarkers is significant, even for Gleason score 7 patients, future studies beyond the scope of this work will be necessary to perform independent validation on much larger sample sets with greater statistical power. Moreover, it is now feasible to perform DASL assays on virtually the entire genome, in an assay that queries 24,526 transcripts derived from the RefSeq database. Future studies will test combined mRNA and miRNA biomarker panels, and query the entire genome to determine whether other biomarker panels can achieve even greater success in prediction of biochemical and clinical recurrence of prostate cancer. Planned larger scale validation studies will determine whether these biomarkers are predictive for Gleason score 7 cases, and their utility at predicting clinical as well as biochemical recurrence.
Acknowledgments
We thank Juliana DaSilva, Malini Krishnamoorthy, Benjamin Barwick, and the Winship Cancer Genomics Shared Resource.
Footnotes
Supported in part by an Emory University Research Committee grant and the PHS Grant UL1 RR025008 from the Clinical and Translational Science Award program, National Institutes of Health, National Center for Research Resources; NIH R01 CA106826 and DOD PCRP IDEA Award PC093328 (to C.S.M); Emory Cancer Genomics Shared Resource and the Winship Cancer Center Support Grant (NIH P30CA138292); and the Canadian Cancer Society Research Institute (grant # 019038).
Supplemental material for this article can be found at http://ajp.amjpathol.org or doi:10.1016/j.ajpath.2011.03.008.
Current address of M.A., Departments of Pathology & Oncology, Jewish General Hospital, Montreal, QC, Canada.
Supplementary data
Clinical data for samples used in the study. 1 = positive, 0 = negative. BCR = biochemical recurrence. L = positive lymph node. M = metastasis.
Custom Prostate DASL panel. Shown are the 522 genes included in the DASL panel.
Complete Custom Prostate DASL and miRNA dataset for 70 cases in the training set.
Complete Custom Prostate DASL and miRNA dataset for 40 cases in the validation set.
Set of 27 DASL probes that were preselected based on a false discovery rate threshold of 0.30 by Cox proportional hazard (PH) model.
References
- 1.American Cancer Society . American Cancer Society; Atlanta: 2009. Cancer Facts & Figures 2009. [Google Scholar]
- 2.Bibikova M., Talantov D., Chudin E., Yeakley J.M., Chen J., Doucet D., Wickham E., Atkins D., Barker D., Chee M., Wang Y., Fan J.B. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol. 2004;165:1799–1807. doi: 10.1016/S0002-9440(10)63435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan J.B., Yeakley J.M., Bibikova M., Chudin E., Wickham E., Chen J., Doucet D., Rigault P., Zhang B., Shen R., McBride C., Li H.R., Fu X.D., Oliphant A., Barker D.L., Chee M.S. A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res. 2004;14:878–885. doi: 10.1101/gr.2167504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abramovitz M., Ordanic-Kodani M., Wang Y., Li Z., Catzavelos C., Bouzyk M., Sledge G.W., Jr., Moreno C.S., Leyland-Jones B. Optimization of RNA extraction from FFPE tissues for expression profiling in the DASL assay. Biotechniques. 2008;44:417–423. doi: 10.2144/000112703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhanasekaran S.M., Barrette T.R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 6.Varambally S., Yu J., Laxman B., Rhodes D.R., Mehra R., Tomlins S.A., Shah R.B., Chandran U., Monzon F.A., Becich M.J., Wei J.T., Pienta K.J., Ghosh D., Rubin M.A., Chinnaiyan A.M. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Lapointe J., Li C., Higgins J.P., Van De Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., Ekman P., DeMarzo A.M., Tibshirani R., Botstein D., Brown P.O., Brooks J.D., Pollack J.R. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaTulippe E., Satagopan J., Smith A., Scher H., Scardino P., Reuter V., Gerald W.L. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 9.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J.E., Shah R.B., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 10.Liu P., Ramachandran S., Ali-Seyed M., Scharer C.D., Laycock N., Dalton W.B., Williams H., Karanam S., Datta M.W., Jaye D., Moreno C.S. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;46:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 11.Bibikova M., Chudin E., Arsanjani A., Zhou L., Garcia E.W., Modder J., Kostelec M., Barker D., Downs T., Fan J.B., Wang-Rodriguez J. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics. 2007;89:666–672. doi: 10.1016/j.ygeno.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.True L., Coleman I., Hawley S., Huang C.Y., Gifford D., Coleman R., Beer T.M., Gelmann E., Datta M., Mostaghel E., Knudsen B., Lange P., Vessella R., Lin D., Hood L., Nelson P.S. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci U S A. 2006;103:10991–10996. doi: 10.1073/pnas.0603678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh D., Febbo P.G., Ross K., Jackson D.G., Manola J., Ladd C., Tamayo P., Renshaw A.A., D'Amico A.V., Richie J.P., Lander E.S., Loda M., Kantoff P.W., Golub T.R., Sellers W.R. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 14.Yu Y.P., Landsittel D., Jing L., Nelson J., Ren B., Liu L., McDonald C., Thomas R., Dhir R., Finkelstein S., Michalopoulos G., Becich M., Luo J.H. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 15.Tomlins S.A., Laxman B., Dhanasekaran S.M., Helgeson B.E., Cao X., Morris D.S., Menon A., Jing X., Cao Q., Han B., Yu J., Wang L., Montie J.E., Rubin M.A., Pienta K.J., Roulston D., Shah R.B., Varambally S., Mehra R., Chinnaiyan A.M. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 16.Tomlins S.A., Mehra R., Rhodes D.R., Smith L.R., Roulston D., Helgeson B.E., Cao X., Wei J.T., Rubin M.A., Shah R.B., Chinnaiyan A.M. TMPRSS2: ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 17.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Goeman J.J. L1 penalized estimation in the Cox proportional hazards model. Biom J. 2010;52:70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- 19.Little R.J.A., Rubin D.B. Statistical Analysis with Missing Data. New York, Wiley. 2002 [Google Scholar]
- 20.Thoma B.S., Vasquez K.M. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol Carcinog. 2003;38:1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- 21.Brignone C., Bradley K.E., Kisselev A.F., Grossman S.R. A post-ubiquitination role for MDM2 and hHR23A in the p53 degradation pathway. Oncogene. 2004;23:4121–4129. doi: 10.1038/sj.onc.1207540. [DOI] [PubMed] [Google Scholar]
- 22.Yanez A.J., Nualart F., Droppelmann C., Bertinat R., Brito M., Concha I.I., Slebe J.C. Broad expression of fructose-1,6-bisphosphatase and phosphoenolpyruvate carboxykinase provide evidence for gluconeogenesis in human tissues other than liver and kidney. J Cell Physiol. 2003;197:189–197. doi: 10.1002/jcp.10337. [DOI] [PubMed] [Google Scholar]
- 23.Hsu H., Xiong J., Goeddel D.V. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 24.Thorburn A. Death receptor-induced cell killing. Cell Signal. 2004;16:139–144. doi: 10.1016/j.cellsig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z., Li Y., Ahmad A., Banerjee S., Azmi A.S., Kong D., Wojewoda C., Miele L., Sarkar F.H. Down-regulation of Notch-1 is associated with Akt and FoxM1 in inducing cell growth inhibition and apoptosis in prostate cancer cells. J Cell Biochem. 2011;112:78–88. doi: 10.1002/jcb.22770. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Wang Z., Li Y., Banerjee S., Kong D., Ahmad A., Nogueira V., Hay N., Sarkar F.H. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. J Cell Biochem. 2010;109:726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 27.Kulik G., Carson J.P., Vomastek T., Overman K., Gooch B.D., Srinivasula S., Alnemri E., Nunez G., Weber M.J. Tumor necrosis factor alpha induces BID cleavage and bypasses antiapoptotic signals in prostate cancer LNCaP cells. Cancer Res. 2001;61:2713–2719. [PubMed] [Google Scholar]
- 28.Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 29.Deyoung M.P., Scheurle D., Damania H., Zylberberg C., Narayanan R. Down's syndrome-associated single minded gene as a novel tumor marker. Anticancer Res. 2002;22:3149–3157. [PubMed] [Google Scholar]
- 30.Halvorsen O.J., Rostad K., Oyan A.M., Puntervoll H., Bo T.H., Stordrange L., Olsen S., Haukaas S.A., Hood L., Jonassen I., Kalland K.H., Akslen L.A. Increased expression of SIM2-s protein is a novel marker of aggressive prostate cancer. Clin Cancer Res. 2007;13:892–897. doi: 10.1158/1078-0432.CCR-06-1207. [DOI] [PubMed] [Google Scholar]
- 31.Arredouani M.S., Lu B., Bhasin M., Eljanne M., Yue W., Mosquera J.M., Bubley G.J., Li V., Rubin M.A., Libermann T.A., Sanda M.G. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res. 2009;15:5794–5802. doi: 10.1158/1078-0432.CCR-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeYoung M.P., Tress M., Narayanan R. Identification of Down's syndrome critical locus gene SIM2-s as a drug therapy target for solid tumors. Proc Natl Acad Sci U S A. 2003;100:4760–4765. doi: 10.1073/pnas.0831000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paweletz C.P., Ornstein D.K., Roth M.J., Bichsel V.E., Gillespie J.W., Calvert V.S., Vocke C.D., Hewitt S.M., Duray P.H., Herring J., Wang Q.H., Hu N., Linehan W.M., Taylor P.R., Liotta L.A., Emmert-Buck M.R., Petricoin E.F., 3rd Loss of annexin 1 correlates with early onset of tumorigenesis in esophageal and prostate carcinoma. Cancer Res. 2000;60:6293–6297. [PubMed] [Google Scholar]
- 34.Patton K.T., Chen H.M., Joseph L., Yang X.J. Decreased annexin I expression in prostatic adenocarcinoma and in high-grade prostatic intraepithelial neoplasia. Histopathology. 2005;47:597–601. doi: 10.1111/j.1365-2559.2005.02300.x. [DOI] [PubMed] [Google Scholar]
- 35.Maschler S., Gebeshuber C.A., Wiedemann E.M., Alacakaptan M., Schreiber M., Custic I., Beug H. Annexin A1 attenuates EMT and metastatic potential in breast cancer. EMBO Mol Med. 2010;2:401–414. doi: 10.1002/emmm.201000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inokuchi J., Lau A., Tyson D.R., Ornstein D.K. Loss of annexin A1 disrupts normal prostate glandular structure by inducing autocrine IL-6 signaling. Carcinogenesis. 2009;30:1082–1088. doi: 10.1093/carcin/bgp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alnemri E.S., Robertson N.M., Fernandes T.F., Croce C.M., Litwack G. Overexpressed full-length human BCL2 extends the survival of baculovirus- infected Sf9 insect cells. Proc Natl Acad Sci U S A. 1992;89:7295–7299. doi: 10.1073/pnas.89.16.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner A.J., Small M.B., Hay N. Myc-mediated apoptosis is blocked by ectopic expression of Bcl-2. Mol Cell Biol. 1993;13:2432–2440. doi: 10.1128/mcb.13.4.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bates S., Rowan S., Vousden K.H. Characterisation of human cyclin G1 and G2: dNA damage inducible genes. Oncogene. 1996;13:1103–1109. [PubMed] [Google Scholar]
- 40.Martinez-Gac L., Marques M., Garcia Z., Campanero M.R., Carrera A.C. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol Cell Biol. 2004;24:2181–2189. doi: 10.1128/MCB.24.5.2181-2189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arachchige Don A.S., Dallapiazza R.F., Bennin D.A., Brake T., Cowan C.E., Horne M.C. Cyclin G2 is a centrosome-associated nucleocytoplasmic shuttling protein that influences microtubule stability and induces a p53-dependent cell cycle arrest. Exp Cell Res. 2006;312:4181–4204. doi: 10.1016/j.yexcr.2006.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho G.W., Kim M.H., Kim S.H., Ha S.A., Kim H.K., Kim S., Kim J.W. TCF/ beta-catenin plays an important role in HCCR-1 oncogene expression. BMC Mol Biol. 2009;10:42. doi: 10.1186/1471-2199-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho G.W., Shin S.M., Namkoong H., Kim H.K., Ha S.A., Hur S.Y., Kim T.E., Chai Y.G., Kim J.W. The phosphatidylinositol 3-kinase/Akt pathway regulates the HCCR-1 oncogene expression. Gene. 2006;384:18–26. doi: 10.1016/j.gene.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Ko J., Lee Y.H., Hwang S.Y., Lee Y.S., Shin S.M., Hwang J.H., Kim J., Kim Y.W., Jang S.W., Ryoo Z.Y., Kim I.K., Namkoong S.E., Kim J.W. Identification and differential expression of novel human cervical cancer oncogene HCCR-2 in human cancers and its involvement in p53 stabilization. Oncogene. 2003;22:4679–4689. doi: 10.1038/sj.onc.1206624. [DOI] [PubMed] [Google Scholar]
- 45.Jung S.S., Park H.S., Lee I.J., Namkoong H., Shin S.M., Cho G.W., Ha S.A., Park Y.G., Lee Y.S., Ko J., Kim J.W. The HCCR oncoprotein as a biomarker for human breast cancer. Clin Cancer Res. 2005;11:7700–7708. doi: 10.1158/1078-0432.CCR-04-2609. [DOI] [PubMed] [Google Scholar]
- 46.Martinelli R., Nardelli C., Pilone V., Buonomo T., Liguori R., Castano I., Buono P., Masone S., Persico G., Forestieri P., Pastore L., Sacchetti L. miR-519d overexpression is associated with human obesity. Obesity (Silver Spring) 2010;18:2170–2176. doi: 10.1038/oby.2009.474. [DOI] [PubMed] [Google Scholar]
- 47.Fan J., Lv J. Sure independence screening for ultrahigh dimensional feature space. J R Stat Soc Ser B Stat Methodol. 2008;70:849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan J., Song R. Sure independence screening in generalized linear models and NP-dimensionality. Ann Stat. 2010;38:3567–3604. [Google Scholar]
- 49.Zhao S.D., Li Y. Harvard University Biostatistics Working Paper Series 2010. Harvard University Department of Biostatistics; Boston: 2010. Principled sure independence screening for Cox models with ultra-high-dimensional covariates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sboner A., Demichelis F., Calza S., Pawitan Y., Setlur S.R., Hoshida Y., Perner S., Adami H.O., Fall K., Mucci L.A., Kantoff P.W., Stampfer M., Andersson S.O., Varenhorst E., Johansson J.E., Gerstein M.B., Golub T.R., Rubin M.A., Andren O. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakagawa T., Kollmeyer T.M., Morlan B.W., Anderson S.K., Bergstralh E.J., Davis B.J., Asmann Y.W., Klee G.G., Ballman K.V., Jenkins R.B. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS ONE. 2008;3:e2318. doi: 10.1371/journal.pone.0002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leong K.G., Gao W.Q. The Notch pathway in prostate development and cancer. Differentiation. 2008;76:699–716. doi: 10.1111/j.1432-0436.2008.00288.x. [DOI] [PubMed] [Google Scholar]
- 53.Wang X.D., Leow C.C., Zha J., Tang Z., Modrusan Z., Radtke F., Aguet M., de Sauvage F.J., Gao W.Q. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev Biol. 2006;290:66–80. doi: 10.1016/j.ydbio.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 54.Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L., Olivi A., McLendon R., Rasheed B.A., Keir S., Nikolskaya T., Nikolsky Y., Busam D.A., Tekleab H., Diaz L.A., Jr., Hartigan J., Smith D.R., Strausberg R.L., Marie S.K., Shinjo S.M., Yan H., Riggins G.J., Bigner D.D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheville J.C., Karnes R.J., Therneau T.M., Kosari F., Munz J.M., Tillmans L., Basal E., Rangel L.J., Bergstralh E., Kovtun I.V., Savci-Heijink C.D., Klee E.W., Vasmatzis G. Gene panel model predictive of outcome in men at highrisk of systemic progression and death from prostate cancer after radical retropubic prostatectomy. J Clin Oncol. 2008;26:3930–3936. doi: 10.1200/JCO.2007.15.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Farrall A.L., Whitelaw M.L. The HIF1alpha-inducible pro-cell death gene BNIP3 is a novel target of SIM2s repression through cross-talk on the hypoxia response element. Oncogene. 2009;28:3671–3680. doi: 10.1038/onc.2009.228. [DOI] [PubMed] [Google Scholar]
- 57.Yoon S.K., Lim N.K., Ha S.A., Park Y.G., Choi J.Y., Chung K.W., Sun H.S., Choi M.J., Chung J., Wands J.R., Kim J.W. The human cervical cancer oncogene protein is a biomarker for human hepatocellular carcinoma. Cancer Res. 2004;64:5434–5441. doi: 10.1158/0008-5472.CAN-03-3665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical data for samples used in the study. 1 = positive, 0 = negative. BCR = biochemical recurrence. L = positive lymph node. M = metastasis.
Custom Prostate DASL panel. Shown are the 522 genes included in the DASL panel.
Complete Custom Prostate DASL and miRNA dataset for 70 cases in the training set.
Complete Custom Prostate DASL and miRNA dataset for 40 cases in the validation set.
Set of 27 DASL probes that were preselected based on a false discovery rate threshold of 0.30 by Cox proportional hazard (PH) model.


